Open Access Article
Open Access ArticleVariable lignin structure revealed in Populus leaves†
Nathan Bryant a,
Nancy Engle
a,
Nancy Engle b,
Timothy Tschaplinski
b,
Timothy Tschaplinski b,
Yunqiao Pu
b,
Yunqiao Pu b and
Arthur J. Ragauskas
b and
Arthur J. Ragauskas *abc
*abc
aDepartment of Chemical and Biomolecular Engineering, University of Tennessee, Knoxville, Tennessee 37996, USA
bBioEnergy Science Center & Center for Bioenergy Innovation, Biosciences Division, University of Tennessee-Oak Ridge National Laboratory Joint Institute for Biological Science, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA
cDepartment of Chemical and Biomolecular Engineering, University of Tennessee, Center for Renewable Carbon, Department of Forestry, Wildlife, and Fisheries, University of Tennessee Institute of Agriculture, Knoxville, TN 37996, USA. E-mail: aragausk@utk.edu
First published on 5th July 2023
Abstract
Lignin has long been a trait of interest, especially in bioenergy feedstocks such as Populus. While the stem lignin of Populus is well studied, foliar lignin has received significantly less consideration. To this end, leaves from 11 field grown, natural variant Populus trichocarpa genotypes were investigated by NMR, FTIR, and GC-MS. Five of these genotypes were sufficiently irrigated, and the other six genotypes were irrigated at a reduced rate (59% of the potential evapotranspiration for the site) to induce drought treatment. Analysis by HSQC NMR revealed highly variable lignin structure among the samples, especially for the syringyl/guaiacyl (S/G) ratio, which ranged from 0.52–11.9. Appreciable levels of a condensed syringyl lignin structure were observed in most samples. The same genotype subjected to different treatments exhibited similar levels of condensed syringyl lignin, suggesting this was not a response to stress. A cross peak of δC/δH 74.6/5.03, consistent with the erythro form of the β-O-4 linkage, was observed in genotypes where significant syringyl units were present. Principle component analysis revealed that FTIR absorbances associated with syringyl units (830 cm−1, 1317 cm−1) greatly contributed to variability between samples. Additionally, the ratio of 830/1230 cm−1 peak intensities were reasonably correlated (p-value < 0.05) with the S/G ratio determined by NMR. Analysis by GC-MS revealed significant variability of secondary metabolites such as tremuloidin, trichocarpin, and salicortin. Additionally, salicin derivatives were found to be well correlated with NMR results, which has been previously hypothesized. These results highlight previously unexplored nuance and variability associated with foliage tissue of poplar.
Lignin is vital for many biological functions such as tissue development, water transport, and stress tolerance.1 The lignin polymer is typically described as having three primary monolignols: sinapyl alcohol, coniferyl alcohol, and p-coumaryl alcohol.2 These phenylpropane units, are produced through the general phenylpropanoid and monolignol specific pathways, and are subsequently incorporated into the lignin polymer as syringyl (S), guaiacyl (G), and p-hydroxyphenyl (H) units, respectively.3
Populus is poised to be a promising resource for renewable biofuels and bioproducts due to favorable characteristic such as fast growth and high cellulose content.4 The cell wall biopolymer lignin from Populus stem tissue has been extensively characterized due its importance in applications such as bioenergy.5 However, less is known about lignin associated with leaf tissue. Most of the studies on this topic describe the lignin content6–8 but not its structure.
Leaves produce a host of secondary metabolites, including lignin. However, from our literature search, there are few, if any, studies which offer detailed examination of the lignin structure from natural variant Populus foliage. This represents a knowledge gap, as there is potentially interesting lignin chemistry occurring in leaves. Indeed, lignification in leaves has recently become a topic of interest.8,9 It is well documented that lignin traits vary between biomass type (i.e., hardwood vs. softwood vs. herbaceous).10 Additionally, differences in lignin content, structure, and enzyme activity have been observed between tissue type, such as between stem and leaf.11,12 In one study, PdeWRKY65-UGT75L28 was found to impact lignification of leaf petioles, which could impact the transport of water or nutrients and ultimately plant growth.13 Several MYB proteins have been implicated in the lignification of stem lignin. Additionally, two MYB proteins have also been shown to impact the lignification of leaves.14,15 Abiotic stress has also been demonstrated to alter lignin structure. For instance, poplar leaves exposed to ozone were observed to produce condensed lignin structure.16 Foliage lignin is also important to ecosystems, as it has been shown to play a significant role in leaf litter decomposition.17,18 While tree stems have long been studied as a biofuel feedstock, tree leaf biomass has also recently become a topic of investigation. Tree leaf biomass has shown promise in bioenergy production, and can especially be beneficial in utilized fallen leaf material urban areas,19,20 Similar results have also been shown with leaf biomass from other species.20 We have therefore characterized foliage samples harvested from 11 Populus trichocarpa natural variant genotypes from a Genome-wide Association Study population at Boardman, OR (3-year-old trees established in 2016) by heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR), Fourier transform infrared (FTIR) spectroscopy, and gas chromatography-mass spectrometry (GC-MS), with an emphasis on examining lignin structure.
Materials and methods
Plant growth and sampling
Foliage samples were harvested from three-year-old, natural variant Populus trichocarpa in July 2018. Samples were named based on the following convention. The first part (i.e., “1121”) denotes the genotype. The second part denotes whether the samples was grown in the north end (“-N”) or south end (“-S”) of the field. The field site near Boardman, OR was established in 2016. During growth, this area experienced a natural environmental drought. The field was segregated into north and south regions. The north end was irrigated and therefore samples from the north end (denoted “-N”) of the field are designated as non-drought (control) samples. The south end (denoted “-S”) was irrigated at 59% of potential evapotranspiration (PET) for Boardman, OR in 2017 and this treatment regime was maintained through 2020, with this treatment designated as the drought samples. Irrigation delivered to the control treatment targeted 100% PET. The south end had been subjected to drought for 2 growing seasons (2017, 2018) before sampling in the middle of the third growing season (July 2018).Metabolite sampling and characterization
One leaf was collected from each tree. To limit environmental variability, leaves collected were of similar in physiological age, position on the tree, and sun exposure. Leaf samples were immediately fast-frozen in the field prior to shipment on dry ice and stored at −80 °C until they were lyophilized and ground to a powder using a SPEX Sample Prep Geno/Grinder. Metabolites were extracted from ∼25 mg of powdered tissue twice overnight with 2.5 mL of 80% ethanol. Sorbitol (75 μL of 1 mg mL−1 aq.) was added as an internal standard to the first day extract as an internal standard. The extracts were combined and a 500 μL aliquot was dried under nitrogen and silylated to generate trimethylsilyl (TMS) derivatives, as described previously.21,22 After 2d, 0.1 μL aliquots were injected into an Agilent Technologies Inc. (Santa Clara, CA, USA) 7890A/5975C inert XL gas chromatograph-mass spectrometer fitted with a Restek Rtx-5MS with Integra-guard (5% diphenyl/95% dimethyl polysiloxane) 30 m × 250 μm × 0.25 μm film thickness capillary column and operated using conditions previously described.21 A large user-created database of mass spectral electron impact ionization (EI) fragmentation patterns of TMS-derivatized compounds as well as the Wiley Registry 10th Edition was used to identify metabolites of interest to be quantified. Metabolite peaks were extracted using a key selected ion, characteristic mass-to-charge (m/z) ratio, scaled back to total ion current using pre-determined scaling factors and normalized to the quantity of internal standard recovered, mass extracted, volume analyzed, and injection volume.Lignin isolation and analysis
After lyophilization and grinding to a powder using a SPEX Sample Prep Geno/Grinder, samples were Soxhlet extracted with toluene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol (2
ethanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v
1, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) overnight to remove extractives. We make the distinction that this toluene
v) overnight to remove extractives. We make the distinction that this toluene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol extraction differs from the 80% ethanol extraction described in the previous section, and aliquots were only subjected to one extraction unique to the intended analysis. After Soxhlet extraction, samples were air dried for at least 24 h. Extractives free samples were then ground on a Retsch planetary ball mill in a zirconium oxide (ZrO2) jar and ZrO2 balls. Grinding was conducted for two hours at 600 RPM at five-minute intervals to avoid overheating (i.e., a 5 minute grinding interval immediately followed by a 5 minute rest interval). The ball milled samples were subjected to enzymatic hydrolysis with a cellulase enzyme for 48 h in a sodium acetate buffer (pH 5.0) to digest polysaccharides. After enzymatic hydrolysis, samples were centrifuged and washed with DI water two times to remove the cellulase enzyme. The resulting enzyme lignin was lyophilized for at least 48 h to remove residual moisture.
ethanol extraction differs from the 80% ethanol extraction described in the previous section, and aliquots were only subjected to one extraction unique to the intended analysis. After Soxhlet extraction, samples were air dried for at least 24 h. Extractives free samples were then ground on a Retsch planetary ball mill in a zirconium oxide (ZrO2) jar and ZrO2 balls. Grinding was conducted for two hours at 600 RPM at five-minute intervals to avoid overheating (i.e., a 5 minute grinding interval immediately followed by a 5 minute rest interval). The ball milled samples were subjected to enzymatic hydrolysis with a cellulase enzyme for 48 h in a sodium acetate buffer (pH 5.0) to digest polysaccharides. After enzymatic hydrolysis, samples were centrifuged and washed with DI water two times to remove the cellulase enzyme. The resulting enzyme lignin was lyophilized for at least 48 h to remove residual moisture.
Enzyme lignin was analyzed via 2D HSQC NMR with a Bruker Avance II 500-MHz spectrometer. Approximately 40 mg of lignin enriched residue was dissolved in DMSO-d6 in a 5 mm NMR tube and subsequently sonicated for 1 h. The standard hsqcetgpsip2.2 Bruker pulse sequence was used with an N2 cryoprobe with the following specifications: 1H spectra width of 12 ppm and 1024 data points; 13C spectra width of 220 ppm with 256 increments and 32 scans. All HSQC spectra were analyzed with Bruker TopSpin 3.5pl6 software. The DMSO-d6 solvent peak at δC/δH 39.5/2.49 was used to calibrate the spectra, and the S2/6 signal was used at an internal standard. The signals used for volume integration are as follows: δC/δH 103.8/6.70 ppm for S2/6, δC/δH 111.0/6.98 ppm for G2, δC/δH 128.2/7.17 ppm for H2/6, δC/δH 130.4/7.62 ppm for PB2/6, δC/δH 128.3/6.45 ppm for Iα, δC/δH 71.8/4.86 for β-O-4, δC/δH 86.8/5.46 for β-5, and δC/δH 84.8/4.65 for β–β. All results are presented on an S + G basis and should be interpreted as abundance per 100 aromatic units. Sample 856-S was selected for a whole cell wall NMR analysis. Approximately 40 mg of toluene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol extracted and ball milled material was dissolved directly in a co-solvent system of DMSO-d6/HMPA-d18 (4
ethanol extracted and ball milled material was dissolved directly in a co-solvent system of DMSO-d6/HMPA-d18 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v
1, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) in an NMR tube and analyzed by the same method discussed above.
v) in an NMR tube and analyzed by the same method discussed above.
FTIR and PCA
Leaf tissue that had been Soxhlet extracted with toluene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol was analyzed via FTIR with a PerkinElmer Spectrum 100 FTIR spectrometer with a universal attenuated total reflection (ATR) accessory. A background scan was completed prior to analyzing samples. Each sample was analyzed from 600 cm−1 to 4000 cm−1 with a 32-scan accumulation and a 2 cm−1 resolution. Data was collected with Spectrum software. All spectra were baseline corrected manually using twelve anchor points. Spectra were then normalized around from [0,1]. The second derivative of baseline corrected, normalized spectra in the 1800–800 cm−1 region were utilized in the principal component analysis (PCA) was performed with Origin Pro 2023 software using the PCA for spectroscopy tool (v1.30).
ethanol was analyzed via FTIR with a PerkinElmer Spectrum 100 FTIR spectrometer with a universal attenuated total reflection (ATR) accessory. A background scan was completed prior to analyzing samples. Each sample was analyzed from 600 cm−1 to 4000 cm−1 with a 32-scan accumulation and a 2 cm−1 resolution. Data was collected with Spectrum software. All spectra were baseline corrected manually using twelve anchor points. Spectra were then normalized around from [0,1]. The second derivative of baseline corrected, normalized spectra in the 1800–800 cm−1 region were utilized in the principal component analysis (PCA) was performed with Origin Pro 2023 software using the PCA for spectroscopy tool (v1.30).
Results and discussion
HSQC NMR
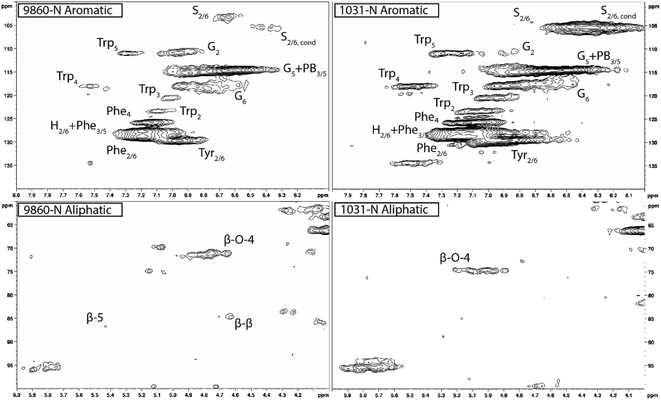
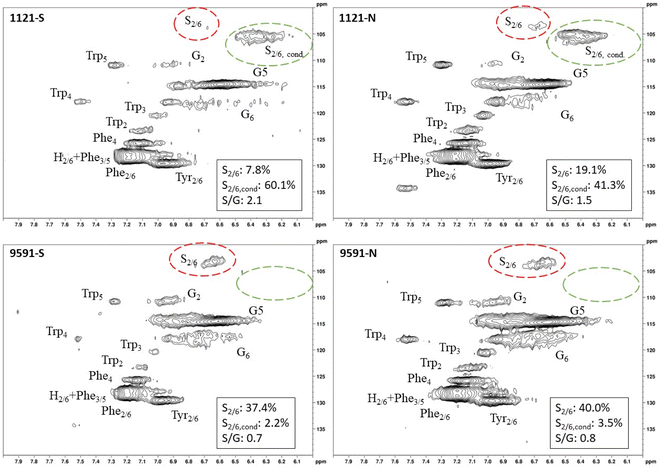
Heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) was utilized to characterize the isolated lignin structure from the leaf tissue. Specifically, the aromatic region (δC/δH 100–140/6.0–8.0) of the spectra was examined for monolignol content, and the aliphatic region (δC/δH 100–60/6.0–4.0) was examined for interunit linkage content. Samples were observed to have appreciable S units as indicated by the S2/6 peak (δC/δH 103.7/6.71). All samples exhibited a signal at (δC/δH 110.7/6.98), indicating the presence of G units. The S/G ratio ranged from 0.41 to 11.9, which is a surprisingly high degree of variability (Fig. 2). For all samples, the β-O-4 aryl ether linkage was the most abundant, as indicated by the signal at (δC/δH 71.8/4.86). The β-O-4 linkage is usually positively correlated with the S/G ratio. However, this relationship was not observed here. However, setting aside the two high S/G samples (856-S and 1031-N), β-O-4 levels were correlated with uncondensed S units with a correlation coefficient (CC) = 0.57. A similar observation has been made in a previous study, where β-O-4 was correlated with S but not the S/G ratio.23 It could be that condensed S units, at least partly, influenced the stereochemistry of linkage formation. This is supported by an observed negative correlation between condensed S units and β-O-4 levels (CC = −0.46). β-5 and β–β linkages were observed at (δC/δH 86.8/5.47) and (δC/δH 84.8/4.64), respectively, but at levels significantly lower than β-O-4 linkages.Most samples also contained condensed S units as indicated by the signal around δC/δH 6.32/106.4. However, condensed G units were not observed, suggesting that S units are selectively condensed. In a previous study, the presence of condensed monolignols was observed through decreased thioacidolysis yields.16 However, thioacidolysis works by cleaving the liable β-O-4 ether bond, and therefore highly condensed lignin structures may bias results. Indeed, Cabané et al.16 reported that the proportion of S units decreased as condensed units increased (i.e., thioacidolysis yield decreased). While the referenced study introduced ozone stress which induced condensed lignin, it would be interesting to evaluate whether only condensed S units were present, or if all monolignols exhibited condensed structures. This observation highlights a unique beneficial utility offered by HSQC NMR.
Initially, there appears to be an abundance of H units as indicated by the signal at (δC/δH 127.9/7.19). However, upon closer inspection, this instead resembles characteristic amino acid contamination that has been described in HSQC NMR spectra.24 To avoid erroneous quantification of H units due to contamination, this signal was not included in the semi-quantitative evaluation of lignin structure. One amino acid that appears prominently in the aromatic region is phenylalanine (Phe). Phenylalanine is a precursor to many secondary metabolites and has been identified as a response to drought stress.25,26 Though it did not interfere with lignin signals, tryptophan (Trp) was also observed in the spectra. Increased levels of tryptophan helps regulate osmotic balance in response to drought stress.27 Tyrosine (Tyr) has also been shown to be associated with drought stress tolerance.28 Strong phenylalanine, tryptophan, and tyrosine signals were present in all samples. These amino acids also play a variety of other roles in plant growth and development. The prominence of amino acid signals in these leaf spectra also illustrate the difference in tissue type, as stem tissue typically registers very low or zero amino acid levels.
HSQC NMR revealed tremendous variability among the 11 samples. Perhaps the most striking observation is the degree of variability exhibited in the S/G ratio. While the actual S/G level may vary by analytical method, the S/G ratio of Populus stem lignin is generally reported in the range of 1.5–2.5.5 These 11 foliage tissue samples exhibited a significantly wider range of S/G ratios, ranging from 0.33 to 9.79. It should be noted that leaf lignin and stem lignin (especially mature wood lignin) are expected to have different properties and should not be directly compared. The authors offer this comparison to illustrate the relatively good agreement of analyses associated with the well-studied stem lignin. S units especially exhibited a high degree of variability. From Fig. 1, the S2/6 signal of sample 9860 (S/G = 0.73) is distinguishable at only slightly higher than background levels. However, in sample 1031 (S/G = 9.79), the S2/6 signal is very prominent and clearly more abundant than the G5 signal. As HSQC NMR is a semi-quantitative technique, these absolute differences are difficult to distinguish here. However, Cabané et al. quantified foliar lignin S/G ratio of Populus tremula x alba to be approximately 0.6 by thioacidolysis,16 which compares well with approximately half of the samples analyzed in this study. The β-O-4 linkages also exhibited striking variability across these foliage samples. In stem lignin the β-O-4 bond is the most abundant linkage, with reported values typically in the range of 60–65%. The foliage samples here exhibited β-O-4 content as low as 26.1% and as high as 73.9%, with only two samples (LILC-26-4-S and 9591-S) exceeding 60%. This is consistent with the findings of Cabané et al.,16 who hypothesized the decreased β-O-4 linkage content was due to the lower S/G ratio. Typically, the β-O-4 linkage content is observed to be positively corrected with the S/G ratio.29 However, a negative trend between these two phenotypes is observed here. It is unclear what factors may be contributing to this observed variability of these natural variant, field-grown samples. Similar variability was observed in both the south (drought) and north (control) sample sets, so treatment is likely not a contributing factor. One potential explanation may be that lignin structure varies by leaf anatomy. It is well documented that lignin can vary by tissue type (i.e., leaf vs. stem). In this case, petiole and/or midrib of the leaf may have a lignin structure different from the lamina (i.e., higher S/G ratio), and varying ratios of petiole![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) midrib
midrib![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lamina could explain some of the observed variability.
lamina could explain some of the observed variability.
One surprising observation is that some samples exhibited differential positions of the Cα–Hα shift in the β-O-4 linkage structure. The Cα–Hα shift of the β-O-4 substructures can vary depending on several factors. For instance, a difference in Cα–Hα chemical shifts has been overserved between G-unit linked β-O-4 substructures and S-unit linked β-O-4 substructures.30 However, the genotype with the lowest S/G ratio (425-N) exhibited a cross peak consistent with the typical β-O-4 assignment in Populus (δC/δH 71.6/4.79). By contrast, the genotype with the highest S/G ratio (1031-N) exhibited a β-O-4 cross peak around (δC/δH 74.6/5.03). Therefore, differences in shifts due to G-linked and S-linked β-O-4 substructures seems unlikely. Differences in Cα–Hα chemical shifts associated with erythro and thero conformations of the β-O-4 linkage have also been reported.31 It has been observed that the erythro form of the β-O-4 dominates in angiosperms, whereas a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mixture of erythro and threo forms are typically present in gymnosperms.32 This is due, at least in part, to the S/G ratio since S units preferentially form the erythro form of the β-O-4 linkage. However, once again examining the genotype 1031-N spectra, the contribution of S units is almost exclusively from condensed S units. Indeed, it was observed that genotypes which exhibited primarily condensed S units also exhibited β-O-4 cross peaks in the δC/δH 74.6/5.03 region. By contrast, genotypes with non-condensed S units exhibited β-O-4 cross peaks in the δC/δH 71.6/4.79 region. Therefore, the observed differences in the chemical shifts of the β-O-4 cross peaks are attributed to the erythro and threo forms of the substructure. Similar findings of unexpected lignin structures in leaves and differences in erythro/thero ratios has also been reported in ginkgo leaf.33 This finding could offer an opportunity for understanding additional factors influencing the erythro/thero ratio of the β-O-4 linkages, which has been shown to impact delignification.34
50 mixture of erythro and threo forms are typically present in gymnosperms.32 This is due, at least in part, to the S/G ratio since S units preferentially form the erythro form of the β-O-4 linkage. However, once again examining the genotype 1031-N spectra, the contribution of S units is almost exclusively from condensed S units. Indeed, it was observed that genotypes which exhibited primarily condensed S units also exhibited β-O-4 cross peaks in the δC/δH 74.6/5.03 region. By contrast, genotypes with non-condensed S units exhibited β-O-4 cross peaks in the δC/δH 71.6/4.79 region. Therefore, the observed differences in the chemical shifts of the β-O-4 cross peaks are attributed to the erythro and threo forms of the substructure. Similar findings of unexpected lignin structures in leaves and differences in erythro/thero ratios has also been reported in ginkgo leaf.33 This finding could offer an opportunity for understanding additional factors influencing the erythro/thero ratio of the β-O-4 linkages, which has been shown to impact delignification.34
While these samples consist of enzyme lignin, another commonly utilized strategy for HSQC NMR analysis is whole cell wall (WCW) analysis, wherein the whole cell wall (after extraction and ball milling) is directly dissolved in the NMR solvent, therefore bypassing the enzymatic hydrolysis step. Sample 856-S was selected for whole cell wall WCW NMR analysis as a comparison to the isolated enzyme lignin (ESI Fig. S1†). The typical S2/6 and G2 signals were noticeably absent from the WCW spectra but were observed to be well resolved in the enzyme lignin spectra. It is expected that these results are influenced by the low lignin content associated with leaf tissue, which has been measured to be approximately 10%.8 The enzymatic hydrolysis procedure allows a more lignin rich residue to be analyzed which improves the corresponding signals. Samples were also subjected to two step acid hydrolysis to determine Klason lignin content. However, this procedure produced suspect results, and similar difficulties with Klason lignin measurement of foliage tissue has been previously documented.35
FTIR
Fourier-transform infrared (FTIR) spectroscopy is an analytical tool widely used to identify the functional groups of many compounds, including lignin. Care must be taken when interpreting FTIR spectra of biomass, as bands can be attributed to multiple biomass components. Fig. 3 displays the FTIR spectra of the toleuene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
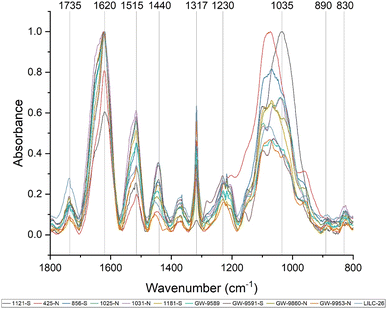
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol extracted samples, which will contain cell wall components besides lignin (i.e., sugars). Prominent bands in the FTIR spectra are summarized in Table 2 and were assigned based on comparison with existing published literature. All samples exhibited a wide band centered around 3290 cm−1, which is associated with O–H stretching. Peaks were also observed at 2920 and 2850 cm−1, corresponding to C–H vibration of CH2 and CH3 functional groups. Fig. 3 displays the absorbance spectra of the fingerprint region of 1800–800 cm−1, as this region typically provides the most information regarding cell wall structure. The peak at approximately 1735 cm−1 represent is typically associated with xylan and has also been associated with stretching vibration of nonconjugated and conjugated ketones. The peak at 1440 cm−1 corresponds to asymmetric O–H deformations in cellulose. The 1515 cm−1 peak is attributed to lignin aromatic skeletal vibrations.
ethanol extracted samples, which will contain cell wall components besides lignin (i.e., sugars). Prominent bands in the FTIR spectra are summarized in Table 2 and were assigned based on comparison with existing published literature. All samples exhibited a wide band centered around 3290 cm−1, which is associated with O–H stretching. Peaks were also observed at 2920 and 2850 cm−1, corresponding to C–H vibration of CH2 and CH3 functional groups. Fig. 3 displays the absorbance spectra of the fingerprint region of 1800–800 cm−1, as this region typically provides the most information regarding cell wall structure. The peak at approximately 1735 cm−1 represent is typically associated with xylan and has also been associated with stretching vibration of nonconjugated and conjugated ketones. The peak at 1440 cm−1 corresponds to asymmetric O–H deformations in cellulose. The 1515 cm−1 peak is attributed to lignin aromatic skeletal vibrations.
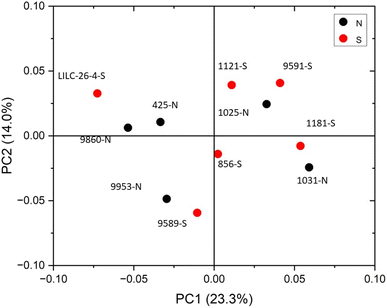
The FTIR spectra were further analyzed by principal component analysis (PCA). PCA is a useful mathematical procedure for analyzing data. The goal of applying PCA to FTIR is to transform a large data set (i.e., thousands of data points from FTIR spectra) into a few key parameters called principal components (PCs). The resulting PCs are typically characterized by the amount of variation they represent, with PC1 accounting for the most variation, PC2 accounting for the second most variation, and so forth. Additionally, PC scores are assigned to each sample, and samples that have similar spectra will be scored similarly. The final result is that PCA reduces the FTIR spectra of many samples down to two dimensions (typically PC1 and PC2), which can then be conveniently represented on a familiar X–Y plot. Samples with similar PC scores will be plotted in close proximity, revealing cluster patterns and allowing differentiation between different groups. The resulting PC1 and PC2 scores from each sample are presented in the score plot in Fig. 4. All spectra were baseline corrected, normalized from [0, 1], and the second derivative of the spectra were taken. Data in the fingerprint region of 1800–800 cm−1 were considered for PCA. PC1 and PC2 accounted for 23.3% and 14.0% of the variability, respectively. The remaining PCs accounted for less than 12% (each) of the variability. No obvious spatial clustering patterns were observed. Specifically, samples from the south (S; drought treatment) and north (N; irrigated treatment) sets are not readily distinguished by PCA at the 95% confidence interval, indicating that treatment was not a significant contributor to the observed variability. Examining the loadings of each PC provides insight as to which structural features (as determined by wavenumbers) contribute most significantly to the observed variation. PC1 is driven primarily by the band around 1317 cm−1, which is typically associated with the C–O stretching of the S unit ring. This aligns with the (semi-quantitative) observation from the NMR spectra, where the S unit signal is more variable than the G unit signal. Wavenumbers that contributed to PC2 include the peak between 1620–1630 cm−1, which is associated with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic skeletal vibrations. The peak around 830 cm−1, associated with C–H bending of syringyl units, also contributed to PC2. Another contributor to PC2 includes wavenumber 1081 cm−1, which is a shoulder of the peak centered around 1035 cm−1 peak associated with C–O stretching of primary alcohols in lignin and polysaccharides. The peak around 1685 cm−1, associated with conjugate carbonate of carboxylic acid and ketone groups, was common to both PC1 and PC2.
C aromatic skeletal vibrations. The peak around 830 cm−1, associated with C–H bending of syringyl units, also contributed to PC2. Another contributor to PC2 includes wavenumber 1081 cm−1, which is a shoulder of the peak centered around 1035 cm−1 peak associated with C–O stretching of primary alcohols in lignin and polysaccharides. The peak around 1685 cm−1, associated with conjugate carbonate of carboxylic acid and ketone groups, was common to both PC1 and PC2.
Various peaks in the FTIR spectrum have been associated with lignin structure, such as syringyl and guaiacyl units. The peak around 1317 cm−1 is associated with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of syringyl units. The peak at 1230 cm−1 associated with C–O, C–C, and C
O stretching of syringyl units. The peak at 1230 cm−1 associated with C–O, C–C, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of guaiacyl units. The ratio of intensities at the 1317 cm−1 peak (syringyl) and 1230 cm−1 peak (guaiacyl) are often used to estimate the S/G ratio of biomass.42 However, the ratio of these two peaks was not well correlated with the S/G ratio. This is likely due to the influence of polysaccharide bands in the FTIR spectra. For instance, a typical lignin peak at around 1594 cm−1, associated with aromatic skeletal vibrations and C
O stretching of guaiacyl units. The ratio of intensities at the 1317 cm−1 peak (syringyl) and 1230 cm−1 peak (guaiacyl) are often used to estimate the S/G ratio of biomass.42 However, the ratio of these two peaks was not well correlated with the S/G ratio. This is likely due to the influence of polysaccharide bands in the FTIR spectra. For instance, a typical lignin peak at around 1594 cm−1, associated with aromatic skeletal vibrations and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching in lignin, was not present. However, the ratio between the 830 cm−1 peak (associated with syringyl units) and the 1230 cm−1 peak (associated with G units) was observed to be well correlated (CC = 0.58, p-value = 0.03) with the S/G ratio determined by NMR. These observations support the S/G measurement obtained by NMR, but also illustrate benefit of utilizing HSQC NMR for analyzing lignin structure.
O stretching in lignin, was not present. However, the ratio between the 830 cm−1 peak (associated with syringyl units) and the 1230 cm−1 peak (associated with G units) was observed to be well correlated (CC = 0.58, p-value = 0.03) with the S/G ratio determined by NMR. These observations support the S/G measurement obtained by NMR, but also illustrate benefit of utilizing HSQC NMR for analyzing lignin structure.
Metabolite profiling
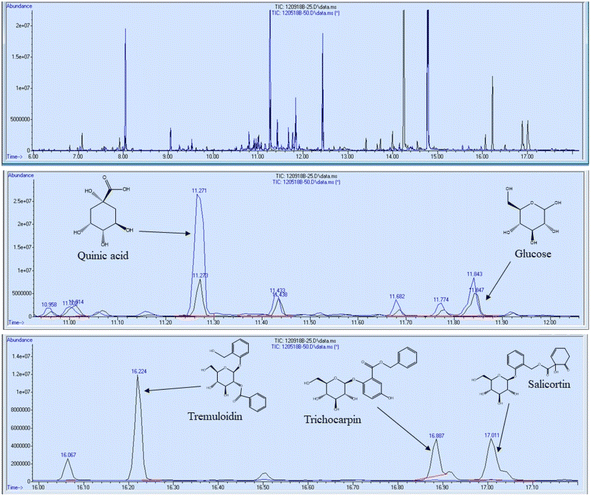
In addition to FTIR and NMR analyses, the 11 foliage samples were subjected to metabolite profiling. Samples were extracted with 80% ethanol, dried under nitrogen, and silylated to generate trimethylsilyl (TMS) derivatives prior to analysis by gas chromatography-mass spectrometry (GC-MS). Sorbitol was used as the internal standard. A summary of some of the most abundant metabolites are reported in Table 3 on the basis of μg metabolite per g dry weight (DW) in sorbitol equivalents. A full list of quantified metabolites is available in the ESI.† Sucrose was by far the most abundant metabolite – approximately 3.5 times more abundant than the second most abundant metabolite (tremuloidin). Other highly abundant metabolites include salicortin, salicin, myo-inositol, quinic acid, glucose, and trichocarpin. Although tremuloidin was the second most abundant metabolite on average, two samples (1025-N, 1031-N) observed to be lacking any measurable tremuloidin, which is a reason these two genotypes were selected for this study. Additionally, sample 1031-N was observed to have low levels of salicin, trichocarpin, catechol, and tremulacin, which were highly abundant in most other samples. This is illustrated in the overlaid total ion chromatograms (TICs) of sample 1031-N and LILC-26-4-S displayed in Fig. 6. From this figure it can be observed that sample LILC-26-4-S had prominent signals at a retention time (RT) of 16.887 and 17.011 corresponding to trichocarpin and salicortin, respectively. However, these peaks were barely discernible in the 1031-N TIC. A similar phenomenon was observed with the signal at RT = 16.224 associated with tremuloidin.As the samples from the north side of the field (denoted with “-N” in the sample name) were irrigated and samples from the south side of the field (denoted with “-S” in the sample name) were subjugated to drought conditions, one may expect to observed established differences in metabolites associated with drought stress. However, this was not necessarily the case. For instance, malic acid has previously been found to increase in response to drought stress,43 though these drought samples has lower malic acid levels (3290 μg g−1 DW) compared to irrigated samples (8751 μg g−1 DW). High variability was observed in levels of tremulacin and salicortin among the samples. On average tremulacin levels were higher in the irrigated samples and salicortin levels were higher in the drought samples. These phenolic glycosides have been linked to roles in herbivore and/or pathogen defense,44 suggesting there may be biotic stressors impacting these samples.
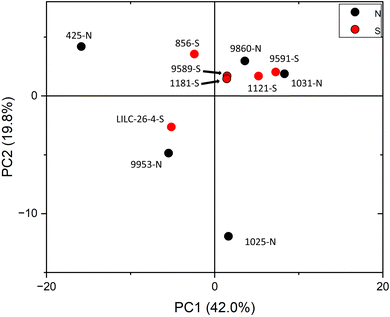
PCA was also utilized to distinguish samples based on the variability of metabolite profiles, with results plotted in Fig. 5. To avoid biasing the results based on the differences in magnitudes of the metabolite concentrations, data were standardized by the standard deviation. PC1 and PC2 accounted for 42.0% and 19.8% of the variability, respectively. Other PCs accounted for less than 14% (each) of the variability. Like the FTIR PCA, 6–7 samples are clustered closer together in (or just outside) quadrant I, whereas 4–5 samples are more dispersed. Loadings for PC2 indicate that it is highly driven by shikimic acid, fructose, glucose, galactose, and raffinose. Loadings for PC1 are less differentiable, but are highly influenced by salicylic acid, catechol, caffeic acid, stearic acid, and maleic acid. PCA did not distinguish samples by treatment at the 95% confidence interval, again indicating that this was not a major contributor to metabolite variability.
The metabolite profiles were also correlated with the lignin traits elucidated by NMR to explore potential relationships between metabolites and lignin structure in leaves. The resulting Pearson correlation coefficients are tabulated in Table 1. One of the ways Populus adapts to a water deficit is through drought tolerance. Drought tolerance mechanisms aim to maintain biological function under stress conditions. For instance, the transcription factor PtoMYB170 was shown to influence drought tolerance and lignin deposition.45 Overexpression of PtroMYB170 induced expression of many lignin biosynthesis genes compared to wild type, including PAL, C4H2, 4Cl5, HCT1, C3H3, CCOAOMOT1, F5H2, CCR2, COMT2, and CAD1. F5H and COMT are responsible for the hydroxylation and methylation, respectively, of coniferyl alcohol and coniferaldehyde, and therefore would be expected to impact the S/G ratio. PtoMYB170 was found to be highly expressed in stem in young leaf tissue, but had low expression in roots, petioles, and mature leaves. In this study, several metabolites were found to be correlated with various lignin traits. Among these correlated metabolites were the salicyloids, salicin and salicylic acid. Specifically, both metabolites were negatively correlated with the S/G ratio. Additional salicin derivatives such as benzyl-salicylic acid-2-O-glucoside, salicyl-coumaroyl-glucoside, and salicyl alcohol also exhibited strong correlations to lignin S/G ratio.
| Sample | S | S, cond. | S, total | G | S/G | β-O-4 | β-5 | β–β |
|---|---|---|---|---|---|---|---|---|
| 9589-S | 1.11 | 59.8 | 60.9 | 39.1 | 1.56 | 22.1 | 4.24 | 1.97 |
| 1181-S | 2.33 | 71.5 | 73.9 | 26.1 | 2.83 | 50.3 | 2.75 | 1.19 |
| 1121-S | 7.82 | 60.1 | 67.9 | 32.1 | 2.12 | 17.9 | 2.47 | 1.73 |
| 856-S | 4.01 | 85.8 | 89.8 | 10.2 | 8.83 | 83.2 | 13.1 | 4.18 |
| LILC-26-4-S | 31.7 | 5.61 | 37.3 | 62.7 | 0.59 | 63.3 | 5.53 | 2.92 |
| 9591-S | 37.4 | 2.2 | 39.6 | 60.4 | 0.70 | 63.6 | 2.9 | 3.5 |
| 9860-N | 23.1 | 11.5 | 34.5 | 65.5 | 0.53 | 26.6 | 5.17 | 1.88 |
| 1031-N | 0.38 | 91.9 | 92.3 | 7.74 | 11.9 | 17.8 | 1.93 | 0.62 |
| 9953-N | 33.8 | 4.08 | 37.8 | 62.2 | 0.61 | 43.5 | 6.15 | 2.87 |
| 1025-N | 26.5 | 20.2 | 46.7 | 53.3 | 0.88 | 74.5 | 10.3 | 2.96 |
| 425-N | 33.0 | 1.22 | 34.2 | 65.8 | 0.52 | 43.5 | 4.28 | 3.67 |
| Observed Peak (cm−1) | Peak assignment | Reference |
|---|---|---|
| 1735 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching in lignin and hemicellulose O stretching in lignin and hemicellulose |
36 |
| 1620 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching O stretching |
37 |
| 1515 | Aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C skeletal vibrations in lignin C skeletal vibrations in lignin |
36 and 38–40 |
| 1440 | O–H in-place deformation in cellulose | 37 |
| 1318 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of syringyl units O stretching of syringyl units |
36, 38 and 40–42 |
| 1230 | C–C, C–O, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of guaiacyl unit O stretching of guaiacyl unit |
36, 40 and 41 |
| 1035 | C–O stretching | 36, 38 and 42 |
| 890 | C–H deformation vibration of cellulose | 42 |
| 830 | C–H bending of syringyl units | 42 |
| Abundance (μg g−1 DW in sorbitol equivalents) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | 1121-S | 1181-S | 9589-S | 9860-N | 9953-N | 1031-N | 9591-S | 1025-N | LILC-26-4-S | 856-S | 425-N |
| Sucrose | 189![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 837 837 |
260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895 895 |
271![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 979 979 |
222![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 162 162 |
457![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 405 405 |
146![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732 732 |
145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 955 955 |
426![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 783 783 |
378![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 962 962 |
224![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151 151 |
407![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 349 349 |
| Tremuloidin | 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 907 907 |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 741 741 |
95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 449 449 |
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 725 725 |
209![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 857 857 |
0 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 878 878 |
0 | 139![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440 440 |
39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 062 |
176![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 411 411 |
| Salicin | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 348 348 |
51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 195 195 |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689 689 |
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 474 474 |
97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 508 508 |
103 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 818 818 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 613 613 |
101![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 144 144 |
44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 351 351 |
94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 061 061 |
| Myo-inositol | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 369 369 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636 636 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 932 932 |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191 191 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 732 732 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 209 209 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249 249 |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 563 563 |
33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 033 033 |
33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 720 |
41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 203 203 |
| Quinic acid | 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 883 883 |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 465 465 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 693 693 |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 131 131 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 392 392 |
55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 523 523 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092 092 |
143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 311 311 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 148 148 |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 405 405 |
8004 |
| Glucose | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 119 119 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 283 283 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 227 227 |
8872 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 190 190 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 444 444 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 488 |
129![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 027 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 380 |
8564 | 7702 |
| Trichocarpin | 2921 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 313 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
8845 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
64 | 3822 | 2127 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094 094 |
6280 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742 742 |
| Malic acid | 2976 | 1542 | 4371 | 5919 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 463 463 |
3062 | 2409 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 002 002 |
7676 | 767 | 5311 |
| Shikimic acid | 4641 | 5524 | 7534 | 5404 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 291 291 |
6718 | 8727 | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191 191 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 158 158 |
5627 | 4326 |
| Citric acid | 2755 | 391 | 5849 | 3718 | 5051 | 1248 | 1349 | 4973 | 4779 | 626 | 4186 |
| 2-Phenethyl-glucoside | 2608 | 4131 | 6153 | 3666 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 858 858 |
4114 | 1798 | 7280 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 685 685 |
7653 | 9432 |
| Catechol | 1932 | 1565 | 1997 | 1816 | 5674 | 18 | 472 | 1499 | 4326 | 2057 | 7035 |
| Fructose | 5528 | 3976 | 4749 | 2477 | 4357 | 4766 | 7720 | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 158 158 |
6884 | 2640 | 1814 |
| 1,2-Cyclohexanediol glucoside | 2993 | 1768 | 1268 | 1763 | 6040 | 222 | 634 | 1965 | 6951 | 5473 | 8631 |
| 5-Oxo-proline | 675 | 1797 | 1427 | 1111 | 3301 | 982 | 975 | 2276 | 2996 | 895 | 2070 |
| Catechin | 1028 | 2323 | 1369 | 907 | 883 | 1013 | 1619 | 5670 | 433 | 3135 | 1702 |
| α-Linolenic acid | 1136 | 1693 | 1293 | 457 | 3071 | 472 | 2120 | 1339 | 8016 | 1657 | 2222 |
| Digalactosylglycerol | 1855 | 3009 | 1509 | 1413 | 2828 | 524 | 3743 | 2656 | 7403 | 2496 | 2164 |
| Tremulacin | 870 | 3474 | 3377 | 800 | 880 | 0 | 310 | 0 | 1412 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667 667 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 895 895 |
| Threonic acid | 632 | 717 | 1060 | 1159 | 3383 | 1360 | 874 | 3809 | 1885 | 550 | 1050 |
| Galactose | 2253 | 952 | 690 | 550 | 2277 | 1597 | 2430 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 068 068 |
1239 | 560 | 526 |
| Salicortin | 215 | 4188 | 3441 | 6307 | 5513 | 164 | 1497 | 622 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 591 591 |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 949 949 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845 845 |
| Monogalactosylglycerol | 1788 | 2005 | 1124 | 725 | 3481 | 188 | 2786 | 1045 | 8974 | 1545 | 1888 |
| Phosphate | 881 | 1011 | 1492 | 311 | 2912 | 1050 | 1557 | 3434 | 1434 | 546 | 475 |
| Glyceric acid | 810 | 763 | 797 | 494 | 3092 | 920 | 508 | 3009 | 1391 | 583 | 369 |
In Populus sp., salicin biosynthesis has a benzoic acid route,46 which can be derived through the shikimate/chorismite pathway47 or the phenylalanine/cinnamate route.48 Current research indicates that the production pathways of salicyloids and lignin are not competitive processes, though tradeoffs between the two processes have been hypothesized.49 The correlation between salicyloids and the lignin S/G ratio would provide weight to this hypothesis, as increased salicyloid content is correlated with increased ratio of guaiacyl units. Although no p-hydroxybenzoate (PB) units were observed in the lignin of these leaf tissues, stem tissue of Populus usually contains ∼5% PB units. While p-bydroxybenzoate has been found to almost exclusive acylate the S unit of lignin, PB has been observed to exhibit a negative correlation with the S units.29 A future study examining salicyloids and benzoate incorporation of lignin may shed additional light on potential trade-offs. Another metabolite found to be correlated with lignin structure was quinic acid (Table 4). Quinic acid can be a precursor of monolignol synthesis.50 However, its incorporation is associated with the phenylpropanoid pathway product p-coumaroyl CoA, which represents a major branchpoint in the pathway and can be directed to various pathways to produce flavonoids, monolignols, or a number of other compounds.51 The flavonoid quercetin, which was also observed to be associated with lignin S/G ratio, is derived by converting p-coumaryl CoA to chalcone, though malonyl CoA is also a substrate.52 A similar relationship has been reported previously, as N-acetylserotonin methyltransferase (MsASMT1) was shown to impact both lignin S/G ratio and glycosides of quercetin when overexpressed in alfalfa.53 Multiple enzymes catalyze reactions of p-coumaroyl CoA. hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) has higher specificity toward shikimic acid and is associated with lignin biosynthesis [reviewed in ref. 51]. Hydroxycinnamoyl CoA: quinate hydroxycinnamoyl transferase (HQT) utilizes quinic acid more efficiently, and is more closely associated with the production of chlorogenic acids [reviewed in ref. 51].
| Metabolite | S | G | S/G | β-O-4 | β-5 | β–β |
|---|---|---|---|---|---|---|
| Sucrose | −0.35 | 0.35 | −0.45 | 0.15 | 0.33 | 0.26 |
| Tremuloidin | −0.59 | 0.59 | −0.48 | 0.48 | 0.53 | 0.55 |
| Salicin | −0.69 | 0.69 | −0.64 | 0.55 | 0.57 | 0.56 |
| Myo-inositol | −0.23 | 0.23 | −0.49 | 0.07 | 0.03 | 0.17 |
| Quinic acid | 0.65 | −0.65 | 0.38 | −0.77 | −0.55 | −0.70 |
| Glucose | 0.39 | −0.39 | 0.17 | −0.51 | −0.25 | −0.41 |
| Trichocarpin | −0.54 | 0.54 | −0.56 | 0.41 | 0.35 | 0.54 |
| α-Linolenic acid | −0.57 | 0.57 | −0.41 | 0.64 | 0.60 | 0.35 |
| Quercetin | −0.55 | 0.55 | −0.59 | 0.46 | 0.51 | 0.41 |
| Salicylic acid | −0.62 | 0.62 | −0.54 | 0.51 | 0.39 | 0.66 |
| Salicyl-salicylic acid-2-O-glucoside | −0.64 | 0.64 | −0.49 | 0.65 | 0.62 | 0.39 |
| Salicyl-coumaroyl-glucoside | −0.66 | 0.66 | −0.55 | 0.57 | 0.42 | 0.52 |
| Caffeic acid | −0.55 | 0.55 | −0.63 | 0.40 | 0.38 | 0.55 |
| Gallocatechin | 0.72 | −0.72 | 0.54 | −0.74 | −0.62 | −0.70 |
In summary, these results provide an extensive characterization of lignin structure and metabolite abundance in Populus foliage tissue, of which there is currently limited knowledge. A surprisingly high degree of variability was observed in the cell wall structure, especially lignin, through HSQC NMR and FTIR. Specifically, condensed syringyl structures were observed in most samples, and these levels appears to be independent of treatment. A high degree of variability of metabolite abundance was also observed by GC-MS analysis, especially in tremuloidin, trichocarpin, and salicortin. These results demonstrate differences between foliage and more well-studied stem tissue, and also highlight previously unexplored nuance and variability associated with foliage tissue of poplar.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Author contributions
Nathan Bryant: investigation, formal analysis, visualization, writing – original draft preparation Nancy Engle: resources, investigation, formal analysis, writing – original draft preparation Timothy Tschaplinski: conceptualization, supervision, writing – review & editing Yunqiao Pu: resources, investigation, writing – review & editing Arthur J. Ragauskas: conceptualization, supervision, writing – review & editing.Acknowledgements
Funding provided by The Center for Bioenergy Innovation both a U.S. Department of Energy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan). The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights.References
- Q. Liu, L. Luo and L. Zheng, Lignins: biosynthesis and biological functions in plants, Int. J. Mol. Sci., 2018, 19(2), 335 CrossRef PubMed.
- H. Hatakeyama and T. Hatakeyama, Lignin structure, properties, and applications, Biopolymers (Advances in Polymer Science), Springer, 2009, pp. 1–63 Search PubMed.
- R. Vanholme, B. Demedts, K. Morreel, J. Ralph and W. Boerjan, Lignin biosynthesis and structure, Plant Physiol., 2010, 153(3), 895–905 CrossRef CAS PubMed.
- A. J. Ragauskas, C. K. Williams, B. H. Davison, G. Britovsek, J. Cairney, C. A. Eckert, et al., The path forward for biofuels and biomaterials. 2006;311(5760):484–489.
- P. Sannigrahi, A. J. Ragauskas and G. A. Tuskan, Poplar as a feedstock for biofuels: a review of compositional characteristics, Biofuels, Bioprod. Biorefin., 2010, 4(2), 209–226 CrossRef CAS.
- N. Richet, K. Tozo, D. Afif, J. Banvoy, S. Legay and P. Dizengremel, et al., The response to daylight or continuous ozone of phenylpropanoid and lignin biosynthesis pathways in poplar differs between leaves and wood, Planta, 2012, 236(2), 727–737 CrossRef CAS PubMed.
- F. A. Rutigliano, A. V. De Santo, B. Berg, A. Alfani and A. Fioretto, Lignin decomposition in decaying leaves of Fagus sylvatica L. and needles of Abies alba Mill, Soil Biol. Biochem., 1996, 28(1), 101–106 CrossRef.
- T. Xu, S. Zhang, K. Du, J. Yang and X. Kang, Insights into the molecular regulation of lignin content in triploid poplar leaves, Int. J. Mol. Sci., 2022, 23(9), 4603 CrossRef CAS PubMed.
- Y. Fu, P. Win, H. Zhang, C. Li, Y. Shen and F. He, et al., PtrARF2. 1 is involved in regulation of leaf development and lignin biosynthesis in poplar trees, Int. J. Mol. Sci., 2019, 20(17), 4141 CrossRef PubMed.
- S. Zhou, Y. Xue, A. Sharma and X. Bai, Lignin valorization through thermochemical conversion: comparison of hardwood, softwood and herbaceous lignin, ACS Sustainable Chem. Eng., 2016, 4(12), 6608–6617 CrossRef CAS.
- M. Bergs, G. Völkering, T. Kraska, R. Pude, X. T. Do and P. Kusch, et al., Miscanthus x giganteus stem versus leaf-derived lignins differing in monolignol ratio and linkage, Int. J. Mol. Sci., 2019, 20(5), 1200 CrossRef CAS PubMed.
- B. Jiang, T. Cao, F. Gu, W. Wu and Y. Jin, Comparison of the structural characteristics of cellulolytic enzyme lignin preparations isolated from wheat straw stem and leaf, ACS Sustainable Chem. Eng., 2017, 5(1), 342–349 CrossRef CAS.
- T. Nvsvrot, X. Yang, Y. Zhang, L. Huang, G. Cai and Y. Ding, et al., The PdeWRKY65-UGT75L28 gene module negatively regulates lignin biosynthesis in poplar petioles, Ind. Crops Prod., 2023, 191, 115937 CrossRef CAS.
- S. Wang, E. Li, I. Porth, J.-G. Chen, S. D. Mansfield and C. Douglas, Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis, Sci. Rep., 2014, 4(1), 1–7 Search PubMed.
- C. Li, X. Wang, L. Ran, Q. Tian, D. Fan and K. Luo, et al., PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa, Plant Cell Physiol., 2015, 56(12), 2436–2446 CrossRef CAS PubMed.
- M. Cabané, J.-C. Pireaux, E. Léger, E. Weber, P. Dizengremel and B. Pollet, et al., Condensed Lignins Are Synthesized in Poplar Leaves Exposed to Ozone, Plant Physiol., 2004, 134(2), 586–594 CrossRef PubMed.
- A. Cuchietti, E. Marcotti, D. E. Gurvich, A. M. Cingolani and N. P. Harguindeguy, Leaf litter mixtures and neighbour effects: low-nitrogen and high-lignin species increase decomposition rate of high-nitrogen and low-lignin neighbours, Appl. Soil Ecol., 2014, 82, 44–51 CrossRef.
- K. T. Steffen, T. Cajthaml, J. Šnajdr and P. Baldrian, Differential degradation of oak (Quercus petraea) leaf litter by litter-decomposing basidiomycetes, Res. Microbiol., 2007, 158(5), 447–455 CrossRef CAS PubMed.
- K. Mudryk, M. Jewiarz, M. Wróbel, M. Niemiec and A. Dyjakon, Evaluation of urban tree leaf biomass-potential, physico-mechanical and chemical parameters of raw material and solid biofuel, Energies, 2021, 14(4), 818 CrossRef CAS.
- N. D. Montiel-Bohórquez and J. F. Pérez, Assessments. Energy valorization strategies of fallen leaves and woody biomass in a based downdraft gasification-engine power plant, Sustain. Energy Technol. Assess., 2022, 49, 101749 Search PubMed.
- T. J. Tschaplinski, R. F. Standaert, N. L. Engle, M. Z. Martin, A. K. Sangha and J. M. Parks, et al., Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog, Biotechnol. Biofuels, 2012, 5(1), 1–15 CrossRef PubMed.
- P. E. Abraham, H. Yin, A. M. Borland, D. Weighill, S. D. Lim and H. C. De Paoli, et al., Transcript, protein and metabolite temporal dynamics in the CAM plant Agave, Nat. Plants, 2016, 2(12), 1–10 Search PubMed.
- A. Guerra, J. P. Elissetche, M. Norambuena, J. Freer, S. Valenzuela and J. Rodriguez, et al., Influence of lignin structural features on Eucalyptus globulus kraft pulping, Ind. Eng. Chem. Res., 2008, 47(22), 8542–8549 CrossRef CAS.
- H. Kim, D. Padmakshan, Y. Li, J. Rencoret, R. D. Hatfield and J. Ralph, Characterization and elimination of undesirable protein residues in plant cell wall materials for enhancing lignin analysis by solution-state nuclear magnetic resonance spectroscopy, Biomacromolecules, 2017, 18(12), 4184–4195 CrossRef CAS PubMed.
- G. L. Barchet, R. Dauwe, R. D. Guy, W. R. Schroeder, R. Y. Soolanayakanahally and M. M. Campbell, et al., Investigating the drought-stress response of hybrid poplar genotypes by metabolite profiling, Tree Physiol., 2014, 34(11), 1203–1219 CrossRef CAS PubMed.
- H. Jia, L. Wang, J. Li, P. Sun, M. Lu and J. Hu, Comparative metabolomics analysis reveals different metabolic responses to drought in tolerant and susceptible poplar species, Physiol. Plant., 2020, 168(3), 531–546 CrossRef CAS PubMed.
- M. Jamil, M. Ahamd, F. Anwar, Z. A. Zahir, M. A. Kharal and F. Nazli, Inducing drought tolerance in wheat through combined use of l-tryptophan and Pseudomonas fluorescens, Pak. J. Agric. Sci., 2018, 55(2), 331–337 Search PubMed.
- N. Khan, A. Bano, M. A. Rahman, B. Rathinasabapathi and M. A. Babar, UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea (Cicer arietinum) metabolome following long-term drought stress, Cell Environ., 2019, 42(1), 115–132 CrossRef CAS PubMed.
- C. G. Yoo, A. Dumitrache, W. Muchero, J. Natzke, H. Akinosho and M. Li, et al., Significance of lignin S/G ratio in biomass recalcitrance of Populus trichocarpa variants for bioethanol production, ACS Sustainable Chem. Eng., 2018, 6(2), 2162–2168 CrossRef CAS.
- B. Jiang, T. Cao, F. Gu, W. Wu and Y. Jin, Comparison of the structural characteristics of cellulolytic enzyme lignin preparations isolated from wheat straw stem and leaf, ACS Sustainable Chem. Eng., 2017, 5(1), 342–349 CrossRef CAS.
- J.-L. Wen, S.-L. Sun, B.-L. Xue and R.-C. Sun, Recent advances in characterization of lignin polymer by solution-state nuclear magnetic resonance (NMR) methodology, Materials, 2013, 6(1), 359–391 CrossRef PubMed.
- M. Hauteville, K. Lundquist and S. von Unge, NMR studies of lignins. VII: 1 H NMR spectroscopic investigation of the distribution of erythro and threo forms of β-0-4 structures in lignins, Acta Chem. Scand., 1986, 40(1), 31–35 CrossRef.
- Z. Xu, H. Nakamura, T. Akiyama, T. Yokoyama, Z. Jin and K. Sasaki, et al., Syringyl ratio and its relation to the erythro ratio of β-O-4-structure in leaf cell walls, J. Wood Chem. Technol., 2021, 41(2–3), 118–127 CrossRef CAS.
- S. Shimizu, T. Yokoyama, T. Akiyama and Y. Matsumoto, Reactivity of lignin with different composition of aromatic syringyl/guaiacyl structures and erythro/threo side chain structures in β-O-4 type during alkaline delignification: As a basis for the different degradability of hardwood and softwood lignin, J. Agric. Food Chem., 2012, 60(26), 6471–6476 CrossRef CAS PubMed.
- Z. Jin, T. Akiyama, B. Y. Chung, Y. Matsumoto, K. Iiyama and S. Watanabe, Changes in lignin content of leaf litters during mulching, Phytochemistry, 2003, 64(5), 1023–1031 CrossRef CAS PubMed.
- H. Yang, C. G. Yoo, X. Meng, Y. Pu, W. Muchero and G. A. Tuskan, et al., Structural changes of lignins in natural Populus variants during different pretreatments, Bioresour. Technol., 2020, 295, 122240 CrossRef CAS PubMed.
- N. Abidi, L. Cabrales and C. H. Haigler, Changes in the cell wall and cellulose content of developing cotton fibers investigated by FTIR spectroscopy, Carbohydr. Polym., 2014, 100, 9–16 CrossRef CAS PubMed.
- Z. Shi, G. Xu, J. Deng, M. Dong, V. Murugadoss and C. Liu, et al., Structural characterization of lignin from D. sinicus by FTIR and NMR techniques, Green Chem. Lett. Rev., 2019, 12(3), 235–243 CrossRef CAS.
- F. Xu, R.-C. Sun, J.-X. Sun, C.-F. Liu, B.-H. He and J.-S. Fan, Determination of cell wall ferulic and p-coumaric acids in sugarcane bagasse, Anal. Chim. Acta, 2005, 552(1–2), 207–217 CrossRef CAS.
- G. Zhou, G. Taylor and A. Polle, FTIR-ATR-based prediction and modelling of lignin and energy contents reveals independent intra-specific variation of these traits in bioenergy poplars, Plant Methods, 2011, 7, 1–10 CrossRef PubMed.
- H. O. Akinosho, C. G. Yoo, A. Dumitrache, J. Natzke, W. Muchero and S. D. Brown, et al., Elucidating the structural changes to Populus lignin during consolidated bioprocessing with Clostridium thermocellum, ACS Sustainable Chem. Eng., 2017, 5(9), 7486–7491 CrossRef CAS.
- L. M. Kline, D. G. Hayes, A. R. Womac and N. Labbé, Simplified determination of lignin content in hard and soft woods via UV-spectrophotometric analysis of biomass dissolved in ionic liquids, BioResources, 2010, 5(3), 1366–1383 CAS.
- A. H. Ahkami, W. Wang, T. W. Wietsma, T. Winkler, I. Lange and C. Jansson, et al., Metabolic shifts associated with drought-induced senescence in Brachypodium, Plant Sci., 2019, 289, 110278 CrossRef CAS PubMed.
- T. P. Clausen, P. B. Reichardt, J. P. Bryant, R. A. Werner, K. Post and K. Frisby, Chemical model for short-term induction in quaking aspen (Populus tremuloides) foliage against herbivores, J. Chem. Ecol., 1989, 15, 2335–2346 CrossRef CAS PubMed.
- C. Xu, X. Fu, R. Liu, L. Guo, L. Ran and C. Li, et al., PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis, Tree Physiol., 2017, 37(12), 1713–1726 CrossRef CAS PubMed.
- T. Ruuhola and R. Julkunen-Tiitto, Trade-off between synthesis of salicylates and growth of micropropagated Salix pentandra, J. Chem. Ecol., 2003, 29(7), 1565–1588 CrossRef CAS PubMed.
- M. C. Wildermuth, Variations on a theme: synthesis and modification of plant benzoic acids, Curr. Opin. Plant Biol., 2006, 9(3), 288–296 CrossRef CAS PubMed.
- J. R. Widhalm and N. Dudareva, A familiar ring to it: biosynthesis of plant benzoic acids, Mol. Plant, 2015, 8(1), 83–97 CrossRef CAS PubMed.
- I. N. Abreu, A. I. Johansson, K. Sokołowska, T. Niittylä, B. Sundberg and T. R. Hvidsten, et al., A metabolite roadmap of the wood-forming tissue in Populus tremula, New Phytol., 2020, 228(5), 1559–1572 CrossRef CAS PubMed.
- J. Barros, H. Serk, I. Granlund and E. Pesquet, The cell biology of lignification in higher plants, Ann. Bot., 2015, 115(7), 1053–1074 CrossRef CAS PubMed.
- N. V. E. Silva, P. Mazzafera and I. Cesarino, Should I stay or should I go: are chlorogenic acids mobilized towards lignin biosynthesis?, Phytochemistry, 2019, 166, 112063 CrossRef CAS PubMed.
- B. Winkel-Shirley, Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology, Plant Physiol., 2001, 126(2), 485–493 CrossRef CAS PubMed.
- H. Cen, T. Wang, H. Liu, H. Wang, D. Tian and X. Li, et al., Overexpression of MsASMT1 promotes plant growth and decreases flavonoids biosynthesis in transgenic alfalfa (Medicago sativa L.), Front. Plant Sci., 2020, 11, 489 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Correlations between S unit content and interunit linkages (Fig. S1). See DOI: https://doi.org/10.1039/d3ra03142j |
| This journal is © The Royal Society of Chemistry 2023 |