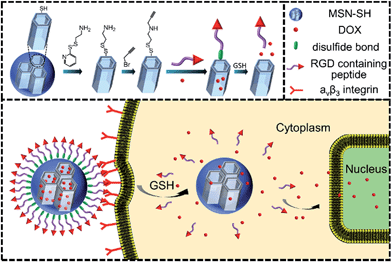
A redox-responsive drug delivery system based on RGD containing peptide-capped mesoporous silica nanoparticles†
Ze-Yong
Li‡
,
Jing-Jing
Hu‡
,
Qi
Xu
,
Si
Chen
,
Hui-Zhen
Jia
,
Yun-Xia
Sun
,
Ren-Xi
Zhuo
and
Xian-Zheng
Zhang
*
Key Laboratory of Biomedical Polymers of the Ministry of Education & Department of Chemistry, Wuhan University, Wuhan 430072, P. R. China. E-mail: xz-zhang@whu.edu.cn
First published on 22nd October 2014
Abstract
In this paper, an intracellular glutathione (GSH) responsive mesoporous silica nanoparticle (MSN-S-S-RGD) was developed as a drug nanocarrier by immobilizing the gatekeeper (RGD containing peptide) onto MSNs using disulfide bonds. The antitumor drug, DOX was loaded onto the porous structure of the MSNs and the DOX@MSN-S-S-RGD system has been proved to be an effective nanocarrier. It was determined that most of the drug could be entrapped with only a slight leakage. After being accumulated in tumor cells via the receptor-mediated endocytosis, the surface peptide layer of DOX@MSN-S-S-RGD was removed to trigger the release of the entrapped drug to kill the tumor cell due to the cleavage of the disulfide bonds by intracellular GSH.
Introduction
Recently, stimuli-responsive mesoporous silica nanoparticles (MSNs) have gained increasing attention as nanodevices for efficient encapsulation and protection of therapeutic agents, as well as the controlled and selective release of them.1–8 Furthermore, these nanodevices can effectively overcome the severe side effects inherently associated with drugs.9,10 The purpose of designing a MSN-based drug delivery system (DDS) is to accurately deliver drugs into diseased tissue without any unwanted premature leakage such that the drug can be effectively released at the target site. In this regard, various MSN-based nanovalves were constructed using a series of gatekeepers, such as inorganic nanoparticles,11–13 macrocyclic compounds,14,15 polymers16 and biomacromolecules17,18 with diverse stimuli responsiveness, including pH, light, redox and enzyme responsiveness. Of these stimuli, redox-responsiveness is frequently investigated as the glutathione (GSH) concentration in different sites tremendously varies. As we know, the GSH molecules are located in the cellular cytoplasm with a concentration of 2–10 mM, which is apparently higher than that in blood (ca. 2 μM).19.20Apart from taking advantage of the enhanced permeation and retention (EPR) effect, in order to further improve the accumulation of MSN-based nanocarriers at the tumor site, its surface is usually functionalized with targeting ligands,21 such as an antibody,22 Arg-Gly-Asp (RGD)23 and folate,24 which have been proved to be able to selectively bind to the overexpressed receptors or antigens on the surface of tumor cells. Generally, the targeting ligand only has the target property and is not involved in the construction of the nanovalve of MSNs.
In this paper, we have tried to integrate the targeting units as the nanovalve of MSNs and the nanovalve is entirely composed of targeting molecules such as RGD. As a proof-of-concept, we report a smart redox-responsive nanocarrier based on RGD containing peptide-capped MSNs as shown in Scheme 1 for tumor targeted delivery of drugs. The RGD containing peptide decorating the surface of MSNs demonstrated the following functions: (a) enhanced tumor target and subsequent tumor cellular uptake and (b) act as an intracellular GSH responsive nanovalve.
Experimental section
Materials
Hexadecyltrimethylammonium bromide (CTAB) and tetraethylorthosilicate (TEOS) were purchased from Shanghai Reagent Chemical Co. (China) and used as received. 3-Mercaptopropyltrimethoxysilane (MPTMS) was purchased from Aladdin Reagent Co. Ltd. (Shanghai, China). Doxorubicin hydrochloride (DOX) was purchased from Zhejiang Hisun Pharmaceutical Co. (China). Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), phosphate buffered saline (PBS), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium-bromide (MTT) and Hochest 33342 were purchased from Invitrogen Co. (America). All the other reagents and solvents were provided by Shanghai Reagent Chemical Co. (China) and used without further treatment or purification.Methods
FTIR spectra were recorded on an AVATAR 360 spectrometer. Samples were pressed into potassium bromide (KBr) pellets. Thermal gravimetric analysis (TGA) was performed on a TGS-2 thermogravimetric analyzer (Perkin-Elmer). Fluorescence spectra were recorded on a RF-5301PC spectrofluorophotometer (Shimadzu). Transmission electron microscopy (TEM) experiments were carried out on a JEM-2100 instrument operating at an acceleration voltage of 80 kV. The surface area was calculated by the Brunauer–Emmett–Teller (BET) approach and the pore size distribution obtained by the Barrett–Joyner–Halenda method (ASAP2020, micromeritics). The ζ potentials of the nanoparticles in DI water were analyzed on a Nano-ZS ZEN3600 particle sizer (Malvern Instruments). Low angle X-ray diffraction (XRD) analysis was performed on an X′ Pert Pro diffractometer (PANalytical).Synthesis of two azidopeptides (N3-GRGDSGRGDS-NH2 and N3-GKKKKKKKKKKK-NH2)
Two azidopeptides were manually synthesized employing standard Fmoc chemistry using 6 equivalents of Fmoc-amino acid or azidobenzoic acid as well as HATU (1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate) and NMM (N-methylmorpholine) as coupling reagents in each step. Finally, the total cleavage was carried out (95.0% trifluoroacetic acid, 2.5% triisopropylsilane, 2.5% DI water, 2.0 h), followed by the precipitation of the peptide in cold ether.Synthesis of 3-mercaptopropyl-functionalized MSN (MSN-SH)
MSN-SH was synthesized by the following procedure: CTAB (1.00 g) was dissolved in 480 mL of deionized water. Sodium hydroxide aqueous solution (2.00 M, 3.50 mL) was added to the solution and heated up to 80 °C. After TEOS (5.00 mL) was added dropwise to the CTAB solution, MPTMS (0.97 mL) was introduced dropwise to the solution. The mixture was vigorously stirred for 2 h to obtain a white precipitate. Finally, the solid product was centrifuged, washed with deionized water and ethanol, and dried at 60 °C. To remove the surfactant template (CTAB), the white solid was refluxed for 16 h in a solution of 9.00 mL of HCl (37%) and 160.00 mL of methanol followed by extensive washing with deionized water and ethanol. The resulting nanoparticles were placed under high vacuum to remove the remaining solvent.Synthesis of S-(2-aminoethylthio)-2-thiopyridine hydrochloride
S-(2-Aminoethylthio)-2-thiopyridine hydrochloride was synthesized according to the reported procedure.25Synthesis of 2-(propyldisulfanyl)ethylamine functionalized MSN (MSN-S-S-NH2)
MSN-SH (100 mg) suspended in methanol (20 mL) was reacted with S-(2-aminoethylthio)-2-thiopyridine hydrochloride (100 mg) at room temperature for 24 h. The resulting nanoparticles were separated by centrifugation, washed four times with ethanol and dried under high vacuum.Synthesis of alkyne functionalized MSN-S-S-NH2 (MSN-S-S-alkyne)
MSN-S-S-NH2 (100 mg) suspended in methanol (20 mL) was reacted with 3 mL of propargyl bromide at room temperature for 24 h. The nanoparticles were separated by centrifugation, washed four times with ethanol and dried under high vacuum.Synthesis of peptide-capped MSNs via disulfide bonds (MSN-S-S-RGD and MSN-S-S-K(Ac)11)
MSN-S-S-alkyne (15 mg), N3-GRGDSGRGDS-NH2 (15 mg), CuBr (5 mg) and THPTA (tris(3-hydroxypropyltriazolylmethyl)amine, 10 mg) were suspended in 6 mL of DMF under an N2 atmosphere. After being stirred for 2 days at room temperature, the resulting nanoparticles (MSN-S-S-RGD) were purified by centrifugation and washed four times with EDTA and deionized water. MSN-S-S-K11 can be obtained following a similar procedure. Next, MSN-S-S-K11 was stirred in the capping solution (5% Ac2O, 6% 2,6-lutidine and 89% DMF) for 1 h. MSN-S-S-K(Ac)11 was separated by centrifugation, washed four times with DMF and deionized water, and dried under high vacuum.Preparation of DOX@MSN-S-S-RGD and DOX@MSN-S-S-K(Ac)11
MSN-S-S-RGD (10 mg) and DOX (5 mg) were suspended in 5 mL of DMF and stirred at room temperature for 24 h. The free DOX was removed by washing with PBS (pH 7.4) to obtain DOX@MSN-S-S-RGD. DOX@MSN-S-S-K(Ac)11 can be obtained following a similar procedure.In vitro release studies
At a concentration of 0.1 mg mL−1, DOX@MSN-S-S-RGD was suspended in different media: PBS (pH 7.4) and PBS (pH 7.4) with 10 mM GSH at 37 °C. At the indicated time intervals, the fluorescence intensity of the solution in the release medium was measured using a RF-5301PC spectrofluorophotometer (Shimadzu). The fluorescence standard curve was employed to calculate the cumulative release amounts of DOX. The cumulative release percentage was obtained on the basis of the released and total amounts of DOX. The emission and excitation slit widths were set at 5 nm with λex = 488 nm.Confocal laser scanning microscopy (CLSM)
U87 MG or COS 7 cells were seeded onto glass-bottom Petri dishes (35 mm × 10 mm). After incubation in a humidified 5% CO2 atmosphere for 24 h, the cells were treated with DOX@MSN-S-S-RGD or DOX@MSN-S-S-K(Ac)11 nanoparticles containing DMEM at the same final DOX concentration of 2 μg mL−1, respectively. After incubation in the dark for 2 h, the cell nuclei were stained with Hoechst 33342 prior to imaging under a confocal laser scanning microscope (NOL-LSM 710).Cellular uptake measured by flow cytometry
U87 MG or COS 7 cells were seeded onto 6-well plates in 1 mL DMEM medium and allowed to grow for 24 h. The media were replaced with DOX@MSN-S-S-RGD or DOX@MSN-S-S-K(Ac)11 nanoparticles containing DMEM at the same final DOX concentration of 2 μg mL−1, respectively. After incubation for 2 h at 37 °C, the cells were washed with PBS and digested by trypsin. The trypsinized cells were harvested and suspended in PBS, and then centrifuged at 1000 rpm for 3 min at 4 °C. The supernatant was discarded and the cell pellets were washed with cold PBS thrice to remove the residual nanoparticles. After that, each sample was quickly analyzed on a flow cytometer (BD FACSAria TM III, USA).In vitro cytotoxicity
U87 MG or COS 7 cells were seeded on a 96 well dish with 200 μL DMEM containing 10% FBS. After incubation (37 °C and 5% CO2) for 24 h, the original culture media in each well was replaced with 200 μL fresh DMEM containing the MSN-S-S-RGD, MSN-S-S-K(Ac)11, DOX@MSN-S-S-RGD, DOX@MSN-S-S-K(Ac)11 nanoparticles or free DOX at the indicated concentrations. After 4 h incubation, the culture media of all the wells was refreshed with culture media (200 μL) and the incubation was continued for 48 h. Next, the media were refreshed with DMEM and 20 μL MTT solution (5 mg mL−1). After incubation for 4 h, 200 μL of DMSO was added to each well, and the plate was shaken at room temperature. The optical density (OD) was obtained and the viability was calculated following a standard protocol.Results and discussion
Preparation and characterization of DOX@MSN-S-S-RGD
The two azidopeptides (N3-GRGDSGRGDS-NH2 and N3-GKKKKKKKKKKK-NH2) used in this study were manually synthesized employing standard Fmoc chemistry (Scheme S1 and S2†).26 The obtained azidopeptides were confirmed by ESI-MS as shown in Fig. S1 and S2.† To prepare the redox-sensitive peptide-functionalized MSNs, we first synthesized a mercaptopropyl-derivatized MSN (MSN-SH) with an average diameter of 100 nm via the reported co-condensation method.11 As depicted in Fig. 1a, the spherical particle shape and the MCM-41 type of hexagonally packed mesoporous structure of MSN was confirmed by TEM. A specific surface area of 1247 m2 g−1 (Fig. 1c) and an average mesopore diameter of 2.64 nm (Fig. 1d) for the MSN were observed by BET and BJH analysis, respectively. The powder X-ray diffraction (XRD) pattern of MSN-SH showed a typical hexagonal array with three low-angle reflections indexed as (100), (110) and (200) Bragg peaks (Fig. S3†), which exhibited the typical diffraction patterns of MCM-41 type mesoporous silica with hexagonal symmetry. As illustrated in Scheme 1, the surface disulfide linker was introduced by reacting MSN-SH with S-(2-aminoethylthio)-2-thiopyridine hydrochloride to yield MSN-S-S-NH2,25 which was then allowed to react with propargyl bromide to prepare MSN-S-S-alkyne. The FTIR spectrum of MSN-S-S-alkyne exhibited an alkyne absorption band at 2100 cm−1 (Fig. S4†) indicating its successful functionalization. Thereafter, N3-GRGDSGRGDS-NH2 was immobilized onto the MSN-S-S-alkyne via “click chemistry” between the azidopeptide and MSN-S-S-alkyne to obtain MSN-S-S-RGD (Fig. 1b), which was verified by FTIR spectroscopy (Fig. S4†) and thermal gravimetric analysis (Fig. S5†). The successful functionalization of the nanoparticles was further confirmed by ζ potential measurements. As shown in Table S1,† the ζ potential of MSN-S-S-NH2 sharply changed from −18.1 mV (MSN-SH) to +30.0 mV, indicating the successful introduction of amino groups to MSN-SH. After the “click reaction”, the ζ potential of MSN-S-S-RGD decreased to +24.3 mV from +36.4 mV (MSN-S-S-alkyne), implying the successful immobilization of the RGD containing peptide to the MSN-S-S-alkyne. The drug loading efficiency (DLE) of DOX in the nanoparticles was determined by fluorescence spectroscopy and the results are as follows: DLE: 3.1% for DOX@MSN-S-S-RGD and 4.9% for DOX@MSN-S-S-K(Ac)11.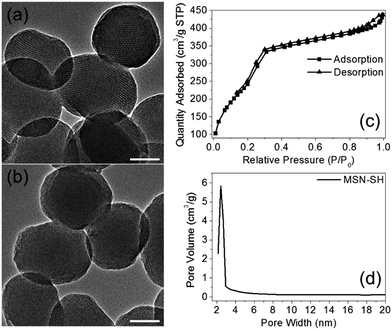 | ||
| Fig. 1 TEM images of MSN-SH (a) and MSN-S-S-RGD (b). Nitrogen adsorption–desorption isotherms (c) and Barrett–Joyner–Halenda pore distribution (d) of MSN-SH. Scale bars: 50 nm. | ||
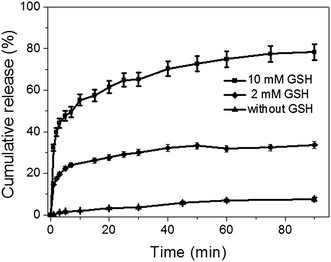
In vitro release studies
The antitumor drug, DOX, was efficiently loaded into the mesopores of silica nanoparticles to produce DOX@MSN-S-S-RGD by stirring MSN-S-S-RGD in a DMF solution of DOX for 24 h. The release of the loaded DOX from DOX@MSN-S-S-RGD in PBS was monitored using fluorescence measurements. In the absence of reductive agents, such as GSH, DOX@MSN-S-S-RGD, was capped by RGD peptides and less than 18% of drug leakage was determined over a period of 24 h (Fig. S6†). Anchoring of the gatekeeper (RGD containing peptide) on the surface of the MSN-S-S-alkyne by the “click reaction” turned out to be an effective procedure to cap the nanoparticles. The addition of reductive agents, GSH, to the PBS suspension of DOX@MSN-S-S-RGD can efficiently trigger the burst release of the entrapped DOX after a fast removal of the RGD peptides by breaking the disulfide bonds. As shown in Fig. 2, the DOX release percentage reached 32% after the introduction of 10 mM GSH in 1 min and over 78% after 90 min. When the concentration of GSH was 2 mM, approximately 33% of DOX was released over 90 min. The DOX cumulative release percentage increased with an increasing concentration of GSH. The results suggest that the incorporated RGD containing peptide can act as an effective nanovalve and most of the DOX could be entrapped in the mesopores of the silica nanoparticles.Cellular uptake measured by CLSM and flow cytometry
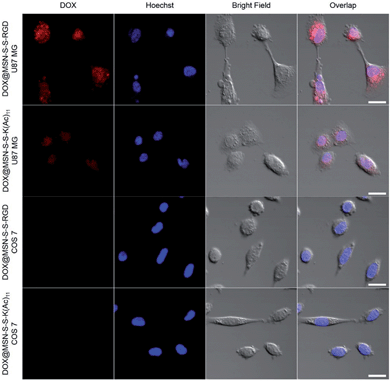
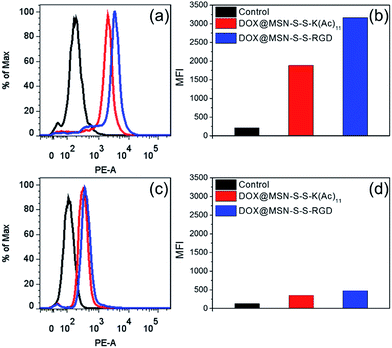
In order to verify the tumor-targeted drug delivery of DOX@MSN-S-S-RGD, the N3-GKKKKKKKKKKK-NH2 peptides were anchored onto the surface of the MSN-S-S-alkyne via “click chemistry”, and then the amino groups were acetylated to prepare MSN-S-S-K(Ac)11 nanoparticle as a control. Herein, acetylation of the surface amino groups is a necessity to exclude the interference that positively charged nanoparticles are more likely endocytosed by cells due to the negatively charged plasma membrane.27 To confirm the RGD receptor-mediated endocytosis of the multifunctional nanoparticles, either DOX@MSN-S-S-RGD or DOX@MSN-S-S-K(Ac)11 nanoparticles were separately incubated with avβ3-integrin-positive tumor cell line (U87 MG) and avβ3-integrin-negative cell line (COS 7) at a DOX concentration of 2 μg mL−1. As shown in Fig. 3, a strong red fluorescence was observed from U87 MG cells incubated with DOX@MSN-S-S-RGD for 2 h. Moreover, the red fluorescence of DOX@MSN-S-S-RGD was apparently stronger than that of DOX@MSN-S-S-K(Ac)11, indicating the targeting attributes of the RGD containing sequence on the DOX@MSN-S-S-RGD surface. For COS 7, a very weak red fluorescence can be detected after 2 h of incubation with nanoparticles, and there was a negligible difference in cellular red fluorescence for the both types of nanoparticles (Fig. 3). It is worth noting that the U87 MG cellular uptake for these two types of nanoparticles were apparently higher compared to COS 7.Flow cytometry analysis was carried out to quantitatively estimate the cellular uptake behavior of DOX@MSN-S-S-RGD and DOX@MSN-S-S-K(Ac)11 in two cell lines. As shown in Fig. 4a, the U87 MG cellular uptake of DOX@MSN-S-S-RGD was obviously higher than that of DOX@MSN-S-S-K(Ac)11 and the mean fluorescence intensity (MFI) value from U87 MG incubating with DOX@MSN-S-S-RGD was 1.7-fold higher than that of DOX@MSN-S-S-K(Ac)11 (Fig. 4b). On the contrary, there was little difference between the MFI values for COS 7 (Fig. 4d). CLSM and flow cytometry results demonstrated that DOX@MSN-S-S-RGD tended to be internalized into U87 MG cells via receptor-mediated endocytosis, which was consistent with that reported in the literature.28
In vitro cytotoxicity
As we know, targeted DOX delivery into tumor cells can efficiently increase the intracellular drug concentration, inhibit cell growth, and ultimately leads to cell death. In addition, a safe drug carrier can effectively eliminate unwanted bystander effects in biological systems.29 It was determined that before drug loading, there was no apparent toxicity for either MSN-S-S-RGD or MSN-S-S-K(Ac)11 towards both cell lines even at a high nanoparticle concentration of 200 mg L−1 (Fig. 5a and b). When we incubated U87 MG cells with DOX@MSN-S-S-RGD and DOX@MSN-S-S-K(Ac)11 individually at different DOX doses for 48 h, it was determined that DOX@MSN-S-S-RGD was more toxic compared to DOX@MSN-S-S-K(Ac)11 (Fig. 5c). However, almost no difference can be observed in COS 7 cells between the two types of DOX-loaded nanoparticles (Fig. 5d). Above results were attributed to the enhanced cellular internalization of nanoparticles via receptor-mediated endocytosis with the aid of the RGD containing sequence for DOX@MSN-S-S-RGD. It worth noting that DOX@MSN-S-S-RGD and DOX@MSN-S-S-K(Ac)11 in U87 MG cells or COS 7 cells exhibited reduced cytotoxicity than free DOX (Fig. 5c and d).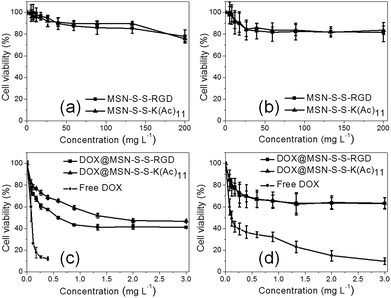 | ||
| Fig. 5 Viability of U87 MG cells (a and c) or COS 7 cells (b and d) incubated with nanoparticles before (a and b), after (c and d) DOX loading and free of DOX (c and d) at different doses. | ||
Conclusions
A novel intelligent drug delivery system, DOX@MSN-S-S-RGD, was constructed by anchoring the gatekeeper, RGD containing peptides on the surface of MSNs using disulfide bonds. It was demonstrated that DOX@MSN-S-S-RGD can be effectively capped by RGD containing peptides. With the aid of the RGD moiety, DOX@MSN-S-S-RGD exhibited enhanced tumor cell uptake. After being taken up by the tumor cells, the anticancer drug, DOX can be released in a burst mode via breaking the disulfide bonds triggered by intracellular GSH. We believe the DOX@MSN-S-S-RGD nanocarrier demonstrated here may have a great potential for cancer therapy.Acknowledgements
We acknowledge the financial support from the Ministry of Science and Technology of China (2011CB606202), the Ministry of Education of China (20120141130003), Natural Science Foundation of Hubei Province of China (2013CFA003) and the Fundamental Research Fund for the Central Universities of China.References
- S. Angelos, Y. W. Yang, K. Patel, J. F. Stoddart and J. I. Zink, Angew. Chem., Int. Ed., 2008, 47, 2222–2226 CrossRef CAS PubMed.
- C. Park, K. Lee and C. Kim, Angew. Chem., Int. Ed., 2009, 48, 1275–1278 CrossRef CAS PubMed.
- L. Xing, H. Zheng, Y. Cao and S. Che, Adv. Mater., 2012, 24, 6433–6437 CrossRef CAS PubMed.
- Z. Y. Li, Y. Liu, X. Q. Wang, J. J. Hu, Q. Xu, L. H. Liu, H. Z. Jia, W. H. Chen, Q. Lei, L. Rong and X. Z. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 14568–14575 CAS.
- H. Meng, M. Xue, T. Xia, Z. Ji, D. Y. Tarn, J. I. Zink and A. E. Nel, ACS Nano, 2011, 5, 4131–4144 CrossRef CAS PubMed.
- A. M. Sauer, A. Schlossbauer, N. Ruthardt, V. Cauda, T. Bein and C. Brauchle, Nano Lett., 2010, 10, 3684–3691 CrossRef CAS PubMed.
- D. Xiao, H. Z. Jia, J. Zhang, C. W. Liu, R. X. Zhuo and X. Z. Zhang, Small, 2014, 10, 591–598 CrossRef CAS PubMed.
- G. F. Luo, W. H. Chen, Y. Liu, Q. Lei, R. X. Zhuo and X. Z. Zhang, Sci. Rep., 2014, 4, 6064 CrossRef PubMed.
- S. Wu, X. Huang and X. Du, Angew. Chem., Int. Ed., 2013, 52, 1–6 CrossRef.
- Z. Y. Li, Y. Liu, X. Q. Wang, L. H. Liu, J. J. Hu, G. F. Luo, W. H. Chen, L. Rong and X. Z. Zhang, ACS Appl. Mater. Interfaces, 2013, 5, 7995–8001 CAS.
- C. Y. Lai, B. G. Trewyn, D. M. Jeftinija, K. Jeftinija, S. Xu, S. Jeftinija and V. S. Li, J. Am. Chem. Soc., 2003, 125, 4451–4459 CrossRef CAS PubMed.
- S. Giri, B. G. Trewyn, M. P. Stellmaker and V. S. Lin, Angew. Chem., Int. Ed., 2005, 44, 5038–5044 CrossRef CAS PubMed.
- F. Muhammad, M. Guo, W. Qi, F. Sun, A. Wang, Y. Guo and G. Zhu, J. Am. Chem. Soc., 2011, 133, 8778–8781 CrossRef CAS PubMed.
- J. Liu, X. Du and X. Zhang, Chem.–Eur. J., 2011, 17, 810–815 CrossRef CAS PubMed.
- Y. L. Sun, B. J. Yang, S. X. Zhang and Y. W. Yang, Chem.–Eur. J., 2012, 18, 9212–9216 CrossRef CAS PubMed.
- D. R. Radu, C. Y. Lai, K. Jeftinija, E. W. Rowe, S. Jeftinija and V. S. Lin, J. Am. Chem. Soc., 2004, 126, 13216–13217 CrossRef CAS PubMed.
- Z. Luo, K. Cai, Y. Hu, L. Zhao, P. Liu, L. Duan and W. Yang, Angew. Chem., Int. Ed., 2011, 50, 640–643 CrossRef CAS PubMed.
- C. Chen, F. Pu, Z. Huang, Z. Liu, J. Ren and X. Qu, Nucleic Acids Res., 2011, 39, 1638–1644 CrossRef CAS PubMed.
- R. Cheng, F. Feng, F. H. Meng, C. Deng, J. Feijen and Z. Y. Zhong, J. Controlled Release, 2011, 152, 2–12 CrossRef CAS PubMed.
- A. N. Koo, H. J. Lee, S. E. Kim, J. H. Chang, C. Park, C. Kim, J. H. Park and S. C. Lee, Chem. Commun., 2008, 6570–6572 RSC.
- E. Wagner, Expert Opin. Biol. Ther., 2007, 7, 587–593 CrossRef CAS PubMed.
- P. M. Friden, L. R. Walus, G. F. Musso, M. A. Taylor, B. Malfroy and R. M. Starzyk, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 4771–4775 CrossRef CAS.
- E. Ruoslahti, Annu. Rev. Cell Dev. Biol., 1996, 12, 697–715 CrossRef CAS PubMed.
- M. Prabaharan, J. J. Grailer, S. Pilla, D. A. Steeber and S. Gong, Biomaterials, 2009, 30, 3009–3019 CrossRef CAS PubMed.
- Y. W. Ebright, Y. Chen, Y. Kim and R. H. Ebright, Bioconjugate Chem., 1996, 7, 380–384 CrossRef CAS PubMed.
- M. Amblard, J. A. Fehrentz, J. Martinez and G. Subra, Mol. Biotechnol., 2006, 33, 239–254 CrossRef CAS PubMed.
- E. C. Cho, J. Xie, P. A. Wurm and Y. Xia, Nano Lett., 2009, 9, 1080–1084 CrossRef CAS PubMed.
- G. F. Luo, W. H. Chen, Y. Liu, J. Zhang, S. X. Cheng, R. X. Zhuo and X. Z. Zhang, J. Mater. Chem. B, 2013, 1, 5723–5732 RSC.
- Q. Zhang, F. Liu, K. T. Nguyen, X. Ma, X. J. Wang, B. G. Xing and Y. L. Zhao, Adv. Funct. Mater., 2012, 22, 5144–5156 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4tb01533a |
| ‡ These authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2015 |