Multiphasic microgel-in-gel materials to recapitulate cellular mesoenvironments in vitro†
Dejan
Husman‡
 ab,
Petra B.
Welzel‡
a,
Steffen
Vogler
c,
Laura J.
Bray
ab,
Petra B.
Welzel‡
a,
Steffen
Vogler
c,
Laura J.
Bray
 ad,
Nicole
Träber
ae,
Jens
Friedrichs
a,
Vincent
Körber
a,
Mikhail V.
Tsurkan
ad,
Nicole
Träber
ae,
Jens
Friedrichs
a,
Vincent
Körber
a,
Mikhail V.
Tsurkan
 a,
Uwe
Freudenberg
a,
Julian
Thiele
a,
Uwe
Freudenberg
a,
Julian
Thiele
 f and
Carsten
Werner
f and
Carsten
Werner
 *abg
*abg
aLeibniz-Institut für Polymerforschung Dresden e.V. (IPF), Max Bergmann Center of Biomaterials Dresden (MBC), Hohe Str. 6, 01069 Dresden, Germany. E-mail: werner@ipfdd.de
bTechnische Universität Dresden, Research Training Group “Nano- and Biotechniques for Electronic Device Packaging”, Helmholtz Str. 18, 01069, Dresden, Germany
cDeutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Arnoldstr. 18, 01307 Dresden, Germany
dQueensland University of Technology (QUT), Institute of Health and Biomedical Innovation, 60 Musk Avenue, Kelvin Grove, 4059 Australia
eTechnische Universität Dresden, Biotechnology Center (BIOTEC), Tatzberg 47/49, 01307 Dresden, Germany
fLeibniz-Institut für Polymerforschung Dresden e.V. (IPF), Department of Nanostructured Materials and Leibniz Research Cluster (LRC), Hohe Str. 6, 01069 Dresden, Germany
gTechnische Universität Dresden, Center for Regenerative Therapies Dresden (CRTD), Tatzberg 47/49, 01307 Dresden, Germany
First published on 25th October 2019
Abstract
Multiphasic in vitro models with cross-scale heterogeneity in matrix properties and/or cellular composition can reflect the structural and compositional complexity of living tissues more faithfully, thereby creating new options for pathobiology and drug development studies. Herein, a new class of tunable microgel-in-gel materials is reported that build on a versatile platform of multifunctional poly(ethylene glycol)-heparin gel types and integrates monodisperse, cell-laden microgels within cell-laden bulk hydrogel matrices. A novel microfluidic approach was developed to enable the high-throughput fabrication of microgels of in situ adjustable diameters, stiffness, degradability and biomolecular functionalization. By choosing structure and composition of the microgel and the bulk gel compartments independently, our microgel-in-gel arrangements provide cross-scale control over tissue-mimetic features and pave the way for culture systems with designed mesoenvironmental characteristics. The potentialities of the introduced approach are exemplarily shown by creating a reductionistic in vitro model of vascularized prostate cancer tissue.
Tunable material platforms mimicking tissue-specific extracellular matrices (ECM) can be instrumental for regenerative therapies and for creating three-dimensional (3D) in vitro tissue and disease models. Various types of hydrogels were shown to be effective in recapitulating specific aspects of cell–matrix and cell–cell interactions.1–12 However, homogeneous materials can not reflect the complexity of living tissues and are limited in supporting locally differing biomolecular and physical requirements of heterocellular cultures. Hydrogel materials with defined spatial distribution of cell-instructive properties as well as spatial separation of co-cultured cell populations produced by 3D printing,13–18 photolithographic patterning,19 microfluidics,20 and other microfabrication techniques21–23 were shown to be advantageous for reconstituting structural features of living tissues more closely. For instance, the provision of growth factors,24 the spatial display of biological ligands,20 and the mechanical heterogeneity of ECM18 have been successfully mimicked with these approaches. However, a more faithful reconstitution of structural and functional features of living tissues by 3D in vitro models requires the combination of multiple functionalities and defined 3D architectures across scales.
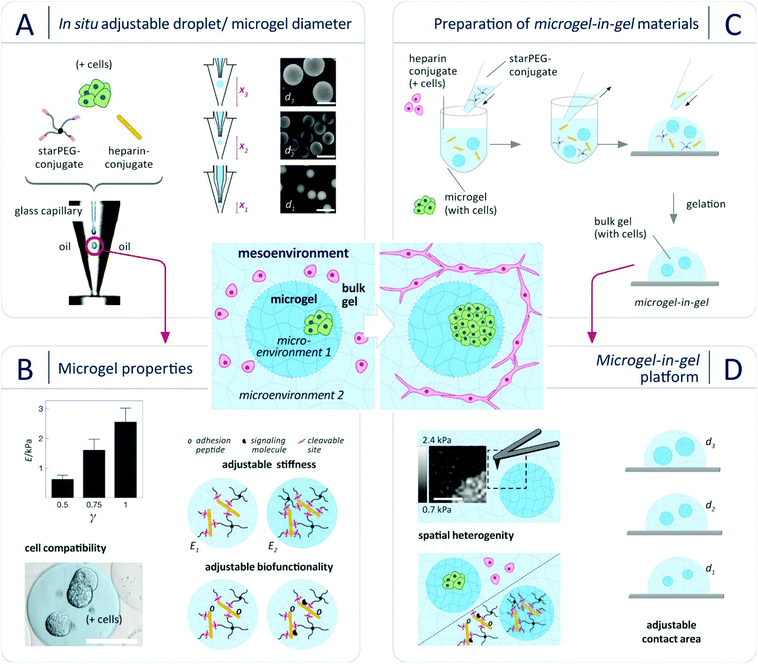
Toward this aim, we herein present a new multiphasic biomaterials design approach (Fig. 1) that relies on integrating cell-laden spherical hydrogel microparticles (referred to as microgels) within cell-laden bulk hydrogel matrices to provide thoroughly tunable microgel-in-gel systems. Building on a recently developed platform of multifunctional glycosaminoglycan-based biohybrid hydrogels,25 this approach allows for independently customizing the microgel and the bulk gel compartments with respect to cell-instructive matrix properties (stiffness, degradability, presentation of adhesion moieties and growth factors) and cellular composition. Moreover, it provides control over the spatial combination of the compartments (microenvironments), i.e. enables the design of cellular mesoenvironments26 (Fig. 1). Micro- and mesoenvironmental parameters can be fine-tuned to reflect fundamental tissue properties in 3D culture. In particular, adjusting the microgel size obviously enables the graduation of interactions between the two engineered compartments (Fig. 1D). To exploit that option, a novel droplet microfluidics scheme was established to produce sets of monodisperse, cell-laden microgels of different sizes through crosslinking peptide-functionalized, four-armed poly(ethylene glycol) (hereafter: starPEG-conjugate or starPEG) and maleimide-functionalized heparin (hereafter: heparin-conjugate or heparin).27 Second, microgel-in-gel systems of spatially gradated stiffness, biofunctionality and/or cellular composition were created by embedding starPEG-heparin microgels in starPEG-heparin bulk hydrogels with different composition and properties.
Finally, the methodology was exemplarily applied to mimic basic patterns of vascularized cancer tissue in a new format that overcomes limitations of 3D in vitro tissue models based on monophasic hydrogel materials: separately engineered hydrogel microenvironments, one supporting tumor spheroid formation of cancer cells and one supporting capillary network formation of vascular endothelial cells, were combined in microgel-in-gel co-cultures to explore the mesoenvironmental interplay of the co-cultured cells.
Our material concept relies on the rational design of modular starPEG-heparin hydrogels25,28 which uniquely allows the decoupling of biomolecular and biophysical matrix cues, creates powerful options for the biomimetic presentation of growth factors and enables the incorporation of adhesive peptide ligands and enzymatically degradable peptide crosslinkers as well as the precise adjustment of mechanical matrix stiffness (easily tuned by altering the molar starPEG to heparin ratio, γ).29–32 Crosslinking of the pre-functionalized polymeric gel precursors (starPEG-conjugate and heparin-conjugate) by a rapid, cyto-compatible Michael-type addition reaction affords cell embedding under very mild conditions, avoiding exposure to UV light or temperature change.7,27,33–35 To tailor droplets of polymer precursor solutions and microgels, respectively, in size, morphology (e.g. spherical, pancake-like or elongated) and compositional heterogeneity (e.g. Janus-like or core–shell), design features of microfluidic devices were to be optimized.36–39 Rapid prototyping by combined photo- and soft lithography, supported by computational fluid dynamics (CFD) simulations, is the standard method of choice for screening the impact of design parameters and microchannel geometry. Yet, as droplet properties can be varied merely to a limited extend by changes in flow ratio and overall flow velocity of disperse and continuous phase given a defined microchannel diameter and droplet-forming nozzle (e.g. flow focusing or T-junction), each change in the microgel properties requires a new microfluidic device.40 We therefore developed a novel variant of droplet microfluidics to effectively vary the microgel diameter d without changing the microfluidic device and even within the course of one experiment (i.e. “in situ”). The approach relies on controlling the droplet diameter only by adjusting the hydraulic diameter (Fig. S1, for details compare ESI†) at constant volumetric flow rates of disperse and continuous phase. For implementing this method, a co-flow type device was constructed from micro-machined polycarbonate blocks to form a cone-shaped channel for the continuous phase with a glass capillary in its centre for the aqueous phase. Due to the cone-formed channel, the hydraulic diameter DH and thus the local flow velocity of the oil phase at the droplet-forming nozzle can be changed at constant oil volumetric flow rate by regulating the relative position of the glass capillary in flow direction (Fig. 1). This principle provides control over droplet diameters across a wide range (200–700 μm) by adjusting the capillary position without altering any other parameter (Fig. 1), as demonstrated in Fig. S2 (ESI†) for water-in-oil (w/o) droplets. For a given capillary position, the resulting (w/o) droplets were found to be monodisperse, with coefficients of variance (CV) of less than 4% for all sizes produced, which is in accordance with previous studies on co-flowing streams.41 Applying our novel method, starPEG-heparin microgels were produced for the first time and in a wide size range from two-component mixtures of the reactive polymeric precursors as shown in Fig. 1: Using three different glass capillary positions at constant volumetric flow rates, monodisperse microgel batches with diameters of 200 μm, 400 μm, and 600 μm were obtained. By changing the molar ratio of the polymeric gel precursors starPEG-conjugate to heparin-conjugate (γ) in the reaction mixture, the Young's modulus of the microgels was tunable from 0.6 kPa–2.5 kPa (Fig. 1B) as confirmed by colloid probe AFM-based nanoindentation (compare ESI†). The determined values are comparable to reference bulk hydrogels of similar composition27,33 and match the stiffness of soft tissues.42 The uniformity of the Young's moduli of microgels from one batch confirmed the effective mixing and reactive network formation within the droplets, which is a challenge for two-component mixtures of reactive polymeric precursors.
Using the accordingly prepared starPEG-heparin microgels, microgel-in-gel systems were fabricated by incorporating sets of microgels into bulk gels of differing characteristics (Fig. 1C). For example, microgels with a molar starPEG to heparin ratio of γ = 1 were embedded within a hydrogel matrix of γ = 0.63. The resulting material was sliced with a vibratome (for details compare ESI†). The cross-sectional slices were probed with AFM-based nanoindentation, providing local differences in Young's Moduli and force-indentation maps (100 × 100 μm, Fig. 1D) displaying the dimensions, shapes and mechanical properties of embedded microgels. The functionality of both hydrogel phases, customized in stiffness by the molar ratio of starPEG to heparin (γ), was further adjusted by incorporation of cell-adhesion mediating (RGD)- as well as matrix metalloprotease (MMP)-sensitive peptide sequences. Similar to the bulk hydrogels,7,27,33 microgels containing enzyme-sensitive linkers were found to be effectively reorganized by embedded cells: Fig. S3 (ESI†) illustrates the decrease in microgel stiffness over the course of 7 days culture from 1.3 to 0.5 kPa by incorporated prostate cancer (PC3) cells (1 × 106 PC3 cells per mL hydrogel).
A few other approaches using microgels incorporated in a hydrogel matrix for co-culture have been recently published.43,44 Lee et al.44 produced gelatin microgels of varied size utilizing a common PDMS flow-focusing junction that does, however, not allow for the in situ variation of droplet size as the device introduced in the present study. Compared to the gelatin-based co-cultures of Lee et al., our starPEG-heparin hydrogel-based system does not only allow for tuning the physical properties of the different microenvironments, but also for the independent and liberal variation of their biomolecular properties. Therefore, we have been able to investigate the mesoenvironmental interplay of cell aggregates/assemblies, whereas Lee et al. explored the interaction of single cells only. Visser et al.43 described an innovative in-air microfluidics methodology for high-throughput one-step printing of larger 3D biomaterials with modular architecture. In contrast to our approach, the applied set-up does not allow for combining cell assemblies, such as spheroids, in customized matrices at different time points, i.e. at different levels of maturation, as both hydrogel phases (microgels and surrounding matrix) are printed simultaneously.
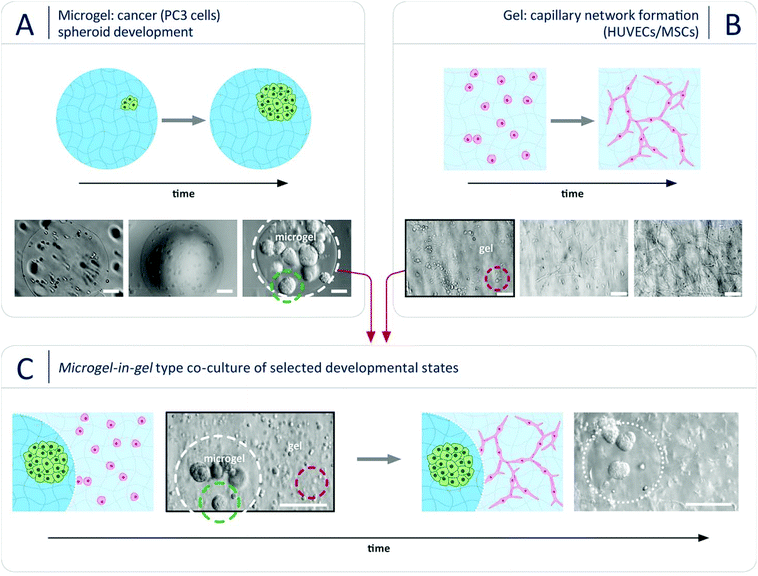
To demonstrate the suitability of our novel microgel-in-gel platform for adjusting cellular mesoenvironments in tissue- or disease-specific ways, a previously established monophasic hydrogel-based 3D culture model of vascularized prostate cancer tumors33 was translated into the microgel-in-gel co-culture format. Thereby, we aimed to overcome drawbacks of the monophasic model that will be discussed below. In vivo, new blood vessels are formed and sprout toward the tumor due to interactions between the cancer cells and endothelial cells. Hereby, the metabolic activities of tumors for further growth is supported, the invasion of cancer cells through the formed blood vessels for metastasis is enabled, and paracrine stimulations for cancer cells against apoptosis is facilitated.21,45,46 Consequently, 3D in vitro models recapitulating the interaction between the cancer cells, capillary network-forming endothelial cells and matrix components can be instrumental to unravel the complex crosstalk underlying cancer growth and to develop and reliably test anti-cancer drugs. In the monophasic 3D prostate cancer model33 mentioned above, human prostate cancer cells (PC3)47 share one common starPEG-heparin hydrogel matrix with a capillary network-forming co-culture of human umbilical cord endothelial cells (HUVECs) and mesenchymal stromal cells (MSCs). The chosen properties of the utilized hydrogel matrix had to be a compromise between the requirements of the two different cell populations. In a delicate balance, the gel stiffness had to be adjusted to keep the incorporated pre-formed prostate cancer spheroids rounded, while allowing the HUVECs and MSCs to remodel the gel matrix to form capillary networks. As invasive cancer cell types, such as PC3 cells, better grow in stiffer microenvironments (most cancers result in a natural increase in ECM stiffness48), whereas soft hydrogels were required for capillary endothelial cell network formation,7 PC3 spheroids had to be pre-formed separately, harvested and co-seeded together with HUVECs and MSCs in the soft hydrogel matrix. This procedure is not ideal, as it alters the cancer-specific ECM formed during natural spheroid formation. Moreover, the transplant of a pre-grown PC3 cell spheroid, once placed inside of the soft hydrogel matrix, will result in a spheroid that will disintegrate into (migratory) single cells, changing the cellular phenotype33 as illustrated in Fig. S4 (ESI†). Additionally, PC3 cells quickly degrade the soft hydrogel leading to structural problems within the hydrogel culture. In contrast, our microgel-in-gel platform offers spatially segregated hydrogel compartments, which can be independently tuned to match the physical and biomolecular triggers of the desired cell organization and function. Using that option, we were growing tumor spheroids in a stiffer hydrogel matrix (microgel) surrounded by a soft hydrogel of functionally distinct properties (gel) suitable to support the co-culture of HUVECs and MSCs in the formation of a capillary endothelial cell network as exemplarily shown in Fig. 2. Due to this spatial (but connected) confinement of the two cell populations the reported system is beneficial: PC3 cells are grown in a stiffer, spheroid-supportive matrix in ways that allow for studying the paracrine effects of these cells on an environmental HUVEC/MSC network that is embedded in a softer matrix.
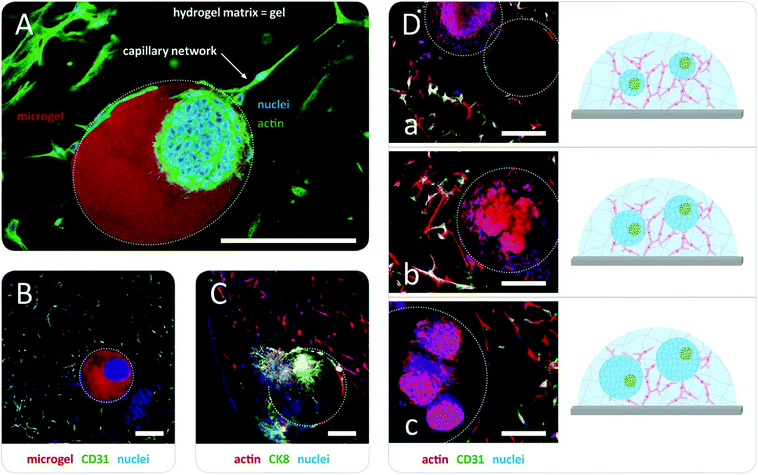
In the first step, prostate cancer cells (PC3; Deutsche Sammlung für Mikroorganismen und Zellkultur GmbH, DSMZ, Braunschweig, Germany) were mixed with a solution of pre-functionalized heparin and co-injected into the co-flow microfluidic device with a solution of peptide-functionalized starPEG (starPEG-conjugate, Fig. 1A); the resulting droplets cured to form microgels with embedded prostate cancer cells which developed into tumor spheroids during 7 days in culture (Fig. 2A). The microgels were produced with a starPEG to heparin ratio of γ = 1 to direct the growth of the PC3 cells into rounded spheroids by stiffer, tumor-like matrix properties33 and contained MMP-cleavable peptide sequences to allow for cellular matrix remodeling. Live/dead staining indicated that encapsulation of the PC3 cells into starPEG-heparin microgels did not restrict cell survival and proliferation (Fig. S5, ESI†). In the second step, microgel-in-gel materials were prepared (Fig. 2C) by embedding microgels containing prostate cancer cells/spheroids after a defined pre-culture period (exemplarily shown for 7 days in Fig. 2A) together with suspended HUVECs (isolated from umbilical veins as previously described49) and MSCs (derived from healthy volunteer donors after informed consent. The use of surplus bone marrow cells for MSC generation was approved by the ethics committee of the Technical University Dresden (Ethics approval ID: EK127042009). Bone marrow-derived MSCs were isolated as previously described50) into a significantly softer bulk hydrogel matrix (gel). In the given example, this gel matrix (Fig. 2B) was formed with a lower molar starPEG to heparin ratio (γ = 0.63) and contained not only MMP-cleavable peptide sequences, but also covalently attached adhesion ligand peptides as well as a combination of pro-angiogenic growth factors at concentrations previously shown to effectively support angiogenesis (vascular endothelial growth factor (VEGF), stromal cell derived factor (SDF-1), and basic fibroblast growth factor (FGF-2)).7,33 After 5 days, the microgel-in-gel culture resulted in vascularized matrix-embedded prostate cancer spheroids (Fig. 2C) and was thoroughly characterized by immunostaining/confocal laser scanning microscopy (CLSM) to confirm the established structures and identify the cellular components within the two distinct hydrogel compartments (Fig. 3A, B and C). The spatially resolved microtissue structures were found to be morphologically accurate, reflecting the in vivo architecture better than dual layer approaches with spatially restricted cancer and vascular cells.51,52
As a particular advantage of the approach, our microgel-in-gel cancer tissue model permits the defined recapitulation of different stages of tumor development in vivo. For that purpose, the culture format allows for combining cell assemblies, such as spheroids, in their customized matrices at different time points, i.e. at different levels of maturation (Fig. 2). Moreover in contrast to dual layer models,51,52 the interfacial area and cross boundary interactions between the prostate cancer cell containing microgels and the surrounding bulk gel matrices containing capillary endothelial cell networks can be easily and systematically varied by the choice of the microgel size (Fig. 1D and 3D).
Applications of the introduced starPEG-heparin microgel-in-gel materials can benefit from the broad range of previously explored hydrogel variants with distinct growth factor signaling, adhesion ligand presentation, susceptibility for enzymatic cleavage, and stiffness. For example, microgel-in-gel materials with gradated mechanical properties of the two hydrogel phases can be used to target stiffness-dependent phenotypes of embedded PC3 cells under otherwise constant conditions: while spheroids (Fig. 2 and 3) were obtained in stiffer microgels, the same cell type appeared more migratory in the surrounding softer hydrogel matrix (Fig. 3C). Independently varying all the above-mentioned matrix properties within the distinct hydrogel phases of our microgel-in-gel materials offers unprecedented options for the identification of their relevance, e.g. in tumor progression, clearly going beyond the previously reported single parameter variations of stiffness53,54 or density.55 However, for detailed biological investigations the novel technology needs further optimization depending on the particular question of interest. For instance, exact control over the spatial arrangement of the cancer spheroids within the microgels is so far limited.
Conclusions
In summary, a powerful novel microfluidic technology was established for creating customized starPEG-heparin microgels that enabled the formation of microgel-in-gel materials containing differently cell-instructive and cell-laden hydrogel types. To demonstrate the potential of our modular design approach, a 3D prostate cancer tissue in vitro model was developed. In this example system, control over mesoenvironmental parameters – cancer stroma thickness, cancer spheroid size as well as microvasculature structure and density – provides unprecedented options for examining the tumor-stroma cross-talk during the early stages of tumor angiogenesis. We anticipate the approach will become instrumental for studying various further aspects of cancer biology, including invasion/metastasis, and tumor interactions with the endothelium, at high levels of precision in vitro. Beyond that, our introduced microgel-in-gel platform is expected to advance various demanding co- and organoid culture schemes, paving the way for an array of more realistic tissue and disease models.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the DFG (Research Training Group “Nano- and Biotechniques for Electronic Device Packaging”). L. J. B. was supported by an Endeavour Fellowship as part of the Australia Awards program. MSCs were kindly gifted by Prof. Martin Bornhäuser (Universitätsklinikum Dresden). J. T. thanks the Federal Ministry of Education and Research (BMBF, “Biotechnologie2020+ Strukturvorhaben: Leibniz Research Cluster, 031A360C), and the DFG (Research Training Group “Hydrogel-based Microsystems” and TH 2037/1).References
- N. A. Peppas, J. Z. Hilt, A. Khademhosseini and R. Langer, Adv. Mater., 2006, 18, 1345–1360 CrossRef CAS.
- N. Huebsch and D. J. Mooney, Nature, 2009, 462, 426–432 CrossRef CAS.
- E. S. Place, N. D. Evans and M. M. Stevens, Nat. Mater., 2009, 8, 457–470 CrossRef CAS.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS.
- K. Y. Lee and D. J. Mooney, Chem. Rev., 2001, 101, 1869–1880 CrossRef CAS.
- E. González-Díaz and S. Varghese, Gels, 2016, 2, 20 CrossRef PubMed.
- K. Chwalek, M. V. Tsurkan, U. Freudenberg and C. Werner, Sci. Rep., 2014, 4, 8 Search PubMed.
- K. Chwalek, L. J. Bray and C. Werner, Adv. Drug Delivery Rev., 2014, 79–80, 30–39 CrossRef CAS.
- A. V. Taubenberger, L. J. Bray, B. Haller, A. Shaposhnykov, M. Binner, U. Freudenberg, J. Guck and C. Werner, Acta Biomater., 2016, 36, 73–85 CrossRef CAS PubMed.
- H. M. Weber, M. V. Tsurkan, V. Magno, U. Freudenberg and C. Werner, Acta Biomater., 2017, 57, 59–69 CrossRef CAS.
- M. Nowak, U. Freudenberg, M. V. Tsurkan, C. Werner and K. R. Levental, Biomaterials, 2017, 112, 20–30 CrossRef CAS.
- C.-Y. Liaw, S. Ji and M. Guvendiren, Adv. Healthcare Mater., 2018, 7, 1701165 CrossRef.
- Y. Yang, X. Song, X. Li, Z. Chen, C. Zhou, Q. Zhou and Y. Chen, Adv. Mater., 2018, 30, 1706539 CrossRef.
- A. Nadernezhad, N. Khani, G. A. Skvortsov, B. Toprakhisar, E. Bakirci, Y. Menceloglu, S. Unal and B. Koc, Sci. Rep., 2016, 6, 33178 CrossRef.
- T. Jiang, J. G. Munguia-Lopez, S. Flores-Torres, J. Grant, S. Vijayakumar, A. D. Leon-Rodriguez and J. M. Kinsella, Sci. Rep., 2017, 7, 4575 CrossRef.
- K. Pataky, T. Braschler, A. Negro, P. Renaud, M. P. Lutolf and J. Brugger, Adv. Mater., 2012, 24, 391–396 CrossRef CAS.
- D. B. Kolesky, R. L. Truby, A. S. Gladman, T. A. Busbee, K. A. Homan and J. A. Lewis, Adv. Mater., 2014, 26, 3124–3130 CrossRef CAS PubMed.
- L. A. Hockaday, K. H. Kang, N. W. Colangelo, P. Y. C. Cheung, B. Duan, E. Malone, J. Wu, L. N. Girardi, L. J. Bonassar, H. Lipson, C. C. Chu and J. T. Butcher, Biofabrication, 2012, 4, 035005 CrossRef CAS.
- C. A. DeForest and K. S. Anseth, Angew. Chem., 2012, 124, 1852–1855 CrossRef.
- S. Kobel and M. P. Lutolf, Curr. Opin. Biotechnol., 2011, 22, 690–697 CrossRef CAS.
- W. Lee and J. Park, Adv. Mater., 2012, 24, 5339–5344 CrossRef CAS.
- M. P. Lutolf, P. M. Gilbert and H. M. Blau, Nature, 2009, 462, 433 CrossRef CAS.
- N. J. S. Darling, E. Sideris, N. Hamada, S. T. Carmichael and T. Segura, Adv. Sci., 2018, 8, 1801046 CrossRef.
- F. Han, F. Zhou, X. Yang, J. Zhao, Y. Zhao and X. Yuan, J. Biomed. Mater. Res., Part B, 2015, 103, 1344–1353 CrossRef CAS.
- U. Freudenberg, Y. K. Liang, K. L. Kiick and C. Werner, Adv. Mater., 2016, 28, 8861–8891 CrossRef CAS.
- The term mesoenvironment (meso – from the Greek word ‘intermediate’) is introduced here to highlight the fact that adjacent cellular microenvironments with discrete characteristics give rise to heterocellular and cell-matrix interactions across spatial scales.
- M. V. Tsurkan, K. Chwalek, S. Prokoph, A. Zieris, K. R. Levental, U. Freudenberg and C. Werner, Adv. Mater., 2013, 25, 2606–2610 CrossRef CAS.
- U. Freudenberg, J. U. Sommer, K. R. Levental, P. B. Welzel, A. Zieris, K. Chwalek, K. Schneider, S. Prokoph, M. Prewitz, R. Dockhorn and C. Werner, Adv. Funct. Mater., 2012, 22, 1391–1398 CrossRef CAS.
- K. Chwalek, K. R. Levental, M. V. Tsurkan, A. Zieris, U. Freudenberg and C. Werner, Biomaterials, 2011, 32, 9649–9657 CrossRef CAS.
- U. Freudenberg, A. Zieris, K. Chwalek, M. V. Tsurkan, M. F. Maitz, P. Atallah, K. R. Levental, S. A. Eming and C. Werner, J. Controlled Release, 2015, 220, 79–88 CrossRef CAS.
- S. Prokoph, E. Chavakis, K. R. Levental, A. Zieris, U. Freudenberg, S. Dimmeler and C. Werner, Biomaterials, 2012, 33, 4792–4800 CrossRef CAS.
- L. Schirmer, P. Atallah, C. Werner and U. Freudenberg, Adv. Healthcare Mater., 2016, 5, 3157–3164 CrossRef CAS.
- L. J. Bray, M. Binner, A. Holzheu, J. Friedrichs, U. Freudenberg, D. W. Hutmacher and C. Werner, Biomaterials, 2015, 53, 609–620 CrossRef CAS.
- L. J. Bray, M. Binner, Y. Körner, M. von Bonin, M. Bornhäuser and C. Werner, Haematologica, 2017, 102, 1215–1226 CrossRef CAS.
- L. J. Bray, C. Secker, B. Murekatete, J. Sievers, M. Binner, P. B. Welzel and C. Werner, Cancers, 2018, 10, 292 CrossRef.
- L. Zhang, K. Chen, H. Zhang, B. Pang, C.-H. Choi, A. S. Mao, H. Liao, S. Utech, D. J. Mooney, H. Wang and D. A. Weitz, Small, 2018, 14, 1702955 CrossRef.
- A. S. Mao, J.-W. Shin, S. Utech, H. Wang, O. Uzun, W. Li, M. Cooper, Y. Hu, L. Zhang, D. A. Weitz and D. J. Mooney, Nat. Mater., 2016, 16, 236–243 CrossRef PubMed.
- Y. Hu, A. S. Mao, R. M. Desai, H. Wang, D. A. Weitz and D. J. Mooney, Lab Chip, 2017, 17, 2481–2490 RSC.
- C.-H. Choi, H. Wang, H. Lee, J. H. Kim, L. Zhang, A. Mao, D. J. Mooney and D. A. Weitz, Lab Chip, 2016, 16, 1549–1555 RSC.
- S. Xu, Z. Nie, M. Seo, P. Lewis, E. Kumacheva, H. A. Stone, P. Garstecki, D. B. Weibel, I. Gitlin and G. M. Whitesides, Angew. Chem., Int. Ed., 2005, 44, 724–728 CrossRef CAS.
- G. F. Christopher and S. L. Anna, J. Phys. D: Appl. Phys., 2007, 40, R319–R336 CrossRef CAS.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS.
- C. W. Visser, T. Kamperman, L. P. Karbaat, D. Lohse and M. Karperien, Sci. Adv., 2018, 4, eaao1175 CrossRef PubMed.
- D. Lee, K. Lee and C. Cha, Adv. Biosyst., 2018, 2, 1800236 CrossRef.
- J. Folkman, N. Engl. J. Med., 1971, 285, 1182–1186 CrossRef CAS.
- R. S. Kerbel, N. Engl. J. Med., 2008, 358, 2039–2049 CrossRef CAS.
- M. E. Kaighn, K. S. Narayan, Y. Ohnuki, J. F. Lechner and L. W. Jones, Invest. Urol., 1979, 17, 16–23 CAS.
- K. Hoyt, B. Castaneda, M. Zhang, P. Nigwekar, P. A. di Sant'Agnese, J. V. Joseph, J. Strang, D. J. Rubens and K. J. Parker, Cancer Biomarkers, 2008, 4, 213–225 Search PubMed.
- J. R. Weis, B. Sun and G. M. Rodgers, Thromb. Res., 1991, 61, 171–173 CrossRef CAS.
- J. Oswald, S. Boxberger, B. Jørgensen, S. Feldmann, G. Ehninger, M. Bornhäuser and C. Werner, Stem Cells, 2004, 22, 377–384 CrossRef.
- R. Janvier, A. Sourla, M. Koutsilieris and C. J. Doillon, Anticancer Res., 1997, 17, 1551–1557 CAS.
- L. C. Roudsari, S. E. Jeffs, A. S. Witt, B. J. Gill and J. L. West, Sci. Rep., 2016, 6, 32726 CrossRef CAS.
- N. Peela, F. S. Sam, W. Christenson, D. Truong, A. W. Watson, G. Mouneimne, R. Ros and M. Nikkhah, Biomaterials, 2016, 81, 72–83 CrossRef CAS.
- S. P. Singh, M. P. Schwartz, E. Y. Tokuda, Y. Luo, R. E. Rogers, M. Fujita, N. G. Ahn and K. S. Anseth, Sci. Rep., 2015, 5, 17814 CrossRef CAS PubMed.
- A. Nyga, M. Loizidou, M. Emberton and U. Cheema, Acta Biomater., 2013, 9, 7917–7926 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, design and performance of the co-flow microfluidic device, cell-induced degradation of microbeads, monophasic 3D prostate cancer model, PC3 cells embedded into microbeads: spheroid formation and live/dead assay data. See DOI: 10.1039/c9bm01009b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: