Open Access Article
Open Access ArticleRaman-encoded microbeads for spectral multiplexing with SERS detection†
Yuming
Lai
abd,
Shuqing
Sun
c,
Tao
He
d,
Sebastian
Schlücker
a and
Yuling
Wang
*a
aFaculty of Chemistry, University of Duisburg-Essen, D-45141, Germany. E-mail: yuling.wang@uni-due.de
bNational Center for Materials Service Safety, University of Science and Technology Beijing, Beijing 100083, China
cLaboratory of Optical Imaging and Imaging, Shenzhen Key Laboratory for Minimally Invasive Medical Technologies, Graduate School at Shenzhen, Tsinghua University, Shenzhen, 518055, China
dLaboratory of Nanodevice, National Center for Nanoscience and Technology, Beijing, 100190, China
First published on 20th January 2015
Abstract
Simultaneous detection of multiple molecular targets can greatly facilitate early diagnosis and drug discovery. Encoding micron-sized beads with optically active tags is one of the most popular methods to achieve multiplexing. Noble metal nanoparticle labels for optical detection by surface-enhanced Raman spectroscopy (SERS) exhibit narrow bandwidths, high photostability and intense Raman signals. In this study, we demonstrate the feasibility of spectral multiplexing by SERS using micron-sized polystyrene (PS) beads loaded with SERS-active nanoparticles. The silica-encapsulated SERS nanotags comprise gold nanocrystals with a self-assembled monolayer (SAM) of aromatic thiols as Raman reporter molecules for spectral identification. SERS microspectroscopic images of single Raman-encoded PS microbeads indicate the homogeneous spatial distribution of the SERS-active nanoparticles on the surface of the beads. By using up to five different Raman reporters, 31 spectrally distinct micron-sized beads were encoded and characterized spectroscopically at the single-bead level.
Introduction
Multiplexed identification of target molecules is particularly crucial for genomics and proteomics,1–3 medical diagnosis4 and drug discovery,5 which demand high throughput analysis of a large amount of biomolecules in a single reaction vessel. Traditional methods for multiplexed detection include multiplex ELISA (enzyme-linked immunosorbant assay)6,7 and microarrays,8,9 which actually suffer from some major drawbacks in data and array reproducibility, slow reaction kinetics due to the planar arrangement of probes, high cost and poor flexibility.10 Encoding strategies based on micron-sized beads have been emerging as attractive alternatives to current techniques for multiplexed detection schemes owing to their great advantages including enhanced reaction kinetics (suspension in homogenous solution), rapidity (shorter incubation and array times, faster analysis), simplicity of use, low cost, higher reproducibility and reliability.11 Typically, this technology is based on encoding the microbeads with a unique identifier. A variety of techniques have been proposed and addressed for encoding. Among these, encoding by quantum dots (QDs)12,13 and molecular fluorophores/dyes14,15 are most frequently used. Self-encoding with polymer particles as beads was also proposed by Fenniri's group, using FT-IR or Raman spectroscopy for read-out.16,17Surface-enhanced Raman spectroscopy (SERS)18 has recently attracted great attention due to its high sensitivity, high photostability, and more importantly, the large multiplexing capability because of the narrow bands/fingerprint of the Raman molecules.19–23 Jun et al. have recently proposed an approach to encode beads by in situ growth of silver nanoparticles as SERS enhancer, followed by Raman reporters encoding, which shows the great potential by using SERS labels as tags for encoding of microbeads.24 However, the co-adsorption of Raman reporter molecules with SiO2 precursors (3-mercaptopropyltrimethoxysilane), leading to a sub-monolayer coverage of Raman reporter molecules on the surface, will lower the SERS intensity. On the other hand, the in situ growth of silver nanoparticles will generate different silver nanostructures, thus leading to poor reproducibility for the Raman enhancement. To improve the reproducibility and obtain maximum SERS signals, Schlücker and co-workers have introduced silica-encapsulated SERS labels with a complete self-assembled monolayer (SAM) of Raman reporters on the surface of noble metal nanoparticles (NPs) including gold (Au).25–28 The maximum surface coverage with Raman reporters on the surface of the AuNPs leads to maximum SERS signals and good reproducibility, while the silica surface enables further chemical modifications for bioconjugation.
Hence, a more general strategy was proposed to address the feasibility for a multiplexed SERS detection by using five different types of silica-encapsulated SAM-based SERS labels (AuNPs-Ra@SiO2) for encoding microbeads. Monodisperse AuNPs-Ra@SiO2 were first synthesized based on approaches reported earlier.25–28 SERS images of PS beads encoded by different SERS labels validate the multiplexing capability of this approach. With five types of SERS labels, all possible combinations (31 different codes) were obtained. We employ a binary notation for characterizing these combinations, where “1” represents the presence and “0” the absence of the corresponding SERS “colour”, respectively. We expect to have five kinds of spectrally distinct codes with one type of SERS label (10000, 01000, 00100, 00010, 00001), 10 different codes with two types of SERS labels (11000, 10100, 10010, 10001, 01100, 01010, 01001, 00110, 00101, 00011), 10 different codes with three types of SERS labels (11100, 11010, 10110, 01110, 11001, 10101, 01101, 10011, 01011, 00111), five different codes with four different SERS labels (01111, 10111, 11011, 11101, 11110), and one code with all five SERS labels (11111).
Experimental
Chemicals
4-Mercaptobenzoic acid (MBA), 5,5′-dithiobis(2-nitrobenzoic) (DTNB), 2,7-mercapto-4-methylcoumarin (MMC), 2-mercapto-4-methyl-5-thiazoleacetic acid (MMTAA), ethanol, isopropanol, tetraethoxyorthosilicate (TEOS), (3-aminopropyl)-trimethoxysilane (APTMS), ammonium hydroxide (30%), N′-(ethylcarbonimidoyl)-N,N-dimethyl-monohydrochloride EDC, N-hydroxysulfosuccinimide sodium salt (s-NHS) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), were purchased from Sigma-Aldrich/Fluka. 2,3,5,6-Tetrafluoro-4-mercaptobenzoic acid (TFMBA) was purchased from Tokyo Chemical Industry (TCI) Co. MNBA was prepared by breaking the disulfide bond in DTNB with NaBH4 as reducing agent before use. Monodisperse polystyrene beads (PS) that contain surface carboxyl groups (PS–COOH) were purchased from micromod Partikeltechnologie GmbH. Ultrapure water (18.2 M cm) was used preparing all aqueous solution.Monodispersed gold nanoparticles (AuNPs) with a diameter of around 70 nm were prepared through the procedure reported by Bastús et al.29
Preparation of SERS nanoparticle labels
Silica-encapsulated SAM-based SERS labels were obtained by encapsulation of Raman reporter-coated AuNPs (AuNPs-Ra) with a silica shell.25,27,28 Briefly, 300 mL colloidal AuNPs with a diameter of about 70 nm (λmax = 548 nm) were centrifuged and redispersed in 100 mL ethanol. The colloid was then incubated overnight with 12 mL Raman reporter solutions (10 mM in ethanol) to form a complete self-assembled monolayer (SAM) on the particle surface. In this study, Raman reporters including MBA, MNBA, TFMBA, MMC and MMTAA (see Materials and methods for the chemical names of the aromatic thiols) were employed as Raman reporter in this proof-of-concept study. SAM-coated AuNPs were centrifuged and redispersed in ethanol (50 mL) to remove the excess Raman reporter molecules. For direct silica encapsulation of the SAM-coated gold nanoparticles (AuNPs–MBA), AuNPs were redispersed in a mixture of isopropanol (45 mL), water (15 mL), and ammonium hydroxide (1.6 mL). TEOS (1 vol% in isopropanol) was then added under vigorous stirring in six portions over 3 h. The thickness of the silica shell was adjusted by the amount of TEOS. For the silica encapsulation of the AuNPs–MNBA, AuNPs–TFMBA, AuNPs–MMC, and AuNPs–MMTAA, APTMS is required before the Stöber synthesis with TEOS. Briefly, 2.7 mL 1 vol% APTMS (in ethanol) were added to the SAM-coated AuNPs. After shaking for 5 min, the colloidal dispersion was centrifuged and redispersed in a mixture of isopropanol (45 mL), water (15 mL), and ammonium hydroxide (1.6 mL). TEOS (1 vol% in isopropanol) was then added under vigorous stirring in six portions over 3 h. The silica-encapsulated SERS labels were centrifuged after stirring overnight to remove excess ammonia and redispersed in 50 mL ethanol. The silica shell was hardened by heating at 65 °C for 5 h in ethanol with 750 rpm shaking, which will greatly increase the stability of AuNPs-Ra@SiO2 according to the report.30Conjugation of SERS NP labels onto the microbead surface
Polystyrene (PS) beads were encoded by SERS NP labels via standard EDC conjugation chemistry. Briefly, silica-encapsulated SERS labels were activated by APTMS to obtain amino-functionalized SERS labels. In a typical experiment, 400 μL SERS label colloid (e.g. AuNPs–TFMBA@SiO2) was washed by ethanol for one more time to remove excess water and then 13 μL 0.5% APTMS together with 13 μL ammonia were added to the SERS colloid under sonication for 5 min and shaking at room temperature. Finally, amino-functionalized SERS labels were washed and redispersed in 400 μL HEPES (pH 5.8) for further conjugation.Monodisperse polystyrene (PS) beads containing surface carboxyl groups were activated by EDC/s-NHS. Briefly, 1.2 mL 0.5 mg mL−1 PS–COOH was centrifuged and redispersed in HEPES buffer (pH 5.8) for activation. 450 μL s-NHS (3 mg in 1.5 mL HEPES buffer) and 450 μL EDC (5 mg in 1.5 mL HEPES buffer) were added into PS–COOH solution and shaked for 20 min to let s-NHS react with carboxyl groups. Then the solution with the PS beads was washed once in HEPES to remove access s-NHS/EDC and redispersed in HEPES. Subsequently, 100 μL amino-functionalized silica-encapsulated SERS labels were added into 50 μL activated PS beads solution under shaking for 1 h. The total volume of SERS labels was kept constant, but the corresponding volume for each type of SERS labels varied, depending on the types of SERS labels encoded on the PS bead. Afterwards, the spectrally encoded micron-sized beads were washed and redispersed in ethanol for SERS microspectroscopic measurements.
Instrumentation
Extinction spectra of AuNPs and AuNPs-Ra@SiO2 were recorded with a Perkin-Elmer Lambda 35 UV-vis absorption spectrometer. Transmission electron microscopy (TEM) images were obtained with a Zeiss EM 902 instrument. Scanning electron microscopy (SEM) images were performed with a FEI Quanta 3D FEG 200/600 scanning electron microscope.Raman experiments were carried out with a WITec alpha 300 R Raman microscope. The 632.8 nm line from a He–Ne laser was used for excitation of Raman scattering. The monochromator comprises a 1800 grooves per mm grating. The acquisition time per Raman spectrum was 0.5 s. The integrated Raman intensity (20 cm−1 “filters”) around the dominant Raman shift of the corresponding Raman reporter molecule was calculated for generating the Raman false colour images. Average spectra were calculated by WITec project software.
Result and discussion
Characterization of SERS labels and PS beads encoded with SERS labels
Five different types of silica-encapsulated SERS labels were synthesized by using five different aryl thiols and characterized by TEM and extinction spectroscopy. Typical TEM images of silica-encapsulated SERS labels with TFMBA as the Raman reporter are displayed in Fig. 1a. Clearly, monodisperse silica-encapsulated AuNPs are observed and the shell thickness is adjusted to be around 12 nm by controlling the amount of TEOS. Silica-encapsulated AuNPs-Ra as SERS labels will therefore ensure the stability of the labels, prevent the interface from the environment and offer a versatile way for surface functionalization. Optical properties of AuNPs and AuNPs-Ra@SiO2 were also investigated by extinction spectroscopy (Fig. S1†), in which the distinct peak located at 548 nm is due to the localized surface plasmon resonance (LSPR) of AuNPs. After the formation of the SAM and the encapsulation by silica, the LSPR of AuNPs was red-shifted to 560 nm owing to the different environmental refractive index. It should be pointed out that the SERS labels are quite stable in water and in ethanol or isopropanol after hardening the silica shell.30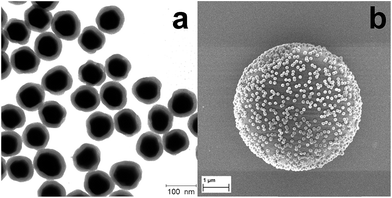 | ||
| Fig. 1 (a) Typical TEM image of SERS labels (AuNPs–TFMBA@SiO2) and (b) SEM image of a single PS bead encoded with SERS nanoparticle labels. | ||
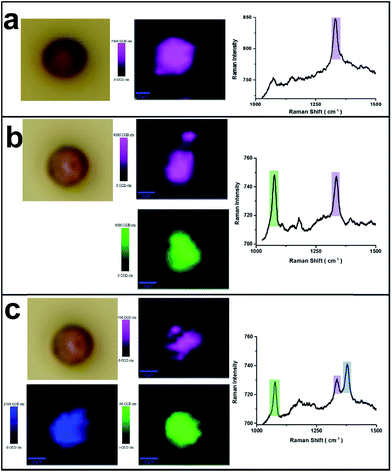
Furthermore, silica-encapsulated AuNPs-Ra were modified by a silane agent (APTMS) to introduce amino groups on the silica surface, which will react with activated carboxyl groups on the surface of the PS beads (∼5 μm). The ratio of SERS labels to PS beads needs to be carefully adjusted. To validate the binding between SERS labels and the PS bead, scanning electron microscopy (SEM) was employed to characterize the SERS labels on the PS beads surface after the reaction as indicated in Fig. 1b: a monolayer of SERS labels was observed clearly on the single PS bead surface. No aggregation or multilayer formation was observed, indicating the strong covalent binding of SERS labels with PS beads.
SERS labels with distinct spectroscopic peaks are needed to achieve multiple spectrally encoded micron-sized beads for optical readout by SERS. As discussed, the small size of aromatic molecules with the uniform orientation on AuNPs surface will enable the multiplexed detection owing to the appearance of only few distinct vibrational Raman peaks. Normalized SERS spectra and the corresponding molecular structures from five types of SERS labels including MBA, MMC, MMTAA, MNBA and TFMBA were obtained as depicted in Fig. 2. Distinct peaks at 1080 cm−1, 1172 cm−1, 1290 cm−1, 1337 cm−1 and 1380 cm−1 were observed, which are characteristic of their own SERS signatures, ensuring an unambiguous spectral identification. The SERS spectrum of each type of SERS label can be easily distinguished because the most intensive peak of each Raman reporter molecule is separated by at least 35 cm−1. Although a mixture of these 5 different SERS labels can also display spectral barcodes, it only shows 5 barcodes, while encoding on PS beads, 31 spectrally distinct SERS signatures (barcodes) can be obtained by mixing up to 5 different Raman reporters.
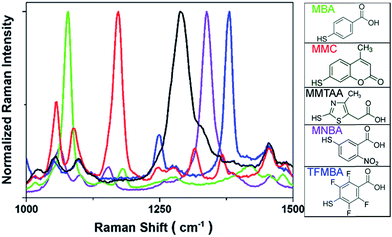 | ||
| Fig. 2 SERS spectra of AuNPs-Ra@SiO2 (left) and molecular structures of Raman reporters employed in this study (right). | ||
Raman mapping of SERS NP-encoded PS beads
The micron-sized PS beads can be easily identified by optical microscopy and then characterized by Raman microscopy. As indicated in Fig. 3, SERS label-encoded PS beads were analysed by SERS microspectroscopy. Fig. 3a shows the results obtained with one type of SERS label encoding the PS bead (binary code: 00010). The false-colour SERS image on a single PS bead was achieved by integrating the Raman intensity between 1320 and 1360 cm−1. It was found that optical and Raman false-colour images showed good correspondence and, importantly, a homogenous distribution of SERS signals across the entire PS bead, demonstrating the uniform coverage of SERS labels on the bead surface. This is very important for SERS applications in multiplexed detection to ensure the reproducibility, reliability and low-cost because the measurement can be performed on a single bead. The average Raman spectrum of the whole bead with SERS labels (MNBA) is shown in the right of Fig. 3a, exhibiting the characteristic peak of the symmetric nitro stretching vibration at 1337 cm−1.SERS images from other types of SERS label-encoded PS beads were characterized accordingly. The corresponding Raman false colours are shown in Fig. 3b and c, representing the combinations 10010 and 10011, respectively. Notably, the average SERS intensity on each type of SERS label-encoded PS bead varies. Typically, the Raman intensity of the characteristic peak from one SERS label decreased with increasing amounts of other SERS labels on the same bead. For instance, the Raman intensity of the characteristic nitro peak at 1337 cm−1 from only one type of SERS label (00010) was ca. 80 counts, while the Raman intensity of the same peak decreased to ca. 40 counts and 20 counts, respectively, in the case of the combination 10010 and 10011, in which two different SERS labels are present on the same PS bead. This can easily be explained by the constant total number of SERS nanoparticle labels on the PS bead surface. Because the encoding of SERS labels on PS bead surface is based on covalent binding and the formation of a monolayer of SERS nanoparticles (as illustrated in the SEM image of Fig. 1b), the total amount of SERS labels on one PS bead is nearly the same. These results further demonstrate the feasibility of spectroscopically encoded beads for multiplexed SERS detection. Overall, 31 different spectral barcodes by SERS label-encoded PS beads can theoretically be achieved (only binary notations, i.e., not including different intensity ratios).
Spectral barcodes from SERS label-encoded PS beads
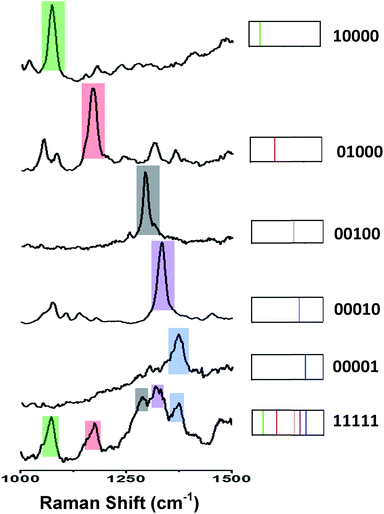
Fig. 4 (left) shows the average Raman spectra from five SERS-NP-coated PS beads, each one spectrally encoded with a different Raman reporter (single “colour”), together with the average Raman spectrum from a PS bead encoded by a mixture of all 5 types of SERS NP labels (five “colours”). Each bead with one “colour” shows a characteristic peak and the bead encoded with five types of SERS labels can be distinguished clearly by 5 distinct peaks, which are from each type of SERS label. The corresponding barcode representations of the Raman spectra are shown in Fig. 4 (right). In the spectral barcode representation, each characteristic Raman peak is represented by a vertical line in a different colour.The other 25 SERS spectra and the corresponding barcodes, which were obtained experimentally by a combination of up to four types SERS labels, are shown in Fig. 5 and 6. Overall, we can differentiate 31 kinds of SERS label-encoded PS beads, which could be very useful in multiplexed identification using single beads.
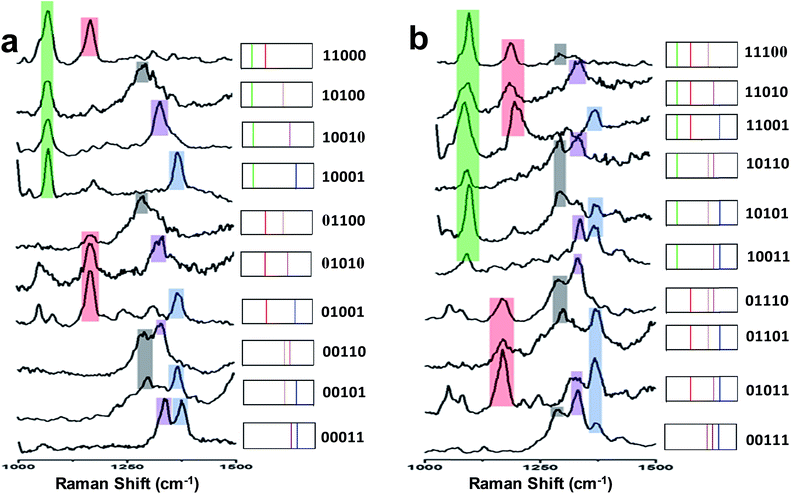 | ||
| Fig. 5 (a) SERS spectra and barcode representation of two types SERS labels on PS bead. (b) SERS spectra and barcode representation of three type SERS labels on PS bead. | ||
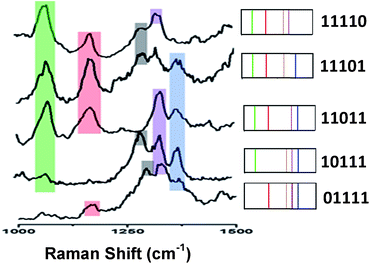 | ||
| Fig. 6 Typical SERS spectra and barcode representation pictures of four type SERS labels on PS bead. | ||
Conclusions
In summary, silica-encapsulated self-assembled monolayers of Raman reporter-coated AuNPs (AuNPs-Ra@SiO2) were synthesized and employed as bright and stable SERS labels for encoding single micron-sized PS beads for multiplexed optical detection. The particles are characterized by extinction spectroscopy, TEM and SEM. Single SERS label-encoded PS beads were further studied by SERS microspectroscopy. The SERS nanoparticles are homogeneously distributed across the single bead. Using up to 5 different Raman reporters, 31 distinct spectral barcodes from single micron-sized bead were obtained, demonstrating the great potential of this approach for future multiplexed biomarker detection schemes based on SERS.Acknowledgements
Financial support from the German Research Foundation (DFG/WA3369/1-1) is acknowledged. Y. M. Lai thanks the China Scholarship Council for the financial support of her research visit at the University of Duisburg-Essen and Fundamental Research Funds for the Central Universities (FRF-TP-14-058A1) for the financial support. T.H. would like to thank National Research Fund for Fundamental Key Projects no. 973 (2011CB933200) for the financial support.References
- G. A. Evans and K. A. Lewis, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 5030 CrossRef CAS.
- P. Ng, J. J. S. Tan, H. S. Ooi, Y. L. Lee, K. P. Chiu, M. J. Fullwood, K. G. Srinivasan, C. Perbost, L. Du, W. K. Sung, C. L. Wei and Y. Ruan, Nucleic Acids Res., 2006, 34, e84 CrossRef PubMed.
- E. Boja, T. Hiltke, R. Rivers, C. Kinsinger, A. Rahbar, M. Mesri and H. Rodriguez, J. Proteome Res., 2011, 10, 66 CrossRef CAS PubMed.
- D. A. A. Vignali, J. Immunol. Methods, 2000, 243, 243 CrossRef CAS.
- K. S. Lam, M. L. Lam and K. Viktor, Chem. Rev., 1997, 97, 411 CrossRef CAS PubMed.
- Quansys Bio multiplex ELISA kit specifications: http://www.Quansysbio.com.
- Ray Biotech multiplex ELISA kit specifications: http://www.raybiotech.com.
- G. L. G. Miklos and R. Maleszka, Nat. Biotechnol., 2004, 22, 615 CrossRef CAS PubMed.
- R. D. Canales, Y. Luo, J. C. Willey, B. Austermiller, C. C. Barbacioru, C. Boysen, K. Hunkapiller and R. V. Jensen, Nat. Biotechnol., 2006, 24, 1115 CrossRef CAS PubMed.
- S. Birtwell and H. Morgan, Integr. Biol., 2009, 1, 345 RSC.
- B. Battersby and M. Trau, Aust. J. Chem., 2007, 60, 343 CrossRef CAS.
- L. C. Mattheakis, J. M. Dias, Y. Choi, J. Gong, M. P. Bruchez, J. Liu and E. Wang, Anal. Biochem., 2004, 327, 200 CrossRef CAS PubMed.
- Y. Zhao, H. C. Shum, H. Chen, L. L. A. Adams, Z. Gu and D. A. Weitz, J. Am. Chem. Soc., 2011, 133, 8790 CrossRef CAS PubMed.
- X. W. Zhao, Z. B. Liu, H. Yang, K. Nagai, Y. H. Zhao and Z.-Z. Gu, Chem. Mater., 2006, 18, 2443 CrossRef CAS.
- B. J. Battersby, D. Bryant, W. Meutermans, D. Matthews, M. L. Smythe and M. Trau, J. Am. Chem. Soc., 2000, 122, 2138 CrossRef CAS.
- H. Fenniri, O. Terreau, S. Chun, S. J. Oh, W. F. Finney and M. D. Morris, J. Comb. Chem., 2006, 8, 192 CrossRef CAS PubMed.
- J. Raez, D. R. Blais, Y. Zhang, R. A. Alvarez-Puebla, J. P. Bravo-Vasquez, J. P. Pezacki and H. Fenniri, Langmuir, 2007, 23, 6482 CrossRef CAS PubMed.
- S. Schlücker, ChemPhysChem, 2009, 10, 1344 CrossRef PubMed.
- S. Schlücker, Angew. Chem., Int. Ed., 2014, 53, 4756 CrossRef PubMed.
- Y. L. Wang and S. Schlücker, Analyst, 2013, 138, 2224 RSC.
- H. Qu, Y. M. Lai, D. Z. Niu and S. Q. Sun, Anal. Chim. Acta, 2013, 763, 38 CrossRef CAS PubMed.
- Y. M. Lai, F. Li and S. Q. Sun, Integr. Ferroelectr., 2013, 146, 88 CrossRef CAS PubMed.
- Y. M. Lai, J. Wang, T. He and S. Q. Sun, Anal. Lett., 2014, 47, 833 CrossRef CAS PubMed.
- B. H. Jun, J. H. Kim, H. Park, J. S. Kim, K. N. Yu, S. M. Lee, H. Choi, S. Y. Kwak, Y. K. Kim, D. H. Jeong, M. H. Cho and Y. S. Lee, J. Comb. Chem., 2007, 9, 237 CrossRef CAS PubMed.
- B. Küstner, M. Gellner, M. Schütz, F. Schöppler, A. Marx, P. Ströbel, P. Adam, C. Schmuck and S. Schlücker, Angew. Chem., Int. Ed., 2009, 48, 1950 CrossRef PubMed.
- M. Gellner, K. Kömpe and S. Schlücker, Anal. Bioanal. Chem., 2009, 394, 1839 CrossRef CAS PubMed.
- M. Schütz, B. Küstner, M. Bauer, C. Schmuck and S. Schlücker, Small, 2010, 6, 733 CrossRef PubMed.
- M. Schütz, M. Salehi and S. Schlücker, Chem.–Asian J., 2014, 9, 2219 CrossRef PubMed.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098 CrossRef PubMed.
- Y. J. Wong, L. Zhu, W. S. Teo, Y. W. Tan, Y. Yang, C. Wang and H. Chen, J. Am. Chem. Soc., 2011, 133, 11422 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra16163g |
| This journal is © The Royal Society of Chemistry 2015 |