Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
N,N′-di-caboxy methyl perylene diimide (PDI) functionalized CuO nanocomposites with enhanced peroxidase-like activity and their application in visual biosensing of H2O2 and glucose†
Miaomiao Chena,
Yanan Dinga,
Yan Gaoa,
Xixi Zhua,
Peng Wanga,
Zhiqiang Shi*b and
Qingyun Liu*a
aCollege of Chemical and Environmental Engineering, Shandong University of Science and Technology, Qingdao 266510, P. R. China. E-mail: qyliu@sdust.edu.cn; Fax: +86 0532 80681197; Tel: +86 0532 86057757
bSchool of Chemistry, Chemical Engineering and Materials, Shandong Normal University, Jinan 250013, P. R. China
First published on 10th May 2017
Abstract
Nanomaterial-based enzyme mimics (nanoenzymes) are a new forefront of chemical research at present. N,N′-Di-carboxy methyl perylene diimide (PDI) functionalized CuO nanobelts with pores (PDI–CuO nanobelts), which were prepared via a facile method and characterized using various analytical techniques, were demonstrated for the first time to possess higher intrinsic peroxidase-like activity towards classical colorimetric substrate 3,3′,5,5′-tetramethylbenzidine (TMB) in the presence of hydrogen peroxide (H2O2), compared with that of pure CuO nanobelts without PDI modification. The underlying reaction mechanism was elucidated by catalyzing the decomposition of H2O2 and generating hydroxyl radicals (˙OH). The improved catalytic activity of PDI–CuO nanobelts for colorimetric reactions could be ascribed to the synergistic effects of CuO and PDI nanobelts. Moreover, the proposed PDI–CuO biosensor platform exhibited a more sensitive response to H2O2 with a low limit of detection (LOD) of 2.38 μM. This convenient sensing platform was further extended to detect glucose in combination with the specificity of glucose oxidase (GOx) for the oxidation of glucose and generation of H2O2. The linear range for glucose detection was from 2 μM to 50 μM with a low detection limit of 0.65 μM. Therefore, besides high sensitivity and selectivity, this colorimetric system also holds considerable potential for biological process research.
1. Introduction
Most natural enzymes are proteins that can catalyze various biochemical reactions with excellent substrate specificity and efficiency under mild conditions, which is of great significance in the aspects of environmental protection, biotechnology, agriculture and clinical diagnosis.1–4 However, the disadvantages of natural enzymes, such as easy denaturation, high cost of preparation and purification, low stability under environmental conditions and some other shortcomings, restrict their further application in practice.5 To overcome the drawbacks of natural enzymes,6 intensive efforts have been made to develop enzyme mimics, due to their easier preparation, stability and cost reduction.7 In recent years, more and more people have focused on studying of nanomaterials as peroxidase-like mimics, since Yan and co-workers found that Fe3O4 nanoparticles possessed an intrinsic enzyme mimetic activity similar to that of naturally occurring horseradish peroxidase (HRP).8 Thus, an incredible number of investigations on nanomaterials possessing peroxidase-like activity have been reported, such as carbon based nanomaterials (graphene oxide,9 carbon nanotubes,10 iron-based metal–organic framework nanomaterials,11 sulfides (CuS,12 FeS13) etc.).With the developing and widening of studies on peroxidase-like mimics, more and more researchers have paid attention to composites such as peroxidase-like mimics, which realize the combination of the respective properties of each component or to achieve cooperatively enhanced performances.14 Generally, the composites can be variously classified into two kinds: one is inorganic–inorganics and the other is organic–inorganics. For example, as for inorganic–inorganics composites with the peroxidase-like activity, scientists have studied bimetallic nanoparticles as peroxidase-like mimics, such as Fe/Co,15 Au/Pb,16 Pd–Ir core–shell nanocubes.17 Moreover, Au/CuS nanocomposites18 and Au–carbon nanotube nanocomplex19 have been found to demonstrate highly peroxidase activity. In addition, Wang and coworkers have prepared highly dispersed CeO2 on TiO2 nanotube and found that the nanocomposites showed superior peroxidase-like activity.20 Liu and coworkers have comparatively studied composites of CeO2, CuS, Ag2S, ZnS based on Montmorillonite as supports and found that these composites possessed peroxidase-like activity and applied in colorimetric detecting for H2O2.21–24
On the other hand, composites based on organic–inorganics have been found to demonstrate peroxidase-like activity. For example, Yu and co-workers have reported that PVP modified MoS2 nanoparticles offer a sensing flatform to achieve efficient detection for H2O2 and glucose.25 Lv and coworkers have prepared bovine serum albumin (BSA)-stabilized MnO2 nanoparticles (NPs) exhibiting highly peroxidase-like activities.26 Huang and coworkers have reported the peroxidase-like activity of chitosan stabilized silver nanoparticles.27 Li and coworkers have reported poly(styrene sulfonate) and Pt bifunctionalized graphene nanosheets as an artificial enzyme to construct a colorimetric chemosensor.28 Nevertheless, in these previous studies, the organics were all employed as powerful supports to prepare inorganics in nanoscale, while not used as organic functional molecules.
Perylene tetracarboxyl diimides (PDI), a kind of organic molecules with conjugated π-electron systems, is one of the most popular visible light absorbing compounds. PDIs are currently being investigated as photoactive materials for use in a variety of fields, due to their excellent photochemical and thermal stability, high luminescence efficiency, and novel optoelectronic properties.29–36 However, to the best of our knowledge, there is no report on studying PDI molecules functionalized CuO nanobelts as enhanced peroxidase-like mimics for the detection of H2O2 and glucose. With these ideas in our mind, if PDI molecules combine with CuO nanobelts to provide a nanocomposite, it will demonstrate a novel peroxidase-like activity.
Herein, we report porous CuO nanobelts modified with N,N′-di-carboxy methyl perylene diimides (PDI) molecules. It is the first time to demonstrate that the composites (PDI–CuO) possess the enhanced intrinsic peroxidase-like activity, compared to the pure CuO nanobelts without PDI modification. PDI–CuO nanocomposites can catalyze the oxidation of peroxidase substrate, 3,3′,5,5′-tetramethylbenzidine (TMB), in the presence of H2O2. Surprisingly, the colorimetric reaction can be observed by the naked eye only in 40 s. The fluorescent experiment verified the rapid reaction mechanism was from the decomposition of H2O2 and generating hydroxyl radicals (˙OH) by adding the PDI–CuO nanocomposites into the colorimetric reaction system (TMB/H2O2). Based on the experimental results, we designed a facile visual colorimetric sensor using PDI–CuO nanocomposites as enzyme mimics for convenient detecting H2O2 and glucose, respectively.
2. Experimental section
2.1 Materials
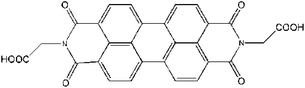
Copper chloride (CuCl2·2H2O), sodium dodecylbenzene sulfonate (SDBS), sodium hydroxide (NaOH), ethanol, N,N-dimethyl formamide (DMF), hydrogen peroxide (30 wt%, H2O2), glucose, fructose, and lactose were purchased from Guangcheng Reagent Co (Tianjin, China). Glucose oxidase (GOx, ≧ 200 U mg−1) was purchased from Sigma-Aldrich and stored in a refrigerator at −18 °C. N,N′-Di-carboxy methyl perylene diimides (PDI) was synthesized according to the previous reports.37 The molecular structure of PDI is shown in Fig. 1. The other regents above were of analytical reagent grade and used without further purification.2.2 Preparation of CuO nanobelts and PDI–CuO nanocomposites
CuO nanobelts were synthesized by a facile hydrothermal method combined with subsequent calcination.38 In a typical procedure, CuCl2·2H2O (2 mmol, 0.341 g) and SDBS (0.696 g) were dissolved in 20 mL distilled water to form a blue solution. NaOH (20 mL, 5 M) aqueous solution was dropwise added to the solution under stirring for 30 min, resulting in a light blue solution. The obtained solution was further aging for 4 h, and then transferred into a teflon-lined stainless steel autoclave reactor for 6 h at 100 °C. After hydrothermal treatment, the system was cooled at room temperature naturally. The precipitate was washed with distilled water and ethanol for several times and dried at 60 °C for 2 h, and finally calcined at 350 °C for 3 h.CuO nanobelts (20 mg) dispersed into DMF solution of PDI (1 mg mL−1) under stirring for 4 h. After that, separated by centrifugation, washed with distilled water and ethanol for several times and dried at 60 °C for 4 h. The target product was obtained for subsequent experiments.
2.3 Characterization
The morphology and size distribution of the samples were characterized by a transmission electron microscope and a high resolution TEM (HRTEM) at an accelerating voltage of 200 kV (TEM JEM-2100, JEOL, Japan). The scan electron microscopy images were obtained by scanning electron microscopy (SEM, JEOL, Japan). The crystalline phases of all catalysts were performed by using a D/Max2500PC-RA diffractometer with Cu Kα (λ = 1.5406 Å) radiation. The 2θ values in the range of 10–90° were collected with a 0.021° step size. X-ray photoelectron spectra (XPS) were recorded on a PHI Quantera SXM spectrometer with an Al Kα = 280.00 eV excitation source, and binding energies were calibrated by referencing the C 1s peak (284.5 eV) to reduce the sample charge effect. The Brunauer–Emmett–Teller (BET) surface area was measured by nitrogen adsorption–desorption at 77 K using a Micromeritics ASAP 2010M instrument.Ultraviolet spectra were recorded on a UV-8000PC spectrophotometer (Shanghai, China). Fluorometric measurement was carried out by a Cary Eclipse spectro fluorophotometer (Varian, Inc, USA).
2.4 Kinetic study
The reaction kinetics for the catalytic oxidation of TMB was measured in time course mode by monitoring the absorbance variation at 652 nm with a 3 min interval. Unless otherwise stated, a typical experience was carried out at 55 °C by using 0.2 mL PDI–CuO (0.03 mg mL−1) in 1.4 mL buffer (HAc–NaAc, pH = 4.0) with 0.2 mL H2O2 and 0.2 mL TMB as substrates, respectively. In the experimental procedure, except for the fixed concentration of TMB (2 mM) and various concentration of H2O2 (0.002–0.1 mM) as well as the fixed concentration of H2O2 (0.25 M) and various concentration of TMB (0.02–0.10 mM). The apparent kinetic parameters based on Lineweaver–Burk plots of the double reciprocal of the Michaelis–Menten equation as described below: 1/v = Km/Vmax (1/[S] + 1/Km), where v is the initial velocity, [S] is the concentration of the substrate, Km is the Michaelis constant and Vm is the maximal reaction velocity.142.5 Reaction mechanism
We suspect that the peroxidase-like activities of the PDI–CuO nanocomposites may due to the decomposition of H2O2 into OH radicals or the mechanism of electron transfer. Thus, a fluorometric spectra method was employed to measure the OH radicals with terephthalic acid as a probe under the optional conditions. The typical experimental procedure is as follows: terephthalic acid (0.5 mM), H2O2 (10 mM), and the PDI–CuO nanocomposites with different concentrations were incubated in acetate buffer (pH = 4.0) at 55 °C for 30 min. Immediately, the solutions were studied for fluorometric measurement.2.6 Colorimetric detection of H2O2 and glucose
For detecting the H2O2 standard solution, PDI–CuO nanobelts were carried out using different concentration of 200 μL H2O2 (0.002–0.1 mM), then mixed with 200 μL TMB of fixed concentration (2 mM) as the substrate in acetate buffer (1.4 mL, pH = 4.0). The resulting mixture was incubated at 55 °C for 3 min. Then, the absorbance of the treated final mixtures were measured at 652 nm.For glucose assay, the catalytic oxidization of glucose by glucose oxidase (GOx) (glucose + O2 → H2O2+ gluconic acid) was coupled with colorimetric detection of H2O2. Briefly, glucose assay was performed as follows: 0.1 mL GOx (0.5 mg mL−1) and 0.5 mL glucose with different concentrations in PBS buffer (pH = 7) were kept for 30 min at 37 °C in a dark condition. Then, 0.2 mL PDI–CuO, 0.2 mL TMB and 0.6 mL above glucose reaction solution were added to 1.0 mL HAc buffer. After 3 min (55 °C) incubation, the UV-vis absorption spectra of solution were recorded.
2.7 Selectivity for colorimetric detection of glucose
For specificity analysis, 5.0 mM fructose, 5.0 mM maltose, 5.0 mM lactose, and 5.0 mM sucrose instead of 1.0 mM glucose were used as control experiences, respectively. The detection process was similar to the standard glucose assay.3. Results and discussion
3.1 Characterization of PDI–CuO nanocomposites
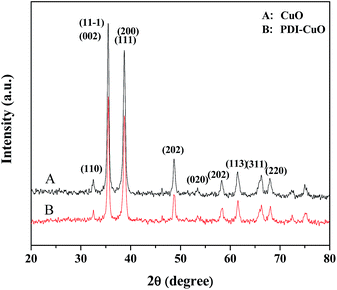
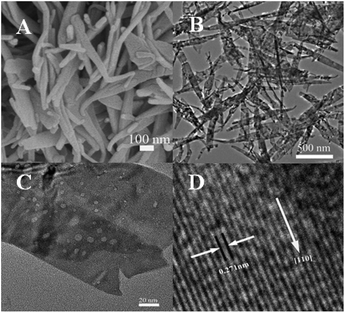
The crystal phases of CuO, PDI–CuO nanocomposite were characterized by X-ray diffraction (XRD), shown in Fig. 2. From Fig. 2A, it can be seen that pure CuO crystal exhibited diffraction peaks (2θ) centered at ca. 32.5°, 35.4°, 35.5°, 38.7°, 38.8° and 48.7°, corresponding to (110), (002), (11−1), (111), (200) and (202) planes, in accordance with the monoclinic crystal structure (JCPDS card no. 48-1548). Compared with the XRD data of CuO, the weak XRD peaks of PDI functionalized CuO nanobelts was found, as shown in Fig. 2B. This demonstrated that the PDI functionalized CuO nanobelts has the same crystal phase as that of the pure CuO, together with no destroying in structure, except for a little weak diffraction peaks.39,40The morphology of the PDI–CuO nanobelts was imaged by SEM and TEM, respectively. From Fig. 3A and B, it can be seen that the PDI–CuO nanocomposites show belt-like morphologies with the average size of 1 μm in length and 100 nm in width. Interestingly, many pores (ca. 5–10 nm) in PDI–CuO nanobelts can be observed, shown in Fig. 3C. The fringe spacing is ca. 0.275 nm, corresponding to (110) crystal plane, shown in Fig. 3D. Theoretically, the large number of nanoporous are able to enlarge the specific surface area, and thus increase the activity sites of materials.41
Then, the composition of PDI–CuO nanocomposites was measured by the EDS mapping images, unambiguously demonstrating the coexistence of C, N, O, Cu elements in the product. Of the different elementary compositions, C and N are from PDI molecules, shown in Fig. S1 (ESI†). Therefore, the EDS mapping images further confirmed that PDI had been successfully distributed on the surface of CuO nanobelts.
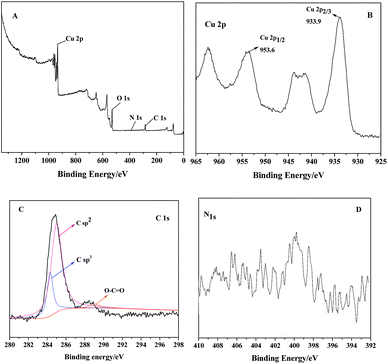
To further determine the interaction between PDI and CuO nanobelts, the XPS spectra of PDI–CuO nanocomposites were used to analyze the chemical composition and surface electronic state of the as-prepared PDI–CuO nanocomposites,42 shown in Fig. 4. As shown in Fig. 4A, all peaks could be contributed to Cu, O, N, C elements. The peaks in Fig. 4B with binding energies at 933.9 and 953.6 eV are from Cu2 + (CuO) due to Cu 2p2/3 and Cu 2p1/2, respectively. C 1s spectrum (Fig. 4C) can be deconvoluted into three peaks at 284.3 eV (C sp2), 285.0 eV (C sp3) and 289.0 eV (O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) with the C sp2 component being dominant.43 As a result, all elements of the PDI–CuO nanocomposites can be found in the XPS data, suggesting that CuO are successfully modified by PDI.
O) with the C sp2 component being dominant.43 As a result, all elements of the PDI–CuO nanocomposites can be found in the XPS data, suggesting that CuO are successfully modified by PDI.
Based on the previous report, the large number of pores are able to enlarge the specific surface area, and thus increase the activity sites.44 The Brunauer–Emmett–Teller (BET) surface area was measured by nitrogen adsorption–desorption at 77.35 K. Before measurement, the samples were degassed at 60 °C for 10 hours. Nitrogen adsorption–desorption isotherms of PDI–CuO nanobelts was shown in Fig. S2 (ESI†). The large surface area (12.947 m2 g−1), pore volume (Vp = 0.044 cc g−1), and small pore diameter (dp = 1.653 nm) of PDI–CuO nanocomposites were found.
3.2 Peroxidase-like activity of PDI–CuO nanocomposites
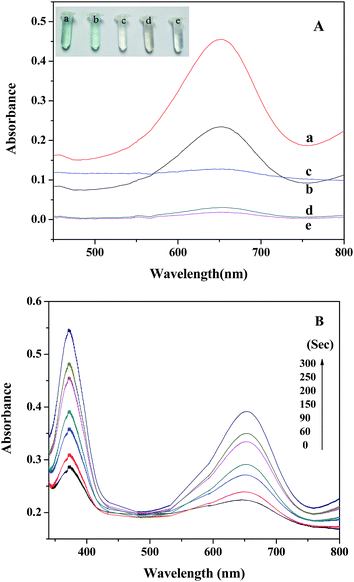
The peroxidase-like activity of PDI–CuO was investigated using the peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (TMB) in the presence of H2O2. TMB is a kind of common chromogenic substrate and is usually used for practical analytical application. Clearly, from Fig. 5A, it can be seen that the absorbance of the reaction system (PDI–CuO + H2O2 + TMB) was much higher than that of the system (CuO + H2O2 + TMB) at 652 nm (curve a and b), indicating the enhanced catalytic activity of PDI–CuO nanocomposites than that of pure CuO nanobelts for the catalytic oxidation of TMB. Surprisingly, we also found that the color of the reaction system containing TMB, H2O2 and PDI–CuO nanocomposites has rapidly been changed from colorless to blue within 40 s. On visual observation, the reaction system of TMB + H2O2 in absence of PDI–CuO nanobelts (Fig. 5A, curve d) or the TMB + PDI–CuO without H2O2 (Fig. 5A, curve e) shows negligible color change, suggesting that both H2O2 and PDI–CuO were necessary for the catalytic oxidation of TMB. Furthermore, it can be also found that the catalytic activity of PDI–CuO nanocomposites with pores is higher than that of PDI–CuO without pores, shown in Fig. S3 (ESI†).Moreover, the experiment results show that the catalytic activity of PDI–CuO nanocomposites was time dependent, shown in Fig. 5B. In addition, the colorimetric reaction of the system is generally required to be incubated for some “time interval” before the corresponding absorption spectra are recognized as a valid measurement.45 We have summarized the required “time interval” for different types of nanomaterials for the detection of H2O2 in this study together with other reported materials as peroxidase mimics, listed in Table 1. Obviously, PDI–CuO nanocomposites could oxidize the substrate TMB with visualized the color change in short time, exhibiting the higher catalytic activity of the nanocomposites, compared with other materials listed in Table 1. In other words, the response time of reaction system containing PDI–CuO nanocomposites observed by the naked eye is shorter than that of other artificial peroxidase-like mimics, such as CuS–MMT,22 PtPd–Fe3O4,46 Fe3O4 NPs,8 Au NPs47 and TCPP–NiO nanomaterials.48
3.3 Optimization of experimental conditions
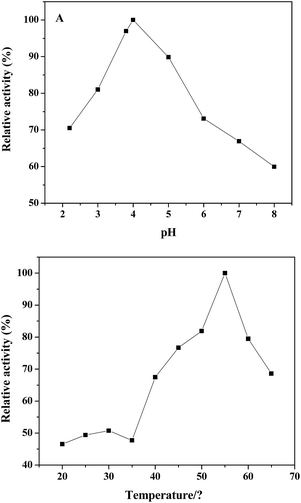
Similar to natural peroxidase, horseradish peroxidase (HRP) and other artificial peroxidase-like mimics based on nanomaterials, the catalytic activity of the PDI–CuO nanocomposites were dependent on pH and temperature, respectively.21,22,48 Likewise, the catalytic activity of PDI–CuO nanocomposites was optimized under a wide scope of experimental conditions, including pH value and temperature. The effect of pH on the peroxidase-like activity of PDI–CuO nanocomposites was measured by varying pH from 2.2 to 8.0 (2.2, 3, 3.8, 4, 5, 6, 7, 8), shown in Fig. 6A. Clearly, from Fig. 6A, it can be seen that the optimal pH was 4.0, similar to Ce NPs, PVP–MoS2 NPs reported in previous publications.25,49 Interestingly, though catalytic activity of PDI–CuO nanocomposites were declined at either lower pH or higher pH values, the relative activity was more than 55% in a wide range of pH. Thus, the result reveals the PDI–CuO nanocomposites displayed an excellent stability than that of HRP in a wide pH scope.8 Fig. 6B shows the temperature-dependant on the peroxidase-like activity of PDI–CuO nanocomposites over a wide range from 20 to 65 °C (20, 25, 30, 35, 40, 45, 50, 55, 60, 65). Generally, the catalytic activity of PDI–CuO nanocomposites was up to the maximum at 55 °C, while the enzymatic activity of HRP sharply decreased over 40 °C due to the denaturalization of the enzyme under high temperature.8 Thus, pH 4.0 and the temperature of 55 °C was selected for the subsequent assays.3.4 Steady-state kinetic of nanoparticles
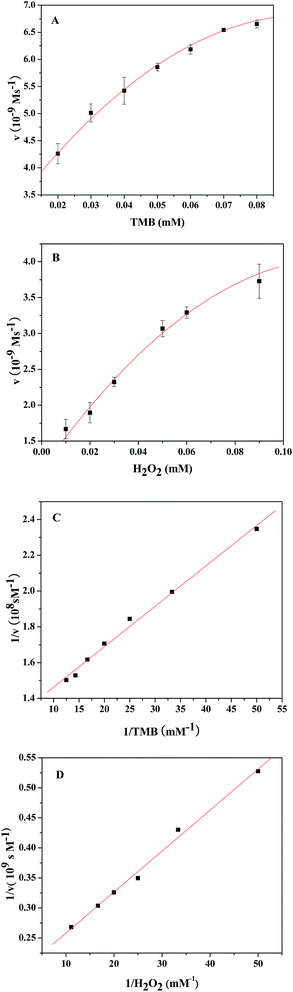
To understand the catalytic performance of the PDI–CuO nanocomposites, the steady-state kinetics was investigated using TMB and H2O2 as substrates under the optimal conditions (pH = 4.0, 55 °C). Fig. 7A and 7B show the typical Michaelis–Menten curves of TMB and H2O2 by plotting the initial reaction rate against concentration. Furthermore, the apparent kinetic parameters (Km and Vm) were calculated according to Lineweaver–Burk double reciprocal plots (Fig. 7C and D). It is well-known that Km represents the affinity of a enzyme toward substrates: the smaller the value of Km, the stronger the affinity between the enzyme (catalysts) and the substrate. As illustrated in Table 2, the Km value for PDI–CuO with TMB as substrate was much lower than horseradish peroxidase (HRP) and peroxides nanomimetics reported previously, indicating that PDI–CuO had a higher affinity towards TMB than HRP and other nanomaterials. However, the Km value of PDI–CuO with H2O2 substrate was 35.74, revealing that a higher concentration of H2O2 was required to obtain maximum catalytic activity.3.5 Mechanism of the peroxidase-like activity of PDI–CuO nanocomposites
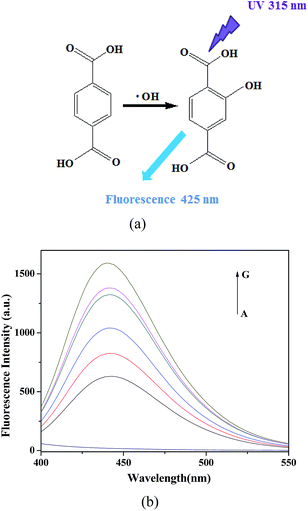
Generally, the catalytic mechanism of peroxidase-like activity of artificial peroxidases can be divided into two kinds. One is the generation of reactive hydroxyl radicals (˙OH), and the other is the electron transfer process.53 In order to shed light on the catalytic mechanism of PDI–CuO nanocomposites, the fluorescent probe, terephthalic acid (TA), was used as a typical ˙OH radical capture reagent in the PDI–CuO + H2O2+TMB system. The reason is that non-luminescent TA could capture ˙OH and generate remarkable fluorescent product, 2-hydroxyterephthalic acid (HTA), at 425 nm (Fig. 8a).54 From Fig. 8b, it can be seen clearly that photoluminescence (PL) intensity gradually increased with increasing of amount of PDI–CuO nanocomposites. In addition, no PL intensity was observed in the absence of H2O2 (Fig. 8b). This fluorescent results revealed that PDI–CuO nanocomposites could catalytically activate H2O2 decomposition to generate ˙OH radicals that subsequently oxidized TMB to form a blue TMB oxide.55 The experience further suggests more ˙OH radicals can be produced with the increase of the amount of PDI–CuO. In other words, the stronger the photoluminescence intensity, the more the generated ˙OH. Therefore, based on the above results, the peroxidase mimic catalytic mechanism of PDI–CuO nanobelts was proposed, shown in Scheme 1.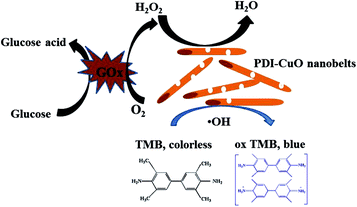 | ||
| Scheme 1 Schematic illustration of colorimetric sensor for H2O2 and glucose detection using PDI–CuO nanobelts as catalysts. | ||
3.6 Colorimetric detection of H2O2 and glucose
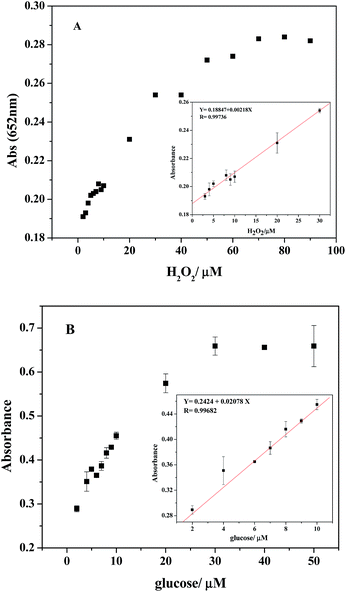
Considering the fact that PDI–CuO possesses the intrinsic peroxidase-like activity, a simple colorimetric method was performed to quantitatively detect H2O2 and glucose, respectively. Fig. 9A shows the typical absorbance at 625 nm with the addition of various concentrations of H2O2 under the optimal conditions (pH 4.0, 55 °C). Obviously, the absorbance of oxidized TMB gradually increased with increasing of concentration H2O2. The homologous linear calibration plot range is from 3 μM to 30 μM with a detection limit of 2.38 × 10−6 M, which was 19 times lower than that of reported H2O2 sensors based on Au@Pt core–shell nanorods (4.5 × 10−5 M).56 It suggested that PDI–CuO nanocomposites can offer a facile and sensitive method to detect H2O2 and other substances which could generate H2O2 in a chemical reaction. As our known, glucose oxidase (GOx) can catalyze the oxidation of glucose to produce H2O2 in the presence of oxygen. The relevant standard curve of the glucose response is shown in Fig. 9B. Clearly, the absorbance was linearly against glucose concentration in the range of 2–50 μM with the low detection limit of 6.533 × 10−7 M.3.7 Selectivity analysis
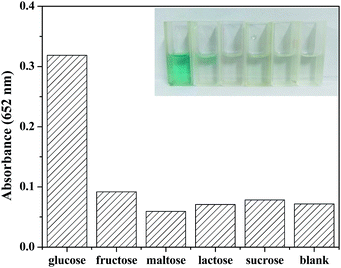
To investigate the selectivity of detection of glucose, some other saccharides including 5.0 mM fructose, 5.0 mM maltose, 5.0 mM lactose, and 5.0 mM sucrose were selected as control samples. As shown in the Fig. 10, there was no obvious absorbance at 652 nm or color change, though the concentration of analogues were 5 times higher than that of glucose. Therefore, it can be concluded that the fabricated colorimetric sensor demonstrates high selectivity and affinity to glucose.4. Conclusions
In summary, we synthesized a new kind of N,N′-di-carboxy methyl perylene diimides functionalized CuO nanocomposites, which demonstrate a intrinsic peroxidase activity. The PDI–CuO nanocomposites could rapidly catalyze the oxidation of the substrate TMB to produce a blue product observed by the naked eye within 40 seconds. The catalysis was shown to coincide with typical Michaelis–Menten kinetics and its catalytic property is strongly dependent on pH and temperature. On the basis of an intrinsic peroxidase-like activity, we designed an colorimetric biosensor platform that can conveniently detect H2O2 and glucose. With the high sensitivity, low limits of detection (H2O2: 2.38 × 10−6 M; glucose: 6.533 × 10−7 M), and rapid response time, the proposed PDI–CuO biosensor platform shows great promise as a low-cost sensor for environmental monitoring, medical diagnostics, immunoassays and biocatalysis.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 21271119) and Innovation Fund of Science & Technology of Graduate Students (SDUST).Notes and references
- L. Han, L. Zeng, M. Wei, C. Li and A. Liu, Nanoscale, 2015, 7, 11678–11685 RSC.
- K. M. Koeller and C. H. Wong, Nature, 2001, 409, 232–240 CrossRef CAS PubMed.
- N. Durán and E. Esposito, Appl. Catal., B, 2000, 28, 83–99 CrossRef.
- I. Y. Sakharov, I. S. Alpeeva and E. E. Efremov, J. Agric. Food Chem., 2011, 59, 809–813 CrossRef PubMed.
- H. Wei and E. Wang, Anal. Chem., 2008, 80, 2250–2254 CrossRef CAS PubMed.
- W. Chen, J. Chen, Y. B. Feng, L. Hong, Q. Y. Chen, L. F. Wu, X. H. Lin and X. H. Xia, Analyst, 2012, 137, 1706–1712 RSC.
- R. Breslow and L. E. Overman, J. Am. Chem. Soc., 1970, 92, 1075–1077 CrossRef CAS PubMed.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. Yang and S. Perrett, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- Y. Song, K. Qu, C. Zhao, J. Ren and X. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- Y. Song, X. H. Wang, C. Zhao, K. Qu, J. Ren and X. Qu, Chem.–Eur. J., 2010, 16, 3617–3621 CrossRef CAS PubMed.
- E. L. Zhou, C. Qin, P. Huang, X. L. Wang, W. C. Chen, K. Z. Shao and Z. M. Su, Chem.–Eur. J., 2015, 21, 11894–11898 CrossRef CAS PubMed.
- W. W. He, H. M. Jia, X. X. Li, Y. Lei, J. Li, H. X. Zhao, L. W. Mi, L. Z. Zhang and Z. Zheng, Nanoscale, 2012, 4, 3501–3506 RSC.
- Z. H. Dai, S. H. Liu, J. C. Bao and H. X. Ju, Chem.–Eur. J., 2009, 15, 4321–4326 CrossRef CAS PubMed.
- Y. L. Dong, H. G. Zhang, Z. U. Rahman, L. Su, X. J. Chen, J. Hu and X. G. Chen, Nanoscale, 2012, 4, 3969–3976 RSC.
- Y. J. Chen, H. Y. Cao, W. B. Shi, H. Liu and Y. M. Huang, Chem. Commun., 2013, 49, 5013–5015 RSC.
- Y. Nangia, B. Kumar, J. Kaushal and C. Raman Suri, Anal. Chim. Acta, 2012, 751, 140–145 CrossRef CAS PubMed.
- X. Xia, J. Zhang, N. Lu, M. J. Kim, K. Ghale, Y. Xu, E. M. Kenzie, J. Liu and H. Ye, ACS Nano, 2015, 9, 9994–10004 CrossRef CAS PubMed.
- Q. Cai, S. K. Lu, F. Liao, Y. Q. Li, S. Z. Ma and M. W. Shao, Nanoscale, 2014, 6, 8117–8123 RSC.
- Y. Zhang, C. Xu, B. Li and Y. Li, Biosens. Bioelectron., 2013, 43, 205–210 CrossRef CAS PubMed.
- H. Zhao, Y. M. Dong, P. P. Jiang, G. L. Wang and J. J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 6451–6461 CAS.
- L. F. Sun, Y. Y. Ding, Y. L. Jiang and Q. Y. Liu, Sens. Actuators, B, 2017, 239, 848–856 CrossRef CAS.
- L. Y. Zhang, M. X. Chen, Y. L. Jiang, M. M. Chen, Y. N. Ding and Q. Y. Liu, Sens. Actuators, B, 2017, 239, 28–35 CrossRef CAS.
- Q. Y. Liu, Y. L. Jiang, L. Y. Zhang, X. P. Zhou, X. T. Lv, Y. Y. Ding, L. F. Sun, P. P. Chen and H. L. Yin, Mater. Sci. Eng., C, 2016, 65, 109–115 CrossRef CAS PubMed.
- Y. Y. Ding, L. F. Sun, Y. L. Jiang, S. X. Liu, M. X. Chen, M. M. Chen, Y. N. Ding and Q. Y. Liu, Mater. Sci. Eng., C, 2016, 67, 188–194 CrossRef CAS PubMed.
- J. Yu, X. Y. Ma, W. Y. Yin and Z. J. Gu, RSC Adv., 2016, 6, 81174–81183 RSC.
- X. Liu, Q. Wang, H. H. Zhao, L. C. Zhang, Y. Y. Su and Yi Lv, Analyst, 2012, 137, 4552–4558 RSC.
- H. Jiang, Z. H. Chen, H. Y. Cao and Y. M. Huang, Analyst, 2012, 137, 5560–5564 RSC.
- J. Chen, J. Ge, L. Zhang, Z. H. Li and L. B. Qu, Sens. Actuators, B, 2016, 233, 438–444 CrossRef CAS.
- M. R. Wasielewski, J. Org. Chem., 2006, 71, 5051–5066 CrossRef CAS PubMed.
- J. A. A. M. Elemans, R. van Hameren, R. J. M. Nolte and A. E. Rowan, Adv. Mater., 2006, 18, 1251–1266 CrossRef CAS.
- K. Y. Law, Chem. Rev., 1993, 93, 449–486 CrossRef CAS.
- H. Langhal, Chim. Acta., 2005, 88, 1309–1343 CrossRef.
- F. Würthner, Chem. Commun., 2004, 1564–1579 RSC.
- C. D. Dimitrakopoulos and P. R. L. Malenfant, Adv. Mater., 2002, 14, 99–117 CrossRef CAS.
- B. A. Jones, M. J. Ahrens, M. H. Yoon, A. Facchetti, T. J. Marks and M. R. Wasielewski, Angew. Chem., Int. Ed., 2004, 43, 6363–6366 CrossRef CAS PubMed.
- F. Yukruk, A. L. Dogan, H. Canpinar, D. Guc and E. U. Akkaya, Org. Lett., 2005, 7, 2885–2887 CrossRef CAS PubMed.
- Y. Shi, H. X. Wu, L. Xue and X. Y. Li, J. Colloid Interface Sci., 2012, 365, 172–177 CrossRef CAS PubMed.
- Z. Y. Fei, P. Lu, X. Z. Feng, B. Sun and W. J. Ji, Catal. Sci. Technol., 2012, 8, 1705–1710 Search PubMed.
- B. Liu and H. C. Zeng, J. Am. Chem. Soc., 2004, 126, 8124–8125 CrossRef CAS PubMed.
- Y. J. Zhang, S. W. Or, X. L. Wang, T. Y. Cui, W. B. Cui, Y. Zhang and Z. D. Zhang, Eur. J. Inorg. Chem., 2009, 1, 168–173 CrossRef.
- T. T. Cao, Z. G. Song, S. B. Wang and J. Xia, Sci. China: Earth Sci., 2015, 58, 510–522 CrossRef CAS.
- Q. Liu, Y. Yang, Q. Jia, P. Chen, H. Li, R. Zhu and C. Gao, Mater. Sci. Eng., C, 2016, 59, 445–453 CrossRef CAS PubMed.
- G. Darabdharaae, B. Sharmaae, M. R. Das, R. Boukherroub and S. Szunerits, Sens. Actuators, B, 2017, 238, 842–851 CrossRef.
- T. T. Cao, Z. G. Song, S. B. Wang and J. Xia, Sci. China: Earth Sci., 2015, 58, 510–522 CrossRef CAS.
- S. S. Pan, W. Lu, Y. H. Zhao, W. Tong, M. Li, L. M. Jin, J. Y. Choi, F. Qi, S. G. Chen, L. F. Fei and S. F. Yu, ACS Appl. Mater. Interfaces, 2013, 5, 12784–12788 CAS.
- X. L. Sun, S. J. Guo, C. S. Chung, W. L. Zhu and S. H. Sun, Adv. Mater., 2013, 25, 132–136 CrossRef CAS PubMed.
- Y. Jv, B. Li and R. Cao, Chem. Commun., 2010, 46, 8017–8019 RSC.
- Q. Y. Liu, Y. T. Yang, H. Li, R. R. Zhu, Q. Shao, S. G. Yang and J. J. Xu, Biosens. Bioelectron., 2015, 64, 147–153 CrossRef CAS PubMed.
- R. C. Guo, Y. R. Wang, S. X. Yu, W. X. Zhu, F. Q. Zheng, W. Liu, D. H. Zhang and J. L. Wang, RSC Adv., 2016, 6, 59939–59945 RSC.
- D. Jampaiah, S. R. Telukutla, A. E. Kandjani, S. Periasamy, Y. M. Sabri, V. E. Coyle, R. Shukla and S. K. Bhargava, J. Mater. Chem. B, 2016, 4, 3874–3885 RSC.
- R. Li, M. Zhen, M. G. Chen, G. Zhang, J. Ge, P. Gong, C. Wang and C. Shu, Biosens. Bioelectron., 2013, 47, 502–507 CrossRef CAS PubMed.
- X. Chen, X. Tian, B. Su, Z. Huang, X. Chen and M. Oyama, Dalton Trans., 2014, 43, 7449–7454 RSC.
- H. Su, D. D. Liu, M. Zhao, W. L. Hu, S. S. Xue, Q. Cao, X. Y. Le, L. N. Ji and Z. W. Mao, ACS Appl. Mater. Interfaces, 2015, 7, 8233–8242 CAS.
- G. W. Yang, T. M. Chen and J. Xiao, RSC Adv., 2016, 6, 70124–70132 RSC.
- L. Li, L. Ai, C. Zhang and J. Jiang, Nanoscale, 2014, 6, 4627–4634 RSC.
- J. B. Liu, X. N. Hu, S. Hou, T. Wen, W. Q. Liu, X. Zhu, J. J. Yin and X. C. Wu, Sens. Actuators, B, 2012, 166–167, 708–714 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04463a |
| This journal is © The Royal Society of Chemistry 2017 |