Short to ultrashort peptide-based hydrogels as a platform for biomedical applications
Nitin
Yadav
ab,
Meenakshi K.
Chauhan
*b and
Virander S.
Chauhan
 *a
*a
aMolecular Medicine Group, International Centre for Genetic Engineering & Biotechnology, Aruna Asaf Ali Marg, New Delhi-110067, India. E-mail: virander@icgeb.res.in
bDelhi Institute of Pharmaceutical Sciences and Research, Mehrauli-Badarpur Road, Sector-3, Pushpvihar, New Delhi-110017, India
First published on 22nd October 2019
Abstract
Short peptides have attracted significant attention from researchers in the past few years due to their easy design, synthesis and characterization, diverse functionalisation possibilities, low cost, possibility to make a large range of hierarchical nanostructures and most importantly their high biocompatibility and biodegradability. Generally, short peptides are also relatively more stable than their longer variants, non-immunogenic in nature and many of them self-assemble to provide an exciting range of nanostructures, including hydrogels. Thus, the development of short peptide-based hydrogels has become an area of intense investigation. Although these hydrogels have a water content of greater than 90%, they are surprisingly highly stable structures, and thus have been used for various biomedical applications, including cell therapeutics, drug delivery, tissue engineering and regeneration, contact lenses, biosensors, and wound healing, by different researchers. Herein, we review the progress of research in the rapidly expanding field of short to ultrashort peptide-based hydrogels and their possible applications. Special attention is paid to address and review this field with regard to the stability of peptide-based hydrogels, particularly to enzymatic degradation.
1. Introduction
Molecular self-assembly, which is the spontaneous arrangement of individual molecules into ordered structures through non-covalent weak interactions, has become an attractive tool for generating different hierarchical structures such as tubes, spheres, films, tapes and fibrils from nano to microscale sizes using monomeric polymer molecules for biomedical applications.1–7 Molecular self-assembly and its application are an outcome of chemistry, materials and polymer science, molecular biology, and engineering. Molecular self-assembly can be classified into two types, intramolecular self-assembly, in which complex polymers assemble from random coil conformations into stable secondary and tertiary structures, and intermolecular self-assembly, in which molecules assemble to form well-ordered supramolecular structures, such as micelle formation by surfactant molecules.6–8 Currently, intermolecular self-assembly is of great interest because scientists are using this tool for generating different structures for biomedical applications including drug delivery, biosensing, tissue engineering, disease diagnosis, and bioimaging.6,9Besides polymers based on high molecular weight natural polymers such as polysaccharides (amylose, chitin, dextran, cellulose and glycosaminoglycans), proteins (gelatin, fibrinogen, silk, and collagen), and polynucleotides (RNA and DNA), and synthetic polymers such as poly(lactic-co-glycolic acid (PLGA), polyglycolic acid (PGA) and polylactic acid (PLA), self-assembling, natural and synthetic peptides have attracted significant attention from researchers in this field.10–19 The advantages of peptide-based self-assembling systems such as ease of synthesis and characterization, high stability, chemical diversity, relatively low cost, inherent biocompatibility, and biodegradability have made them attractive candidates for applications in drug delivery, tissue engineering, diagnostics, biosensing, and drug development (Fig. 1). In the last few decades, the finding that relatively short peptides (di-, tri- and tetra-peptides) can self-assemble into ordered nanostructures, such as nanospheres, nanotubes and fibrillar gels, has made this area of research even more active and exciting.12,20–25
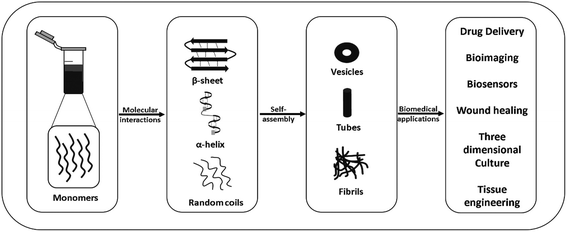 | ||
| Fig. 1 Schematic representation of the self-assembly of peptides into different nanostructures and their possible applications. | ||
Hydrogels, a network of three dimensional (3D) self-assembled polymer fibrillar chains, are soft semi-solid-like materials containing large amounts (>90%) of water. The self-assembly of peptides and proteins into fibrils in in vitro and in vivo conditions is well known and has been the subject of many reviews.12,14–17,20,22–24 Based on the aggregation of proteins in in vivo conditions such as the amyloid beta polypeptide (Aβ42), tau proteins, TAR (trans-activator regulatory) DNA-binding protein-43 (TDP-43), and islet amyloid polypeptide (IAPP) indicated in Alzheimer's disease, researchers have identified many small sequences responsible for the assembly these proteins in their native form.26–44 The shortest peptide sequence identified from these studies was a dipeptide, Phe-Phe, corresponding to the 19th and 20th residues of the central hydrophobic cluster of the highly amyloidogenic peptide Aβ42.27,45,46 Although many small peptide sequences have been identified in various disease conditions, the development of peptide-based therapeutics has remained a challenge primarily due to their inherent susceptibility to proteases. Considering that more than 500 putative proteases are present in the human body, peptide-based drugs and drug delivery systems generally suffer from very low bioavailability.47–54 Thus, researchers have tried many strategies in peptide design to resolve this problem, such as the introduction of conformationally restricted amino acids, D-amino acids and non-protein amino acids.55–62 In this review, we focus on the peptides that readily form hydrogels and their possible applications in the biomedical field.
2. Peptide based hydrogels
Many short peptides have been found to self-assemble into hydrogel-like structures. The self-assembly of short peptide sequences (>8 amino acids) can be categorized into two major classes, i.e. amphiphilic peptides and self-complementary peptides. Ultrashort peptides (<8 amino acids) have also been found to self-assemble into different nanostructures due to the amphiphilicity and/or self-complementarity of their peptide sequences. Hydrophobic stacking interactions (π–π interactions) due to aromatic residues and bulky protecting groups such as fluorenylmethyloxycarbonyl (Fmoc) have also been found to play a crucial role in their self-assembly.63–722.1 Self-assembly of short peptides into hydrogels
In the case of ultrashort peptides containing aromatic residues, the π–π interactions among their aromatic residues have been found to greatly influence the self-assembly of these peptides.77 As some important examples emphasizing the role of π–π interactions in the formation of stable hydrogels, Janmey et al. reported the formation of hydrogels from Fmoc derivatives of different dipeptides, i.e. Fmoc-Leu-Asp, Fmoc-Ala-Asp and Fmoc-Ile-Asp,78 Gazit et al. used Fmoc-Phe-Phe-COOH, Boc-Phe-Phe-COOH and Z-Phe-Phe-COOH27 and Chauhan et al. used Phe-ΔPhe and Leu-ΔPhe.79,80 A study carried out by Ulijn et al. also showed the presence of a high degree of π–π stacking of fluorenyl groups upon hydrogelation of Fmoc-Phe-Phe.81
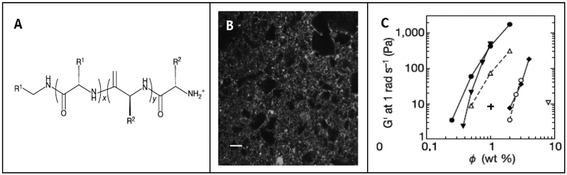
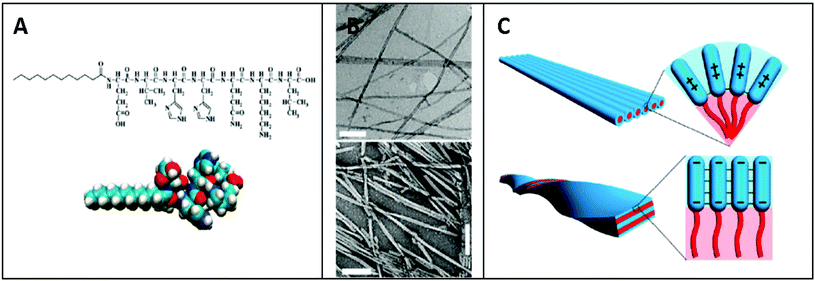
He et al. investigated the self-assembly of two peptide amphiphiles based on the Aβ(1–40) polypeptide associated with Alzheimer's disease, i.e. 2C12-Lys-Aβ(12–17) and C12-Aβ(11–17)-C12, and showed the importance of the alkyl chain positions in their self-assembly behaviour and stability.88,89 2C12-Lys-Aβ(12–17) was found to self-assemble into long fibrillar structures with micrometer length and 25 nm diameter in aqueous solution, while C12-Aβ(11–17)-C12 self-assembled into short ribbons and lamellae with a length of ∼100 nm (Fig. 3). These results clearly indicate that small but subtle changes in the hydrophobic chain positions and charge of peptide sequences can control self-assembly behaviour of peptide amphiphiles into different nanostructures.89
Similarly, Hartgerink et al. investigated the self-assembly of 12 variants of the peptide amphiphile [CCCCGGGS(PO4)RGD] with changes in its alkyl tail length, polymerizable regions, and C-terminal extensions through pH changes. They also showed an alternative mode of peptide self-assembly triggered by divalent ions such as calcium to promote self-assembly at physiological pH, which may be relevant for biomedical applications where gel formation at physiological pH is required.90
From the above described studies, it is quite clear that based on the general principle of designing peptides containing charge entities and hydrophobic regions it is possible to generate a wide variety of self-assembled peptide-based hydrogels. Further, given the wide choice of diverse amino acids, with both charged as well as hydrophobic residues, and a wide variety of hydrophobic alkyl groups, it should be possible to develop different types of hydrogels with specific designer properties. Likewise, using design principles from the principles that emerged from the examples described above under ionic peptide, a variety of hydrogels may be generate based on charge–charge interaction.
2.2 Self-assembly of ultrashort peptides into hydrogels (less than 8 amino acids)
In search of ultrashort self-assembling synthetic peptides, Chauhan et al. demonstrated the self-assembling properties of short peptide sequences derived from EDLIIKG and MNFGAFS, the shortest fibrillogenic peptide sequences from the TDP-43 protein (a 414-residue DNA/RNA binding nuclear protein) indicated in neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobar degeneration. C-terminal and N-terminal deletions of amino acid residues from MNFGAFS and EDLIIKG resulted in a series of peptides (Table 1), and their self-assembling properties were investigated using static light scattering studies, thioflavin T fluorescence assay, TEM, CD and rheology. Among the peptides, a tetrapeptide, DLII, and a pentapeptide, NFGAF, were found to form self-supportive translucent hydrogels (Fig. 4).43,103 CD spectroscopy of the peptides showed clear secondary structure changes with time during the self-assembly process, changing from α-helix to β-sheet post hydrogelation in 24 to 48 h.| S. no. | Peptides derived from TDP-43 (246–255), i.e. MNFGAFSINP | Peptides derived from TDP-43 (311–320), i.e. EDLIIKGISV |
|---|---|---|
| 1. | MNFGAFS | EDLIIKG |
| 2. | MNFGAF | EDLIIK |
| 3. | MNFGA | EDLII |
| 4. | NFGAF | DLII |
| 5. | FGAF | LII |
| 6. | GAF | II |
| 7. | AFGAF | ALII |
| 8. | NAGAF | KLII |
| 9. | NFGAA | NLII |
| 10. | — | YLII |
| 11. | — | LIID |
| 12. | — | DAII |
| 13. | — | DLAI |
| 14. | — | DLIA |
Banerjee et al. reported the self-assembly of a tetrapeptide, GAIL (peptide 1), leading to hydrogel formation. Hydrogelation of peptide-1 occurred at a very high peptide concentration i.e. 17% w/v. An analogue of peptide-1 was prepared by replacing alanine at the 3rd position in the peptide-1 sequence with phenylalanine, which formed a hydrogel at a relatively low concentration of 2.65% w/v (peptide-2).104 CD, FTIR and X-ray diffraction studies showed the presence of anti-parallel β-sheet arrangements in both gels. X-ray diffraction studies also showed the presence of π–π packing interactions in the peptide-2 gel. The anticancer drug doxorubicin was entrapped in both hydrogels and the release kinetics was determined. Both gels showed sustained drug release up to 45 h. However, no in vivo studies have been reported by these authors.104
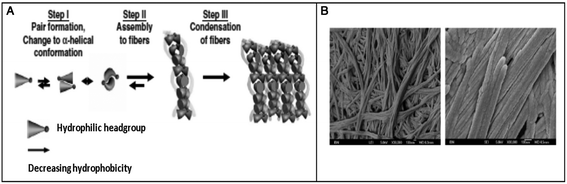
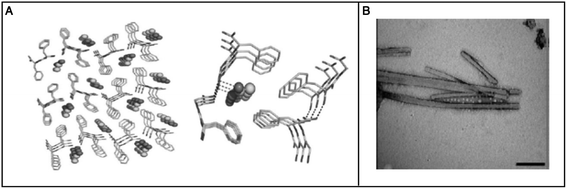
Hauser et al. reported the self-assembly of small aliphatic peptides of 3–6 amino acids long into s hydrogel. These designed peptides consisted of a hydrophilic head with charged nonaromatic amino acids (polar) at the C-terminal, followed by a tail of aliphatic chain of amino acids at their N-terminal. Both, the length of the peptide sequence and the polarity of the peptide head group influenced hydrogel formation. A decrease in non-polar character from the N- to C-terminal in the peptides led to the formation of cone-like structures, which were prone to self-assemble in a parallel antiparallel stacking pattern (Fig. 5).105 These authors also showed that the ultrashort sequences can also form α-helical conformations from random coils upon crossing a threshold concentration. CD and X-ray diffraction studies revealed the presence of a combination of α-helical and poly-L-proline-like helical conformations. Among the peptides investigated, two hexapeptides (LIVAGD and AIVAGD) and a tripeptide (IVD) were found to from stable hydrogels at a relatively low concentration and were investigated extensively.105 Following leads from the above described work, Floudas et al. synthesized a series of peptides based on the theoretical calculation of fold specificity and association affinity of new peptide sequences. Among the computationally predicted peptide analogs, two peptides, namely VIE and MYD, readily formed hydrogels and the number of their sequence variants was investigated.106 Consequently, the authors of this study were able to introduce a computational de novo peptide design strategy for self-associating peptides, which may help researchers to design new self-associating peptides and understanding the mechanism of their self-assembly process.
Gazit et al. investigated the self-assembly of short fragments of human calcitonin, a 32 amino acid polypeptide hormone (hCT), hCT15–19 (DFNKF), hCT16–19 (FNKF), hCT15–18 (DFNK), and hCT15–17 (DEN) together with hCT15–19 (F→A) (DANKA) into amyloid fibrillar structures.44 Among them, the pentapeptide DFNKF formed fibrillar structures similar to the native hCT polypeptide. The tetrapeptide, DFNK, also self-assembled into fibrils, but its fibrillar structures were less dense compared to that of the pentapeptide, DFNKF. Since amyloid formation in hCT occurs under acidic pH conditions,44 the authors suggested that the negatively charged residues present in hCT15–19 undergo protonation, resulting in amyloid formation. The presence of two phenylalanine in hCT15–19 also plays a key role in its self-assembly. DFNKF showed clear and strong green birefringence in the Congo Red birefringence study, one of the properties of amyloid fibrils. The Fourier transform infrared (FTIR) spectrum of DFNKF showed double minima at 1639 and 1669 cm−1, which indicated the presence of antiparallel sheet-like conformations, similar to the amyloid-forming hexapeptide fragment of human islet amyloid polypeptide (hIAPP22–27: NFGAIL).33 Although it is generally believed that amyloidosis in proteins occurs due to the exposure of the hydrophobic patches embedded in the hydrophobic core of the protein structure, the above studies demonstrated that the short hydrophilic sequences in the protein sequence can also induce amyloidosis as efficiently as the hydrophobic sequences. Taken together, these studies have made a major contribution in understanding the fibrilization mechanisms of various amyloidogenic proteins and the design of other fibrillar hydrogel-forming peptides.43,44,103,105,106
The development of hydrogels based on ultrashort peptide sequences is a rapidly expanding field of research. Many single amino acids, dipeptides and tripeptides conjugated with bulky aromatic groups such as fluorenylmethyloxycarbonyl (Fmoc), pyrene, naphthalene, cinnamoyl, carboxybenzyl and phenothiazine have been reported to self-assemble into various structural assemblies, especially fibrillar hydrogels, and have received much attention recently.63,64,66–69,71,72,81,107–109 However, these bulky, mostly aromatic groups are generally toxic in nature, increase the flexibility of the peptide backbone and block N- or C- or both the terminals of the peptides, especially the N-terminal, which hamper their functionalizability.110
Vegners et al. demonstrated for the first time in 1995 that three Fmoc-based dipeptides, Fmoc-Leu-Asp, Fmoc-Ala-Asp and Fmoc-Ile-Asp, formed viscoelastic hydrogels by boiling peptide solutions at 100 °C in 10 mM phosphate buffer (pH 7.4) followed by vigorous stirring and cooling.78 The Fmoc-Leu-Asp hydrogel was used as an adjuvant to deliver antiviral adamantanimine derivatives, 3,5-dimethyl-adamantylamine hydrochloride and 5-methyl-1-adamantine 3-carboxylic acid, to produce specific antibodies against these drugs, which remained non-antigenic unless injected with the hydrogel adjuvant or coupled to protein carriers. The antiviral drugs entrapped in the peptide hydrogels when injected in rabbits showed the generation of antibody titers as high as in animals immunized with bovine serum albumin (BSA) conjugated drug molecules using Freund's adjuvant, which indicated the possible use of these ultrashort peptide hydrogels for a number of practical applications in biomolecular delivery.78
Xu et al. showed the formation of a self-assembled hydrogel using a mixture of three individual components having two Fmoc-based amino acid derivatives, i.e. Fmoc-Leu-OH, Fmoc-lys-OH with a bisphosphonate drug, and pamidronate. Heating the mixture at 70 °C and then cooling to room temperature resulted in the formation of a fibrillar hydrogel in aqueous media at pH 9–10.4. Fmoc-Leu-OH and Fmoc-lys-OH were used as hydrogelators, while pamidronate was reported to act as both a hydrogen acceptor and donor to promote hydrogelation though hydrogen bonding. Application of this hydrogel matrix in mice bearing uranium-infected skin wounds resulted in significant recovery.111
In an outstanding study, Gazit et al. identified the shortest fibrillogenic sequence, a dipeptide, Phe-Phe, of the amyloid polypeptide Aβ-42 and showed that it formed nanotubes similar to Aβ-42 itself.27 Further, in 2006, Gazit and co-workers showed the gelation of a derivative of diphenylalanine, Fmoc-diphenylalanine, into a hydrogel and used it as a scaffold for three-dimensional culture of Chinese Hamster Ovary (CHO) cells for tissue engineering applications. The cells grown in the hydrogel matrix were alive, healthy and similar to the cells grown in the control two-dimensional non-hydrogel-coated surfaces.63 Inspired by these studies on Phe-Phe, Groot et al. substituted the N-terminal Phe of the dipeptide with 19 amino acids and investigated its effect on fibrilization. All the substitutions except isoleucine substitution resulted in low aggregation propensities compared to the native peptide. In a further study, de Groot et al. found that Ile-Phe self-assembled into a transparent and thermoreversible fibrillar hydrogel.45 The earlier hypothesis that head-to-tail interactions between the amino and carboxyl-terminus of peptides stabilize the overall peptide assembly may not explain self-assembly of Ile-Phe since an analogous dipeptide, Val-Phe, with a small difference in the side chain of the N-terminal did not self-assemble, even at high concentrations.45 Clearly, even small changes in the size and hydrophobicity of the N-terminal of X-Phe (X = Ile, Val, etc.) can significantly affect its self-assembly behaviour. Yan et al. designed a hydride hydrogel using Fmoc-FF and C60 pyrrolidine tris-acid-based fullerene derivative nanoparticles for photodynamic antibacterial applications. Incorporation of fullerene nanoparticles in the peptide hydrogel resulted in improved mechanical hydrogel properties. Enhanced photodynamic antibacterial activity was observed in in vitro and in vivo conditions for the hybrid hydrogel.112 In a recent study, Yan et al. also showed the self-assembly of a cyclic-dipeptide, Leu-Phe (CLP), into strong hydrogels in both physiological and harsh conditions, including water, Dulbecco's Modified Eagle's Medium (DMEM), phosphate buffer saline (PBS), aqueous solutions of different pH (pH 3, 5, 9 and 11) and trypsin. The gelation of CLP was driven by hydrogen bonding, which was confirmed by FTIR and computation simulation analysis.113
During the last two decades, a large number of studies have suggested that ultrashort peptides, particularly di- and tri-peptides, can form hydrogels as stable and useful as many longer peptides. However, the ease of synthesis of ultrashort peptides, their relatively higher stability to enzymatic degradation and high biocompatibility make them much more attractive candidates for creating hydrogels for multiple biomedical applications. Further, these small peptides can easily be prepared in laboratories that are not necessarily equipped for peptide synthesis, thereby opening the door for other chemists to enter this exciting field of nanomaterial science.
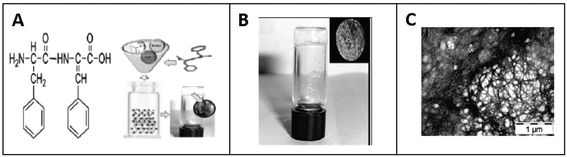
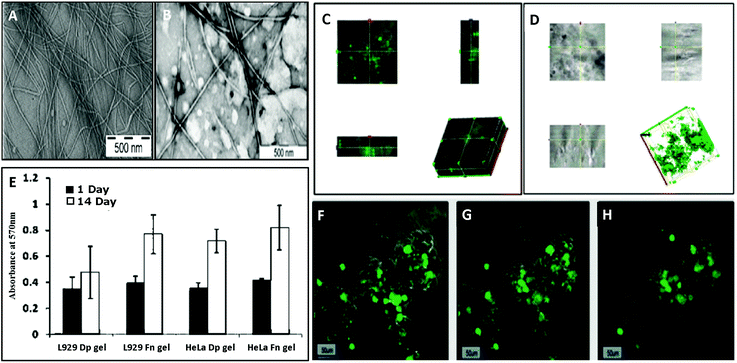
They also found that Phe-ΔPhe can self-assemble into highly stable, and mechanically strong hydrogels at higher peptide concentrations, 0.2–1.0% w/v, in 0.8 M acetate buffer at pH-7 upon boiling and cooling the solution to room temperature (Fig. 8).79 The dipeptide hydrogel had significantly higher mechanical strength of 209 kPa (storage modulus) compared to other peptide-based hydrogels such as MAX1 (2.5 kPa),126 Fmoc-Phe-Phe (10 kPa),63 and tetrapeptide peptide-2 hydrogel.127 The TEM images of the Phe-ΔPhe hydrogel revealed the presence of a network of long fibrils with a micrometer length and 15–30 nm diameter (Fig. 8). The thermal circular dichroism (CD) spectra of the hydrogel showed no change with temperature up to 50 °C, indicating the stability of the hydrogel at physiological temperatures. The gel could entrap a number of drug-like molecules of different molecular weights and physicochemical properties and release them in a sustained gradual manner. pH-Sensitive drug release (∼3 times increased) from the hydrogel was observed when the environmental pH of the gel was changed from neutral (pH 7) to acidic (pH 2) and basic (pH 10).79 The dipeptide, Phe-ΔPhe, hydrogel was used as a scaffold for the three-dimensional (3D) growth of cells.128 Due to the availability of free N- and C-terminal groups (–NH2 and –COOH) in the Phe-ΔPhe hydrogel, the gel was easily functionalized covalently with a peptide ligand, RGDGG, containing the well-known cell adhesion motif, RGD, the shortest sequence of fibronectin, which binds to αVβ3 integrin proteins on the surface of cells. The functionalized dipeptide hydrogel showed enhanced cell growth of cervical cancer cells, HeLa and subcutaneous areolar and adipose tissue cells, L292, in its matrix compared to the non-functionalized gel and two-dimensional (2D) culture of cells (Fig. 9).128 These characteristics of the Phe-ΔPhe hydrogel highlight its suitability for possible use in drug delivery and tissue engineering applications.
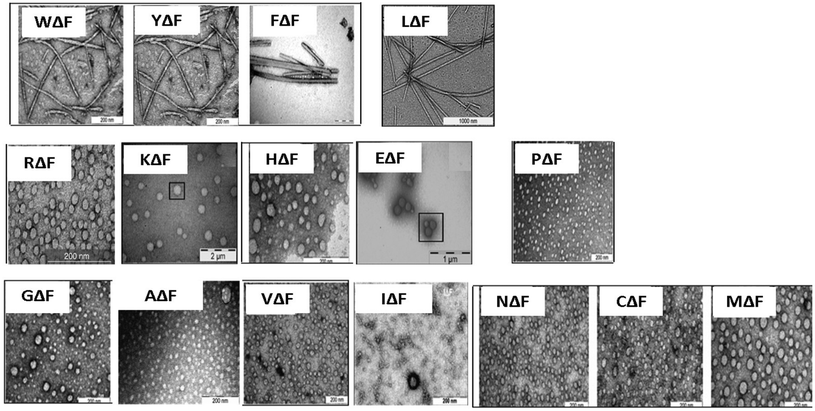
As a follow up of the Phe-ΔPhe study, the same research group screened a set of 16 dipeptides (Table 2) containing different N-terminal L-amino acids together with ΔPhe at their C-terminal, for self-assembly into hydrogels. They found that only two dipeptides, Trp-ΔPhe and Leu-ΔPhe, formed gel-like structures under physiological conditions (Fig. 10).80 However, the gel formed by Trp-ΔPhe was significantly weaker than that of Leu-ΔPhe, which self-assembled instantaneously into a highly stable and biocompatible hydrogel under physiological conditions. The Leu-ΔPhe hydrogel showed a peptide concentration-dependent increase in mechanical strength (5.46 ± 0.58 kPa at 0.4 wt% to 136.0 ± 29.8 kPa at 1.0 wt%) in the rheology studies. Also, the hydrogel showed self-healing properties after structural disruption, indicating its injectable nature, a highly desirable characteristic required for the development of hydrogels as platforms for various biomedical applications. The hydrogel entrapped a number of drug-like molecules with different physicochemical properties and released them in a much more sustained manner than other polymer and peptide-based hydrogels.79,80,129 A change in environmental pH, from neutral to acidic and basic resulted in faster drug release, which indicated stimuli-responsive drug release from the hydrogel matrix. Furthermore, the hydrogel was completely non-toxic in in vitro and in vivo conditions. An increase in the regression of tumor size (tumor volume) was observed when the hydrogel entrapped with an anticancer drug, mitoxantrone, was injected in mice bearing B16F10 melanoma tumor.80 The Leu-ΔPhe hydrogel exhibited all the expectations of an ideal hydrogel system and its gelation represented an unusual and intriguing example of peptide-based self-assembly considering the fact that very similar dipeptides, Ile-ΔPhe and Val-ΔPhe, did not form hydrogels under the same conditions.
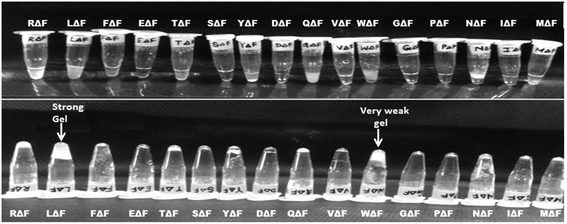 | ||
| Fig. 10 Tube inversion test showing the self-assembly of a set of modified dipeptides, XΔPhe, into a hydrogel. A dipeptide LeuΔPhe, instantaneously self-assembled into a self-supporting hydrogel.80 | ||
| S. no. | Peptide name | Peptide sequence |
|---|---|---|
| 1. | Arginine-α,β-dehydrophenylalanine | RΔF |
| 2. | Leucine-α,β-dehydrophenylalanine | LΔF |
| 3. | Phenylalanine-α,β-dehydrophenylalanine | FΔF |
| 4. | Glutamic acid-α,β-dehydrophenylalanine | EΔF |
| 5. | Threonine-α,β-dehydrophenylalanine | TΔF |
| 6. | Serine-α,β-dehydrophenylalanine | SΔF |
| 7. | Tyrosine-α,β-dehydrophenylalanine | YΔF |
| 8. | Aspartic acid-α,β-dehydrophenylalanine | DΔF |
| 9. | Glutamine-α,β-dehydrophenylalanine | QΔF |
| 10. | Valine-α,β-dehydrophenylalanine | VΔF |
| 11. | Tryptophan-α,β-dehydrophenylalanine | WΔF |
| 12. | Glycine-α,β-dehydrophenylalanine | GΔF |
| 13. | Proline-α,β-dehydrophenylalanine | PΔF |
| 14. | Asparagine-α,β-dehydrophenylalanine | NΔF |
| 15. | Isoleucine-α,β-dehydrophenylalanine | IΔF |
| 16. | Methionine-α,β-dehydrophenylalanine | MΔF |
These studies suggest that the use of non-protein amino acids, particularly those modified at the α-carbon of their amino acids, may offer additional attractive choices in the toolbox of researchers interested in designing novel self-assembled peptide-based structures, particularly hydrogels.
3. Strategies to enhance the biostability of peptide-based hydrogels
Peptide-based drugs and delivery vehicles are attracting increasing interest because of their easy design, synthesis and characterization, low cost, diverse functionalisation possibilities, possibility to make a large range of hierarchical nanostructures and their inherent biocompatibility. However, a major hurdle in the development of these peptides as possible drug and/or drug delivery vehicles is their inherent susceptibility to proteolytic degradation, which limits their biomedical application. Thus, there have been huge efforts by researchers to stabilize peptides against enzymatic degradation, including the use of Cα amino acids with the D-configuration, i.e.D-amino acids, Cα substituted amino acids such as Aib, α,β-dehydro amino acids, Cα-fluoroalkyl substituted amino acids, N-alkyl-Cα amino acids (peptoids), polymer linked peptides and cyclic peptides.130All amino acids, except glycine, due to the chirality at its Cα carbon, exist in two isomeric forms, i.e. D and L forms, and almost all naturally occurring peptides, proteins and synthetic peptides made via the recombinant method contain L-amino acids. In biological systems, enzymes recognise only L-amino acids as their substrate for peptide or protein degradation purposes. D-Amino acids can easily be incorporated at defined positions in any peptide using standard chemical peptide synthesis methods. Xu et al. reported the self-assembly of a D-tripeptide (PheD-PheD-TyrD) by enzymatic dephosphorylation and utilized this hydrogel to inhibit the growth of cancer cells (HeLa cells).55,131–133 Ding et al. also showed the self-assembly of a D-peptide (8 residues, Nap-GDFDFDYGRGD) into biostable hydrogels, which were used as delivery vehicles for an anticancer drug, 10-hydroxycamtothecin, for cancer therapy.134 However, although the stability of these peptides to enzymatic degradation was enhanced, there was significantly reduced cellular uptake in case of these peptides.55,131,135
Peptides containing α-substituted amino acids or α,α-disubstituted glycine have shown stability towards proteolytic degradation. Many naturally occurring peptides have been identified, which contain Cα substituted amino acids such as antimicrobial peptide-alamethicin.130,136 The introduction of Cα-substituted amino acids such as Aib introduces conformational constraint in the peptide backbone and provides stability against enzymatic degradation.137–140 Recently, Chaudhary et al. described the self-assembly of different dipeptides, including Asn-Gly, Aib-ProD, and ProD-Gly. They demonstrated drug (doxorubicin) entrapment and the three-dimensional growth of cells (rat pancreatic cell line, RIN-5F and human embryonic kidney cells, HEK-293) in the peptide hydrogel matrices, but did not compare their enzymatic stability with their natural L-amino acid-containing analogs.141
Chauhan and co-workers showed that the introduction of a noncoded α,β-dehydro amino acid, namely α,β-dehydrophenylalanine, induced conformational constrain both in the peptide backbone and the side chain as well as enhanced resistance to enzymatic degradation. Using these design principles, this research group developed conformationally restricted and biologically active peptides with various possible therapeutic applications.118,119,125,142 They also showed the self-assembly of a series of dipeptides containing ΔPhe at their C-terminal (XΔPhe) into different hierarchical nanostructures and investigated their possible use in biomolecular delivery and three-dimensional growth of cells. Out of a panel of twenty dipeptides, two dipeptides, namely Phe-ΔPhe and LeuΔPhe, self-assembled into mechanically strong hydrogels with enhanced enzymatic stability. Drug delivery and tissue engineering applications were also investigated by this group.79,80 The above described studies have attracted keen interest and enhanced efforts in developing more stable hydrogels.
4. Biomedical applications of peptide-based hydrogels
As mentioned earlier, peptide-based hydrogels, especially short peptides, due to their high biocompatibility, biodegradability and ease of synthesis have attracted significant interest in the biomedical field and have been used for different applications including drug delivery, scaffolds for wound healing, bioimaging, bioprinting and tissue engineering.4.1 Drug delivery
Administration of drugs conventionally generally suffers from various drawbacks, including solubility issues, burst release, low bioavailability due to high clearance or metabolism, high degradation and nonspecific distribution. To overcome these problems, many researchers have tried drug delivery techniques to deliver drugs using different polymer-based nanostructures. Both hydrophilic and hydrophobic drugs can be encapsulated in polymeric nanostructures depending on their properties, which protect them from the outer environment and deliver them in a controlled manner. Peptide-based nanostructures, especially injectable hydrogels, have attracted great interest in this field and many researchers have used them as delivery vehicles. Banerjee et al. showed the entrapment and sustained release of an anticancer drug, doxorubicin, from tetrapeptide (GAIL and GFIL) hydrogels in in vitro studies.104 Xu et al. showed the controlled delivery of a drug, pamidronate, using a hydrogel formed from two Fmoc-based amino acid derivatives, Fmoc-Leu-OH and Fmoc-Lys-OH. They showed significant recovery of uranium-infected skin wounds in mice treated with the hydrogel matrix entrapped with pamidronate.111 Chauhan et al. showed the entrapment and sustained release of a number of drugs, both hydrophilic and hydrophobic, from two chemically modified dipeptide hydrogels, Phe-ΔPhe and Leu-ΔPhe. These hydrogels showed pH-sensitive drug release. Mitoxantrone, an anticancer drug, entrapped with Leu-ΔPhe hydrogel, when injected in tumor-bearing mice showed significant regression of the tumors.80 Vegners et al. used an Fmoc-Leu-Asp hydrogel to deliver antiviral adamantanimine derivatives, 3,5-dimethyl-adamantylamine hydrochloride and 5-methyl-1-adamantine 3-carboxylic acid, to produce specific antibodies against these drugs and showed the generation of antibody titers as high as in rabbits immunized with bovine serum albumin (BSA) conjugated drug molecules.784.2 Three-dimensional growth and tissue engineering
Hydrogels mimicking the extracellular matrix of cells are promising materials for tissue engineering and 3D cell culture. There is much evidence to support the fact that the extracellular matrix plays a key role in regulating cell behaviour, and therefore many researchers have used different hydrogel matrices to culture cells in the 3D environment of hydrogels.143 Cells cultured in hydrogels provide the growth of cells in all directions, allow multiple cell–cell and cell–matrix interactions, normal cell cycle, 3D fluid perfusion, complete cellular polarization and protein and gene expression similar to that in in vivo conditions.8,9,20 Zhang et al. used EAK16- and RAD16-based hydrogels for cell attachment and growth.101 Both peptide hydrogels were found to support the attachment of diverse cell types, including hepatocytes, keratinocytes and fibroblasts. In addition to supporting cell growth, hydrogels have been shown to maintain differentiated morphologies and functions of cells.101 Many analogues of EAK16 were made and tested for 3D cell culture. Holmes et al. used RAD16-I- and RAD16-II-, new analogues of EAK16, based hydrogels as scaffolds for the culture of neuronal cells. Both hydrogels supported cell attachment and differentiation with extensive neurite outgrowth.102 Pilocci et al. showed an enzyme, P. cepacian, catalysed the hydrogelation of a tripeptide, Fmoc-Phe-Phe-Phe, and used this peptide hydrogel as a 3D matrix for the growth of microglial cells.144 Puramatrix, a commercially available polypeptide hydrogel, is also intended for use in various 3D growth and tissue engineering applications.145 Chauhan et al. used a dipeptide, Phe-ΔPhe, hydrogel for 3D cell culture and functionalized it with a ligand, the Arg-Gly-Asp motif, the shortest sequence of fibronectin required for attachment to cell surface integrins, to enhance cell attachment and growth (Fig. 9). The authors showed the enhanced cellular growth of HeLa (ovarian cancer) and L929 (mouse fibroblast) cells in the functionalized hydrogel and indicated its potential for further development for tissue engineering applications.128 In a recent study, Chauhan et al. used two ultrashort peptide-based hydrogels, a dipeptide, Leu-ΔPhe, and a tetrapeptide, Asp-Leu-Leu-Ile, for the 3D growth of cells and used them as scaffolds for tibial bone defects and showed that the healing of bone defects was significantly higher in rabbits treated with peptide hydrogels compared to the PBS treatment group (unpublished). Hauser et al. showed the self-assembly of two hexapeptides, ILVAGK and LIVAGK, into dense fibrillar structures and made hydrogel patches (25 mm in diameter) for their easy applicability on skin wounds. They showed the significantly higher recovery of partial thickness burn wounds in rats treated with the peptide hydrogels compared to the PBS and Mepitel, a silicone-coated polyamide matrix, groups.146 Hauser et al. also used peptide, LIVAGKC and LK6C, hydrogels as dressings for full-thickness excision wounds in mice and found that the wounds treated with the hydrogel dressing potentiated complete re-epithelialization compared to the control non-treated group.147 These studies clearly suggest that peptide-based hydrogels, especially ultrashort peptides, can further be developed as platforms for tissue engineering and regeneration, bioinks and scaffolds for wound healing applications in clinic.4.3 Antimicrobials and wound healing
Presently, microbial infections are among the most prevalent cause of death worldwide. Disease conditions such as diabetic foot ulcers, where healing is very slow and chances of getting an infection are very high, are very difficult to treat. Also, the excessive use of antibiotics has resulted in the occurrence of drug-resistant microbial strains and several pharmaceutical industries have now stopped their antibiotic research, which has made this problem even more serious. Many naturally occurring and synthetically designed antimicrobial peptides have been identified by researchers in the past few years.148 Peptide-based antimicrobials due to their unique mechanism of action seem promising candidates for treating multidrug resistant infections.149 Antimicrobial peptide hydrogels can be applied to injured areas as would dressings, implant coating and/or creams and have been used by many researchers. Christman et al. developed a synthetic cell-adhesive polypeptide hydrogel, poly(lys)60(Ala)60 and its conjugated version with polyethylene glycol-amide succinimidyl glutarate, with antibacterial activities.150 Recently Li et al. investigated the self-assembly of a series of cationic peptide amphiphiles (PA) into pH-responsive hydrogels with a varying number of lysine (PA-Kn, where n is the number of lysine, which was 0–3). An increase in the number of lysine resulted in reduced self-assembling properties, and as expected, the lysine-containing PA showed higher antibacterial activity than the control PA without lysine.1514.4 Biosensors
The development of peptide hydrogel-based biosensors is a rapidly expanding field of research due to the low biocompatibility and difficult and costly synthesis of other polymer-based hydrogels. Yang et al. used a self-assembled hydrogel of the Fmoc-Phe-Phe dipeptide to construct a smart biointerface, which was used for enzyme-based electrochemical biosensing and cell monitoring purposes. The authors used this hydrogel as a host for horseradish peroxidase and showed the detection of H2O2 levels released from HeLa cells in in vitro conditions.152 Similarly, Park et al. utilized Fmoc-Phe-Phe dipeptide hydrogel encapsulating enzyme bioreceptors (e.g., glucose oxidase or horseradish peroxidase) and fluorescent reporters (e.g., CdTe and CdSe quantum dots) to make enzyme-based optical biosensors. The authors reported the detection of different levels of glucose and phenolic compounds using the dipeptide-based hydrogel biosensor.153 Alves et al. also used an Fmoc-Phe-Phe hydrogel containing an antigen, i.e. Leishmania infantum chagasi, for the detection of Leishmaniasis disease in patient samples.154 Miller et al. developed an octapeptide-based hydrogel in combination with a DNA aptamer as a biosensor to identify single or multiple nucleotide polymorphisms, aiming to recognise susceptibility to certain monogenic or complex diseases in patients.155Many peptide-based hydrogels such as Puramatrix and Matrigel are already in use and commercially available, while many others are in clinical trials.156,157 It is clear from the reviewed literature that in general, peptide-based, short and ultrashort, hydrogels represent nanostructures having better chemical, biophysical, biocompatible characteristics than synthetic organic and inorganic polymers with potential applications in drug delivery, tissue engineering, bioprinting, bioimaging and many other biomedical applications. Considering the significant interest in this field of research, it will not be surprising to see many novel peptide-based hydrogels being developed for various biomedical purposes.
5. Current challenges and the way forward
Molecular self-assembly has become an attractive tool for generating different hierarchical structures such as tubes, spheres, films, tapes and fibrils from nano to microscale sizes for biomedical applications. Peptide-based self-assembled nanostructures are inherently more attractive due to their easy and inexpensive synthesis, easy functionalisation with site-specific ligands, biocompatibility and biodegradability. The findings that ultrashort peptides (<8 residue) including even di- and tri-peptides can self-assemble to form nanostructures such as hydrogels have made this area of research more active and exciting. Ultrashort self-assembled peptide-based hydrogels are being developed for various biomedical applications, including drug delivery, scaffolds for wound healing, bioimaging, bioprinting and tissue and engineering. However, although these developments are very exciting, several issues such as resemblance of extracellular matrices in tissue engineering, lack of information on the pharmacokinetics of peptide-based hydrogels in in vivo conditions, issues of desirable mechanical properties in the case of their use as biomolecular delivery and tissue engineering platforms, and careful study of their in vivo toxicity have yet to be resolved. As these hydrogels are further developed for their practical use in biomedical applications, their long-term stability will also need to be established. Also, although the preparation, characterization and in vitro use of peptide-based hydrogels and their possible use in biomedical applications have been described, there is little information on their clinical trials in humans. Thus, there is an urgent need to take some of the promising peptide-based hydrogels on a translational path and to carry out careful studies under GLP conditions to finally develop them for their intended use.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We acknowledge the Department of Biotechnology (DBT), Government of India (Grant No. BT/HRD/35/07/VSC/2015 and BT/PR2573/NNT/28/534/2011) and Department of Science and Technology (DST), Government of India (Grant No. SB/S2/JCB-41/2014) for the financial support. Nitin Yadav thanks Centre for Cellular and Molecular Platforms (C-Camp) and Biotechnology Ignition Grant (BIG), Biotechnology Industry Research Assistance Council (BIRAC), for financial support (Grant No-BIRAC/CCAMP0770/BIG-13/18).References
- T. Ohtake, Bottom-up approaches for material and device designing using practical aspects of self-assembled molecular architectures, Mol. Syst. Des. Eng., 2018, 3(5), 804–818 RSC.
- S. Zhang, Fabrication of novel biomaterials through molecular self-assembly, Nat. Biotechnol., 2003, 21(10), 1171 CrossRef CAS.
- G. M. Whitesides, J. P. Mathias and C. T. Seto, Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures, Science, 1991, 254(5036), 1312–1319 CrossRef CAS.
- S. Zhang, Emerging biological materials through molecular self-assembly, Biotechnol. Adv., 2002, 20(5–6), 321–339 CrossRef CAS.
- D. Yan, Y. Zhou and J. Hou, Supramolecular self-assembly of macroscopic tubes, Science, 2004, 303(5654), 65–67 CrossRef CAS.
- K. Subramani and W. Ahmed, Self-Assembly of Proteins and Peptides and Their Applications in Bionanotechnology and Dentistry, in Emerging Nanotechnologies in Dentistry, Elsevier, 2012, pp. 209–224 Search PubMed.
- K. Subramani, A. Khraisat and A. George, Self-assembly of proteins and peptides and their applications in bionanotechnology, Curr. Nanosci., 2008, 4(2), 201–207 CrossRef CAS.
- J. A. Pelesko, Self assembly: the science of things that put themselves together, Chapman and Hall/CRC, 2007 Search PubMed.
- M. Shibata, et al., Intramolecular folding or intermolecular self-assembly of amphiphilic random copolymers: on-demand control by pendant design, Macromolecules, 2018, 51(10), 3738–3745 CrossRef CAS.
- T. J. Deming, Synthetic polypeptides for biomedical applications, Prog. Polym. Sci., 2007, 32(8–9), 858–875 CrossRef CAS.
- H. Acar, et al., Self-assembling peptide-based building blocks in medical applications, Adv. Drug Delivery Rev., 2017, 110, 65–79 CrossRef.
- E. Gazit, Self-assembled peptide nanostructures: the design of molecular building blocks and their technological utilization, Chem. Soc. Rev., 2007, 36(8), 1263–1269 RSC.
- M. Reches and E. Gazit, Molecular self-assembly of peptide nanostructures: mechanism of association and potential uses, Curr. Nanosci., 2006, 2(2), 105–111 CrossRef CAS.
- J. Lu and X. Wang, Biomimetic Self-Assembling Peptide Hydrogels for Tissue Engineering Applications, in Biomimetic Medical Materials, Springer, 2018, pp. 297–312 Search PubMed.
- T. Sawada, H. Mihara and T. Serizawa, Peptides as New Smart Bionanomaterials: Molecular-Recognition and Self-Assembly Capabilities, Chem. Rec., 2013, 13(2), 172–186 CrossRef CAS.
- S. Kyle, et al., Recombinant self-assembling peptides as biomaterials for tissue engineering, Biomaterials, 2010, 31(36), 9395–9405 CrossRef CAS.
- K. E. Inostroza-Brito, et al., Cross-linking of a biopolymer-peptide co-assembling system, Acta Biomater., 2017, 58, 80–89 CrossRef CAS PubMed.
- N. Habibi, et al., Self-assembled peptide-based nanostructures: Smart nanomaterials toward targeted drug delivery, Nano Today, 2016, 11(1), 41–60 CrossRef CAS.
- C. J. Edwards-Gayle and I. W. Hamley, Self-assembly of bioactive peptides, peptide conjugates, and peptide mimetic materials, Org. Biomol. Chem., 2017, 15(28), 5867–5876 RSC.
- K. H. Chan, et al., Systematic Moiety Variations of Ultrashort Peptides Produce Profound Effects on Self-Assembly, Nanostructure Formation, Hydrogelation, and Phase Transition, Sci. Rep., 2017, 7(1), 12897 CrossRef.
- K. H. Chan, et al., C-Terminal Residue of Ultrashort Peptides Impacts on Molecular Self-Assembly, Hydrogelation, and Interaction with Small-Molecule Drugs, Sci. Rep., 2018, 8(1), 17127 CrossRef.
- M. Ni and S. Zhuo, Applications of self-assembling ultrashort peptides in bionanotechnology, RSC Adv., 2019, 9(2), 844–852 RSC.
- W. Y. Seow and C. A. Hauser, Short to ultrashort peptide hydrogels for biomedical uses, Mater. Today, 2014, 17(8), 381–388 CrossRef CAS.
- J. J. Panda and V. S. Chauhan, Short peptide based self-assembled nanostructures: implications in drug delivery and tissue engineering, Polym. Chem., 2014, 5(15), 4418–4436 RSC.
- S. Bera, et al., Rigid helical-like assemblies from a self-aggregating tripeptide, Nat. Mater., 2019, 1 CAS.
- K. Rajagopal and J. P. Schneider, Self-assembling peptides and proteins for nanotechnological applications, Curr. Opin. Struct. Biol., 2004, 14(4), 480–486 CrossRef CAS.
- M. Reches and E. Gazit, Casting metal nanowires within discrete self-assembled peptide nanotubes, Science, 2003, 300(5619), 625–627 CrossRef CAS.
- L. Adler-Abramovich, et al., Self-assembled arrays of peptide nanotubes by vapour deposition, Nat. Nanotechnol., 2009, 4(12), 849 CrossRef CAS.
- C. A. Hauser and S. Zhang, Designer self-assembling peptide nanofiber biological materials, Chem. Soc. Rev., 2010, 39(8), 2780–2790 RSC.
- T. Scheibel, et al., Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(8), 4527–4532 CrossRef CAS PubMed.
- E. Kasotakis, et al., Design of metal-binding sites onto self-assembled peptide fibrils, Pept. Sci., 2009, 92(3), 164–172 CrossRef CAS.
- G. T. Westermark, U. Engström and P. Westermark, The N-terminal segment of protein AA determines its fibrillogenic property, Biochem. Biophys. Res. Commun., 1992, 182(1), 27–33 CrossRef CAS.
- K. Tenidis, et al., Identification of a penta-and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties, J. Mol. Biol., 2000, 295(4), 1055–1071 CrossRef CAS PubMed.
- M. Reches and E. Gazit, Amyloidogenic hexapeptide fragment of medin: homology to functional islet amyloid polypeptide fragments, Amyloid, 2004, 11(2), 81–89 CrossRef CAS.
- Y. Mazor, et al., Identification and characterization of a novel molecular-recognition and self-assembly domain within the islet amyloid polypeptide, J. Mol. Biol., 2002, 322(5), 1013–1024 CrossRef CAS.
- M. L. de la Paz, et al., De novo designed peptide-based amyloid fibrils, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(25), 16052–16057 CrossRef CAS.
- F. T. Senguen, et al., Clarifying the influence of core amino acid hydrophobicity, secondary structure propensity, and molecular volume on amyloid-β 16–22 self-assembly, Mol. BioSyst., 2011, 7(2), 497–510 RSC.
- T. M. Doran, et al., Role of amino acid hydrophobicity, aromaticity, and molecular volume on IAPP (20–29) amyloid self-assembly, Proteins: Struct., Funct., Bioinf., 2012, 80(4), 1053–1065 CrossRef CAS.
- J. J. Balbach, et al., Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR, Biochemistry, 2000, 39(45), 13748–13759 CrossRef CAS.
- R. Azriel and E. Gazit, Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide an experimental support for the key role of the phenylalanine residue in amyloid formation, J. Biol. Chem., 2001, 276(36), 34156–34161 CrossRef CAS.
- E. Gazit, Mechanisms of amyloid fibril self-assembly and inhibition: Model short peptides as a key research tool, FEBS J., 2005, 272(23), 5971–5978 CrossRef CAS.
- L. M. Igaz, et al., Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies, J. Biol. Chem., 2009, 284(13), 8516–8524 CrossRef CAS.
- A. Saini and V. S. Chauhan, Delineation of the core aggregation sequences of TDP-43 C-terminal fragment, ChemBioChem, 2011, 12(16), 2495–2501 CrossRef CAS.
- M. Reches, Y. Porat and E. Gazit, Amyloid fibril formation by pentapeptide and tetrapeptide fragments of human calcitonin, J. Biol. Chem., 2002, 277(38), 35475–35480 CrossRef CAS.
- N. S. de Groot, et al., Ile-Phe dipeptide self-assembly: clues to amyloid formation, Biophys. J., 2007, 92(5), 1732–1741 CrossRef.
- Y. Song, et al., Synthesis of peptide-nanotube platinum-nanoparticle composites, Chem. Commun., 2004,(9), 1044–1045 RSC.
- L. Di, Strategic approaches to optimizing peptide ADME properties, AAPS J., 2015, 17(1), 134–143 CrossRef CAS.
- L. Otvos Jr. and J. D. Wade, Current challenges in peptide-based drug discovery, Front. Chem., 2014, 2, 62 Search PubMed.
- B. J. Bruno, G. D. Miller and C. S. Lim, Basics and recent advances in peptide and protein drug delivery, Ther. Delivery, 2013, 4(11), 1443–1467 CrossRef CAS.
- R. Böttger, R. Hoffmann and D. Knappe, Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum, PLoS One, 2017, 12(6), e0178943 CrossRef.
- H. H. Gorris, et al., Rapid profiling of peptide stability in proteolytic environments, Anal. Chem., 2009, 81(4), 1580–1586 CrossRef CAS PubMed.
- J. L. Lau and M. K. Dunn, Therapeutic peptides: Historical perspectives, current development trends, and future directions, Bioorg. Med. Chem., 2018, 26(10), 2700–2707 CrossRef CAS.
- M. L. English and C. H. Stammer, The enzyme stability of dehydropeptides, Biochem. Biophys. Res. Commun., 1978, 83(4), 1464–1467 CrossRef CAS.
- M. Gupta, et al., Self-assembly of a dipeptide-containing conformationally restricted dehydrophenylalanine residue to form ordered nanotubes, Adv. Mater., 2007, 19(6), 858–861 CrossRef CAS.
- M. Melchionna, K. E. Styan and S. Marchesan, The unexpected advantages of using D-amino acids for peptide self-assembly into nanostructured hydrogels for medicine, Curr. Top. Med. Chem., 2016, 16(18), 2009–2018 CrossRef CAS.
- R. Tugyi, et al., Partial D-amino acid substitution: Improved enzymatic stability and preserved Ab recognition of a MUC2 epitope peptide, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(2), 413–418 CrossRef CAS PubMed.
- J. S. Khara, et al., Unnatural amino acid analogues of membrane-active helical peptides with anti-mycobacterial activity and improved stability, J. Antimicrob. Chemother., 2016, 71(8), 2181–2191 CrossRef.
- C.-L. Towse, et al., Nature versus design: the conformational propensities of D-amino acids and the importance of side chain chirality, Protein Eng., Des. Sel., 2014, 27(11), 447–455 CrossRef CAS.
- A. Rozek, et al., Structure-based design of an indolicidin peptide analogue with increased protease stability, Biochemistry, 2003, 42(48), 14130–14138 CrossRef CAS.
- G. H. Bird, et al., Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(32), 14093–14098 CrossRef CAS PubMed.
- A. A. Strömstedt, et al., Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37, Antimicrob. Agents Chemother., 2009, 53(2), 593–602 CrossRef.
- R. Swanson, Long live peptides—evolution of peptide half-life extension technologies and emerging hybrid approaches, Drug Discovery World, 2014, 15, 57–61 Search PubMed.
- A. Mahler, et al., Rigid, self-assembled hydrogel composed of a modified aromatic dipeptide, Adv. Mater., 2006, 18(11), 1365–1370 CrossRef CAS.
- Z. Yang, et al., Enzymatic formation of supramolecular hydrogels, Adv. Mater., 2004, 16(16), 1440–1444 CrossRef CAS.
- S. Sutton, et al., Controlled release from modified amino acid hydrogels governed by molecular size or network dynamics, Langmuir, 2009, 25(17), 10285–10291 CrossRef CAS.
- S. Roy and A. Banerjee, Amino acid based smart hydrogel: formation, characterization and fluorescence properties of silver nanoclusters within the hydrogel matrix, Soft Matter, 2011, 7(11), 5300–5308 RSC.
- Z. Yang, et al., Phenyl groups in supramolecular nanofibers confer hydrogels with high elasticity and rapid recovery, J. Mater. Chem., 2010, 20(11), 2128–2132 RSC.
- G. Cheng, et al., Hydrogelation of self-assembling RGD-based peptides, Soft Matter, 2011, 7(4), 1326–1333 RSC.
- S. Debnath, et al., Hydrogelation through self-assembly of Fmoc-peptide functionalized cationic amphiphiles: potent antibacterial agent, J. Phys. Chem. B, 2010, 114(13), 4407–4415 CrossRef CAS.
- V. Castelletto, et al., Slow-release RGD-peptide hydrogel monoliths, Langmuir, 2012, 28(34), 12575–12580 CrossRef CAS.
- C. Tang, R. V. Ulijn and A. Saiani, Effect of glycine substitution on Fmoc–diphenylalanine self-assembly and gelation properties, Langmuir, 2011, 27(23), 14438–14449 CrossRef CAS.
- C. Tang, et al., Fmoc-diphenylalanine self-assembly mechanism induces apparent p K a shifts, Langmuir, 2009, 25(16), 9447–9453 CrossRef CAS.
- L. Wang, et al., Controlling the self-assembly of biomolecules into functional nanomaterials through internal interactions and external stimulations: a review, Nanomaterials, 2019, 9(2), 285 CrossRef CAS.
- J. Wang, et al., Peptide self-assembly: thermodynamics and kinetics, Chem. Soc. Rev., 2016, 45(20), 5589–5604 RSC.
- A. S. Mahadevi and G. N. Sastry, Cooperativity in noncovalent interactions, Chem. Rev., 2016, 116(5), 2775–2825 CrossRef CAS.
- T. Fan, X. Yu, B. Shen and L. Sun, Peptide Self-Assembled Nanostructures for Drug Delivery Applications, J. Nanomater., 2017, 2017, 4562474 Search PubMed.
- B. Rybtchinski, Adaptive supramolecular nanomaterials based on strong noncovalent interactions, ACS Nano, 2011, 5(9), 6791–6818 CrossRef CAS.
- R. Vegners, I. Shestakova, I. Kalvinsh, R. M. Ezzell and P. A. Janmey, Use of a gel-forming dipeptide derivative as a carrier for antigen presentation, J. Pept. Sci., 1995, 1(6), 371–378 CrossRef CAS.
- J. J. Panda, et al., Stimuli responsive self-assembled hydrogel of a low molecular weight free dipeptide with potential for tunable drug delivery, Biomacromolecules, 2008, 9(8), 2244–2250 CrossRef CAS.
- C. K. Thota, N. Yadav and V. S. Chauhan, A novel highly stable and injectable hydrogel based on a conformationally restricted ultrashort peptide, Sci. Rep., 2016, 6, 31167 CrossRef CAS PubMed.
- A. M. Smith, R. J. Williams, C. Tang, P. Coppo, R. F. Collins, M. L. Turner, A. Saiani and R. V. Ulijn, Fmoc-diphenylalanine self assembles to a hydrogel via a novel architecture based on π–π interlocked β-sheets, Adv. Mater., 2008, 20(1), 37–41 CrossRef CAS.
- F. Qiu, et al., Amphiphilic peptides as novel nanomaterials: design, self-assembly and application, Int. J. Nanomed., 2018, 13, 5003 CrossRef CAS.
- K. Kornmueller, et al., Peptides at the interface: self-assembly of amphiphilic designer peptides and their membrane interaction propensity, Biomacromolecules, 2016, 17(11), 3591–3601 CrossRef CAS.
- D. Lombardo, M. A. Kiselev, S. Magazù and P. Calandra, Amphiphiles Self-Assembly: Basic Concepts and Future Perspectives of Supramolecular Approaches, Adv. Condens. Matter Phys., 2015, 2015, 151683 Search PubMed.
- B. Findlay, G. G. Zhanel and F. Schweizer, Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold, Antimicrob. Agents Chemother., 2010, 54(10), 4049–4058 CrossRef CAS.
- P. Berndt, G. B. Fields and M. Tirrell, Synthetic lipidation of peptides and amino acids: monolayer structure and properties, J. Am. Chem. Soc., 1995, 117(37), 9515–9522 CrossRef CAS.
- A. P. Nowak, et al., Rapidly recovering hydrogel scaffolds from self-assembling diblock copolypeptide amphiphiles, Nature, 2002, 417(6887), 424 CrossRef CAS.
- M. Deng, et al., Self-assembly of Peptide− Amphiphile C12− Aβ (11− 17) into Nanofibrils, J. Phys. Chem. B, 2009, 113(25), 8539–8544 CrossRef CAS.
- C. He, et al., Self-assembly of Aβ-based peptide amphiphiles with double hydrophobic chains, Langmuir, 2012, 28(7), 3391–3396 CrossRef CAS.
- J. D. Hartgerink, E. Beniash and S. I. Stupp, Peptide-amphiphile nanofibers: a versatile scaffold for the preparation of self-assembling materials, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(8), 5133–5138 CrossRef CAS PubMed.
- P. Chen, Self-assembly of ionic-complementary peptides: a physicochemical viewpoint, Colloids Surf., A, 2005, 261(1–3), 3–24 CrossRef CAS.
- M. Kabiri, et al., Toward a mechanistic understanding of ionic self-complementary peptide self-assembly: role of water molecules and ions, Biomacromolecules, 2013, 14(11), 3943–3950 CrossRef CAS.
- L. Sun, C. Zheng and T. J. Webster, Self-assembled peptide nanomaterials for biomedical applications: promises and pitfalls, Int. J. Nanomed., 2017, 12, 73 CrossRef CAS.
- Y. Hong, et al., Critical self-assembly concentration of an ionic-complementary peptide EAK16-I, J. Adhes., 2004, 80(10–11), 913–931 CrossRef CAS.
- B. He, X. Yuan and D. Jiang, Molecular self-assembly guides the fabrication of peptide nanofiber scaffolds for nerve repair, RSC Adv., 2014, 4(45), 23610–23621 RSC.
- M. Rivas, et al., Peptide Self-Assembly into Hydrogels for Biomedical Applications Related to Hydroxyapatite, Gels, 2019, 5(1), 14 CrossRef CAS.
- S. Y. Fung, H. Yang and P. Chen, Sequence effect of self-assembling peptides on the complexation and in vitro delivery of the hydrophobic anticancer drug ellipticine, PLoS One, 2008, 3(4), e1956 CrossRef PubMed.
- H. Yang, et al., Modification of hydrophilic and hydrophobic surfaces using an ionic-complementary peptide, PLoS One, 2007, 2(12), e1325 CrossRef.
- S. Zhang, et al., Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane, Proc. Natl. Acad. Sci. U. S. A., 1993, 90(8), 3334–3338 CrossRef CAS.
- S. Jun, et al., Self-assembly of the ionic peptide EAK16: the effect of charge distributions on self-assembly, Biophys. J., 2004, 87(2), 1249–1259 CrossRef CAS.
- S. Zhang, et al., Self-complementary oligopeptide matrices support mammalian cell attachment, Biomaterials, 1995, 16(18), 1385–1393 CrossRef CAS.
- T. C. Holmes, et al., Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(12), 6728–6733 CrossRef CAS.
- A. Saini and V. S. Chauhan, Self-assembling properties of peptides derived from TDP-43 C-terminal fragment, Langmuir, 2014, 30(13), 3845–3856 CrossRef CAS.
- J. Naskar, G. Palui and A. Banerjee, Tetrapeptide-based hydrogels: for encapsulation and slow release of an anticancer drug at physiological pH, J. Phys. Chem. B, 2009, 113(35), 11787–11792 CrossRef CAS.
- C. A. Hauser, et al., Natural tri-to hexapeptides self-assemble in water to amyloid β-type fiber aggregates by unexpected α-helical intermediate structures, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(4), 1361–1366 CrossRef CAS.
- J. Smadbeck, et al., De novo design and experimental characterization of ultrashort self-associating peptides, PLoS Comput. Biol., 2014, 10(7), e1003718 CrossRef PubMed.
- R. Orbach, et al., Self-assembled Fmoc-peptides as a platform for the formation of nanostructures and hydrogels, Biomacromolecules, 2009, 10(9), 2646–2651 CrossRef CAS.
- V. Jayawarna, et al., Introducing chemical functionality in Fmoc-peptide gels for cell culture, Acta Biomater., 2009, 5(3), 934–943 CrossRef CAS.
- T. Liebmann, et al., Self-assembling Fmoc dipeptide hydrogel for in situ 3D cell culturing, BMC Biotechnol., 2007, 7(1), 88 CrossRef.
- S. Maude, E. Ingham and A. Aggeli, Biomimetic self-assembling peptides as scaffolds for soft tissue engineering, Nanomedicine, 2013, 8(5), 823–847 CrossRef CAS.
- Z. Yang, et al., Self-assembly of small molecules affords multifunctional supramolecular hydrogels for topically treating simulated uranium wounds, Chem. Commun., 2005,(35), 4414–4416 RSC.
- Y. Zhang, et al., An injectable dipeptide–fullerene supramolecular hydrogel for photodynamic antibacterial therapy, J. Mater. Chem. B, 2018, 6(44), 7335–7342 RSC.
- M. Yang, et al., A versatile cyclic dipeptide hydrogelator: Self-assembly and rheology in various physiological conditions, Colloids Surf., A, 2019, 572, 259–265 CrossRef CAS.
- I. Tarasenko, et al., Amphiphilic polypeptides with prolonged enzymatic stability for the preparation of self-assembled nanobiomaterials, RSC Adv., 2018, 8(60), 34603–34613 RSC.
- S. Parween, et al., Self-assembled dipeptide nanotubes constituted by flexible β-phenylalanine and conformationally constrained α, β-dehydrophenylalanine residues as drug delivery system, J. Mater. Chem. B, 2014, 2(20), 3096–3106 RSC.
- R. Jain, et al., First observation of left-handed helical conformation in a dehydro peptide containing two L-Val residues. Crystal and solution structure of Boc-L-Val-ΔPhe-ΔPhe-ΔPhe-L-Val-OMe, J. Am. Chem. Soc., 1997, 119(14), 3205–3211 CrossRef CAS.
- U. Ramagopal, et al., Crystal structure of Boc-LAla-ΔPhe-ΔPhe-ΔPhe-ΔPhe-NHMe: a left-handed helical peptide, J. Pept. Res., 1998, 52(3), 208–215 CrossRef CAS PubMed.
- U. A. Ramagopal, et al., De novo design and characterization of an apolar helical hairpin peptide at atomic resolution: Compaction mediated by weak interactions, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(3), 870–874 CrossRef CAS.
- P. Mathur, S. Ramakumar and V. Chauhan, Peptide design using α, β-dehydro amino acids: From β-turns to helical hairpins, Pept. Sci., 2004, 76(2), 150–161 CrossRef CAS.
- K. K. Bhandary and V. Chauhan, Peptide design 310-helical conformation of a linear pentapeptide containing two dehydrophenylalanines, Boc-Gly-ΔZPhe-Leu-ΔZPhe-Ala-NHCH3, Biopolymers, 1993, 33(2), 209–217 CrossRef CAS.
- R. Jain and V. S. Chauhan, Conformational characteristics of peptides containing α, β-dehydroamino acid residues, Pept. Sci., 1996, 40(1), 105–119 CrossRef CAS.
- K. Rajashankar, et al., Observation of Water-Mediated Helix-Terminating Conformation in a Dehydrophenylalanine Peptide: Crystal and Solution Structure of the Octapeptide Ac-. DELTA. Phe-Val-. DELTA. Phe-Phe-Ala-Val-. DELTA. Phe-Gly-OMe, J. Am. Chem. Soc., 1995, 117(47), 11773–11779 CrossRef CAS.
- K. Rajashankar, et al., Role of two consecutive α, β-dehydrophenylalanines in peptide structure: Crystal and molecular structure of Boc-Leu-ΔPhe-ΔPhe-Ala-Phe-NHMe, Biopolymers, 1997, 42(3), 373–382 CrossRef CAS.
- A. Mishra, et al., Nanovesicles based on self-assembly of conformationally constrained aromatic residue containing amphiphilic dipeptides, Langmuir, 2008, 24(9), 4571–4576 CrossRef CAS.
- M. Gupta and V. S. Chauhan, De novo design of α, β-didehydrophenylalanine containing peptides: From models to applications, Biopolymers, 2011, 95(3), 161–173 CrossRef CAS.
- J. P. Schneider, et al., Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide, J. Am. Chem. Soc., 2002, 124(50), 15030–15037 CrossRef CAS.
- A. K. Das, et al., Low molecular weight organogelators from self-assembling synthetic tripeptides with coded amino acids: morphological, structural, thermodynamic and spectroscopic investigations, Supramol. Chem., 2006, 18(8), 645–655 CrossRef CAS.
- J. J. Panda, et al., 3D cell growth and proliferation on a RGD functionalized nanofibrillar hydrogel based on a conformationally restricted residue containing dipeptide, ACS Appl. Mater. Interfaces, 2010, 2(10), 2839–2848 CrossRef CAS.
- J. Liu, et al., Controlled release of paclitaxel from a self-assembling peptide hydrogel formed in situ and antitumor study in vitro, Int. J. Nanomed., 2011, 6, 2143 CrossRef CAS.
- L. Gentilucci, R. De Marco and L. Cerisoli, Chemical modifications designed to improve peptide stability: incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization, Curr. Pharm. Des., 2010, 16(28), 3185–3203 CrossRef CAS.
- J. Li, et al., Dephosphorylation of D-peptide derivatives to form biofunctional, supramolecular nanofibers/hydrogels and their potential applications for intracellular imaging and intratumoral chemotherapy, J. Am. Chem. Soc., 2013, 135(26), 9907–9914 CrossRef CAS.
- Y. Kuang, et al., Pericellular hydrogel/nanonets inhibit cancer cells, Angew. Chem., Int. Ed., 2014, 53(31), 8104–8107 CrossRef CAS.
- Z. Feng and B. Xu, Inspiration from the mirror: D-amino acid containing peptides in biomedical approaches, Biomol. Concepts, 2016, 7(3), 179–187 CAS.
- J. Liu, J. Liu, L. Chu, Y. Zhang, H. Xu, D. Kong, Z. Yang, C. Yang and D. Ding, Self-assembling peptide of D-amino acids boosts selectivity and antitumor efficacy of 10-hydroxycamptothecin, ACS Appl. Mater. Interfaces, 2014, 6(8), 5558–5565 CrossRef CAS.
- M. Sela and E. Zisman, Different roles of D-amino acids in immune phenomena, FASEB J., 1997, 11(6), 449–456 CrossRef CAS.
- T. Degenkolb and H. Brückner, Peptaibiomics: towards a myriad of bioactive peptides containing Cα-dialkylamino acids?, Chem. Biodiversity, 2008, 5(9), 1817–1843 CrossRef CAS.
- C. Toniolo, et al., Structures of peptides from α-amino acids methylated at the α-carbon, Biopolymers, 1993, 33(7), 1061–1072 CrossRef CAS.
- I. L. Karle and P. Balaram, Structural characteristics of. alpha.-helical peptide molecules containing Aib residues, Biochemistry, 1990, 29(29), 6747–6756 CrossRef CAS.
- P. Maity and B. Koenig, Enantio-and diastereoselective syntheses of cyclic Cα-tetrasubstituted α-amino acids and their use to induce stable conformations in short peptides, Pept. Sci., 2008, 90(1), 8–27 CrossRef CAS PubMed.
- P. Maity, M. Zabel and B. König, Tetrahydrofuran Cα-tetrasubstituted amino acids: two consecutive β-turns in a crystalline linear tripeptide, J. Org. Chem., 2007, 72(21), 8046–8053 CrossRef CAS.
- D. Datta, V. Kumar, S. Kumar, R. Nagaraj and N. Chaudhary, Limpid hydrogels from β-turn motif-connected tandem repeats of Aβ16–22, Soft Matter, 2019, 15, 4827–4835 RSC.
- P. C. Dewan, et al., Antimicrobial action of prototypic amphipathic cationic decapeptides and their branched dimers, Biochemistry, 2009, 48(24), 5642–5657 CrossRef CAS PubMed.
- S. R. Caliari and J. A. Burdick, A practical guide to hydrogels for cell culture, Nat. Methods, 2016, 13(5), 405 CrossRef CAS.
- L. Chronopoulou, S. Sennato, F. Bordi, D. Giannella, A. Di Nitto, A. Barbetta, M. Dentini, A. R. Togna, G. I. Togna, S. Moschini and C. Palocci, Designing unconventional Fmoc-peptide-based biomaterials: Structure and related properties, Soft Matter, 2014, 10(12), 1944–1952 RSC.
- K. Liang, K. H. Bae and M. Kurisawa, Recent advances in the design of injectable hydrogels for stem cell-based therapy, J. Mater. Chem. B, 2019, 7, 3775–3791 RSC.
- Y. Loo, et al., Ultrashort peptide nanofibrous hydrogels for the acceleration of healing of burn wounds, Biomaterials, 2014, 35(17), 4805–4814 CrossRef CAS.
- W. Y. Seow, G. Salgado, E. B. Lane and C. A. Hauser, Transparent crosslinked ultrashort peptide hydrogel dressing with high shape-fidelity accelerates healing of full-thickness excision wounds, Sci. Rep., 2016, 6, 32670 CrossRef CAS.
- X. Kang, et al., DRAMP 2.0, an updated data repository of antimicrobial peptides, Sci. Data, 2019, 6(1), 1–10 CAS.
- J.-K. Lee, T. Luchian and Y. Park, New antimicrobial peptide kills drug-resistant pathogens without detectable resistance, Oncotarget, 2018, 9(21), 15616 CrossRef.
- A. Song, A. A. Rane and K. L. Christman, Antibacterial and cell-adhesive polypeptide and poly (ethylene glycol) hydrogel as a potential scaffold for wound healing, Acta Biomater., 2012, 8(1), 41–50 CrossRef CAS PubMed.
- Y. Wan, et al., pH-responsive peptide supramolecular hydrogels with antibacterial activity, Langmuir, 2017, 33(13), 3234–3240 CrossRef CAS.
- M. Lian, et al., Self-assembled peptide hydrogel as a smart biointerface for enzyme-based electrochemical biosensing and cell monitoring, ACS Appl. Mater. Interfaces, 2016, 8(38), 25036–25042 CrossRef CAS PubMed.
- J. H. Kim, et al., Self-assembled, photoluminescent peptide hydrogel as a versatile platform for enzyme-based optical biosensors, Biosens. Bioelectron., 2011, 26(5), 1860–1865 CrossRef CAS.
- S. F. Souza, E. R. Silva and W. A. Alves, Nanostructured antigen-responsive hydrogels based on peptides for leishmaniasis detection, J. Braz. Chem. Soc., 2017, 28(9), 1619–1629 CAS.
- P. J. King, A. Saiani, E. V. Bichenkova and A. F. Miller, A de novo self-assembling peptide hydrogel biosensor with covalently immobilised DNA-recognising motifs, Chem. Commun., 2016, 52(40), 6697–6700 RSC.
- S. Koutsopoulos, Self-assembling peptides in biomedicine and bioengineering: Tissue engineering, regenerative medicine, drug delivery, and biotechnology, in Peptide Applications in Biomedicine, Biotechnology and Bioengineering, Elsevier, 2018, pp. 387–408 Search PubMed.
- M. Silvia, Recent Patents on Peptide Self-Assembled Hydrogels as Nanostructured Biomaterials, Recent Pat. Nanomed., 2014, 4(2), 77–81 Search PubMed.
| This journal is © The Royal Society of Chemistry 2020 |