Poly(glycoamidoamine) brush nanomaterials for systemic siRNA delivery in vivo†
X.
Luo‡
a,
W.
Wang‡
bc,
J. R.
Dorkin
d,
O.
Veiseh
bce,
P. H.
Chang
bc,
I.
Abutbul-Ionita
f,
D.
Danino
f,
R.
Langer
bceg,
D. G.
Anderson
*bceg and
Y.
Dong
*ahij
aDivision of Pharmaceutics and Pharmaceutical Chemistry, College of Pharmacy, The Ohio State University, Columbus, OH 43210, USA. E-mail: dong.525@osu.edu
bDavid H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. E-mail: dgander@mit.edu
cDepartment of Anesthesiology, Children's Hospital Boston, Harvard Medical School, Boston, MA 02115, USA
dDepartment of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
eDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
fDepartment of Biotechnology and Food Engineering, Technion Institute of Technology and the Russell Berrie Nanotechnology Institute, Haifa 32000, Israel
gInstitute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
hDepartment of Biomedical Engineering, The Ohio State University, Columbus, OH 43210, USA
iThe Center for Clinical and Translational Science, The Ohio State University, Columbus, OH 43210, USA
jThe Comprehensive Cancer Center, The Ohio State University, Columbus, OH 43210, USA
First published on 6th December 2016
Abstract
Delivery is the key challenge for siRNA based therapeutics. Here, we report the development of new poly(glycoamidoamine) brush nanomaterials for efficient siRNA delivery. GluN4C10 polymer brush nanoparticles, a lead material, demonstrated significantly improved delivery efficiency for siRNA against factor VII (FVII) in mice compared to poly(glycoamidoamine) brush nanomaterials reported previously.
Small interfering RNA (siRNA) has been extensively applied for biological and therapeutic purposes in the past two decades.1–6 Clinical results demonstrated the potential of siRNA for treating a wide variety of diseases.5,7,8 Although tremendous efforts have been made to improve the delivery of siRNA, systemic and effective delivery of siRNA remains a challenging issue for its broad therapeutic applications.6,9–15 Here, we report the design, synthesis, and characterization of new poly(glycoamidoamine) brush nanomaterials for efficient siRNA delivery both in vitro and in vivo.
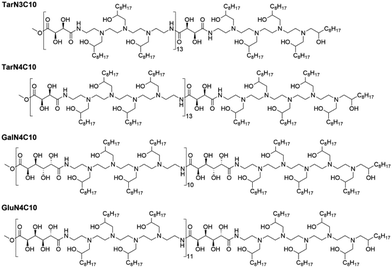
Previously, we reported a class of poly(glycoamidoamine) brush materials and evaluated their efficiency for siRNA and mRNA delivery.16 Analysis of structure–activity relationships indicated that an increased number of amines in the monomer and short alkyl tails facilitated RNA delivery.16 Based upon these design criteria, we synthesized three new materials (Fig. 1).16 Three modified poly(glycoamidoamine) polymers consisting of tartarate (Tar), galactarate (Gal), or glucarate (Glu) sugars were first obtained using the method reported by Reineke.17–22 1,2-Epoxydecane then underwent ring-opening reactions with these polymers to afford the designed poly(glycoamidoamine) brush materials. The structures of the polymer brush materials were confirmed by 1H NMR.
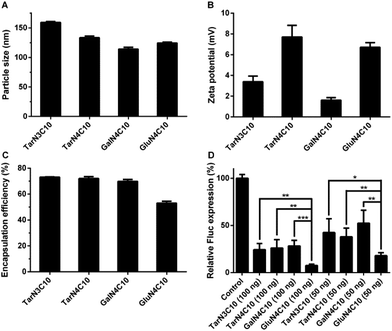
Polymer brush materials were subsequently formulated with DSPC, cholesterol (Chol), DMG-PEG2000, and siRNA against Fluc into polymer–siRNA nanoparticles. Then, we characterized these nanoparticles:16,23 particle size ranged from 114 nm to 159 nm; surface charge was neutral or slightly positive; and siRNA encapsulation efficiency was between 53% and 73% (Fig. 2a–c). In order to evaluate the siRNA delivery efficiency of these formulations in vitro, dual-HeLa cells expressing both firefly and Renilla luciferase were treated with polymer brush nanoparticles.24,25 As shown in Fig. 2d, the formulation GluN4C10 silenced Fluc expression 93% at a siRNA dose of 100 ng and 82% at a siRNA dose of 50 ng, which was significantly more effective compared to other formulations including TarN3C10, a lead material reported previously.16 Consequently, GluN4C10 was selected for further studies.
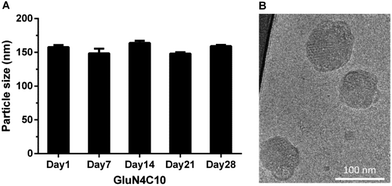
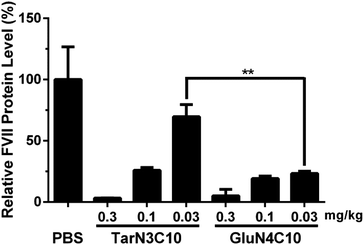
We then characterized GluN4C10 nanoparticles for their stability and morphology. The particle size was measured weekly by dynamic light scattering (DLS). The results indicated that this formulation was stable at 4 °C for at least 4 weeks (Fig. 3a). TarN3C10, TarN4C10, and GalN4C10 nanoparticles showed a similar stability at the same time course (Fig. S1†). We also observed apparent cellular uptake of the GluN4C10, TarN4C10 and GalN4C10 nanoparticles using Alexa 647-labelled siRNA (Fig. S2†). The Cryo-TEM image revealed the morphology of the GluN4C10 nanoparticles with particle size consistent with the measurements from DLS (Fig. 3b). Given the promising results of the GluN4C10 nanoparticles in vitro, we evaluated the delivery efficiency of GluN4C10 for siRNA against FVII in vivo. We then injected the GluN4C10-FVII siRNA nanoparticles into mice through their tail vain at three different doses: 0.3 mg kg−1, 0.1 mg kg−1, and 0.03 mg kg−1. TarN3C10 nanoparticles served as a positive control. As shown in Fig. 4, both TarN3C10 and GluN4C10 polymer brush nanoparticles displayed dose-dependent silencing of FVII. At a siRNA dose of 0.3 mg kg−1, GluN4C10 showed effective and comparable FVII silencing activity (up to 95%) compared to TarN3C10. At a lower siRNA dose of 0.03 mg kg−1, GluN4C10 displayed a significantly higher FVII silencing than TarN3C10 (77% versus 30% at 0.03 mg kg−1). Reflecting the results above, the GluN4C10 polymer brush nanoparticles were capable of efficiently delivering siRNA molecules in vivo.
Conclusions
In summary, we designed and synthesized three new polymer brush materials based on the design criteria established previously. The formulation GluN4C10 nanoparticles demonstrated efficient siRNA delivery both in vitro and in vivo. We speculate that sugar units may play a more critical role in this series, and thereby improve delivery efficiency. Most importantly, GluN4C10 were capable of silencing 77% of FVII expression at a dose of 0.03 mg kg−1, significantly more potent than TarN3C10. Therefore, GluN4C10 polymer brush nanomaterials are promising siRNA delivery vehicles and merit further development for therapeutic applications.All procedures used in animal studies conducted at MIT were in compliance with Massachusetts laws or guidelines, were approved by the Institutional Animal Care and Use Committee (IACUC) and were also consistent with local, state, and federal regulations as applicable.
Acknowledgements
This work was supported by the National Cancer Institute Center of Cancer Nanotechnology Excellence at MIT-Harvard (U54-CA151884), the National Heart, Lung, and Blood Institute, the National Institutes of Health (NIH), as a Program of Excellence in Nanotechnology (PEN) Award, contract #HHSN268201000045C, as well as by the NIH Grants R01-EB000244–27, 5-R01-CA132091–04, and R01-DE016516–03. O. V. was supported by the Department of Defense Congressionally Directed Medical Research Program (DOD/CDMRP) postdoctoral fellowships (grant W81XWH-13-1-0215, respectively). Y. D. acknowledges supports from the Bayer Hemophilia Awards Program for the Early Career Investigator Award, the National PKU Alliance for Research Awards, the AAPS Foundation for New Investigator Grant, the National Institute of General Medical Sciences for the Maximizing Investigators’ Research Award 1R35GM119679 as well as the start-up fund from the College of Pharmacy at The Ohio State University.References
- M. E. Davis, Mol. Pharm., 2009, 6, 659–668 CrossRef CAS PubMed.
- D. H. Kim, M. A. Behlke, S. D. Rose, M. S. Chang, S. Choi and J. J. Rossi, Nat. Biotechnol., 2005, 23, 222–226 CrossRef CAS PubMed.
- M. E. Davis, J. E. Zuckerman, C. H. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
- R. Juliano, J. Bauman, H. Kang and X. Ming, Mol. Pharm., 2009, 6, 686–695 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- J. C. Burnett, J. J. Rossi and K. Tiemann, Biotechnol. J., 2011, 6, 1130–1146 CrossRef CAS PubMed.
- S. A. Barros and J. A. Gollob, Adv. Drug Delivery Rev., 2012, 64, 1730–1737 CrossRef CAS PubMed.
- S. J. Tan, P. Kiatwuthinon, Y. H. Roh, J. S. Kahn and D. Luo, Small, 2011, 7, 841–856 CrossRef CAS PubMed.
- L. Yin, N. Zheng and J. Cheng, Methods Mol. Biol., 2016, 1364, 37–47 Search PubMed.
- Y. Wang, L. Miao, A. Satterlee and L. Huang, Adv. Drug Delivery Rev., 2015, 87, 68–80 CrossRef CAS PubMed.
- M. A. Islam, E. K. G. Reesor, Y. Xu, H. R. Zope, B. R. Zetter and J. Shi, Biomater. Sci., 2015, 3, 1519–1533 RSC.
- W. Liao, W. Li, T. Zhang, M. Kirberger, J. Liu, P. Wang, W. Chen and Y. Wang, Biomater. Sci., 2016, 4, 1051–1061 RSC.
- S. Krishnamurthy, R. Vaiyapuri, L. Zhang and J. M. Chan, Biomater. Sci., 2015, 3, 923–936 RSC.
- E. Keles, Y. Song, D. Du, W.-J. Dong and Y. Lin, Biomater. Sci., 2016, 4, 1291–1309 RSC.
- Y. Dong, J. R. Dorkin, W. Wang, P. H. Chang, M. J. Webber, B. C. Tang, J. Yang, I. Abutbul-Ionita, D. Danino, F. DeRosa, M. Heartlein, R. Langer and D. G. Anderson, Nano Lett., 2016, 16, 842–848 CrossRef CAS PubMed.
- M. Tranter, Y. Liu, S. He, J. Gulick, X. Ren, J. Robbins, W. K. Jones and T. M. Reineke, Mol. Ther., 2012, 20, 601–608 CrossRef CAS PubMed.
- N. P. Ingle, B. Malone and T. M. Reineke, Trends Biotechnol., 2011, 29, 443–453 CrossRef CAS PubMed.
- Y. Liu and T. M. Reineke, Biomacromolecules, 2010, 11, 316–325 CrossRef CAS PubMed.
- P. M. McLendon, K. M. Fichter and T. M. Reineke, Mol. Pharm., 2010, 7, 738–750 CrossRef CAS PubMed.
- Y. Liu, L. Wenning, M. Lynch and T. M. Reineke, ACS Symp. Ser., 2006, 923, 217–227 CrossRef CAS.
- Y. Liu, L. Wenning, M. Lynch and T. M. Reineke, J. Am. Chem. Soc., 2004, 126, 7422–7423 CrossRef CAS PubMed.
- B. Li, X. Luo, B. Deng, J. Wang, D. W. McComb, Y. Shi, K. M. Gaensler, X. Tan, A. L. Dunn, B. A. Kerlin and Y. Dong, Nano Lett., 2015, 15, 8099–8107 CrossRef CAS PubMed.
- T. Love Kevin, P. Mahon Kerry, G. Levins Christopher, A. Whitehead Kathryn, W. Querbes, J. R. Dorkin, J. Qin, W. Cantley, L. Qin Liu, T. Racie, M. Frank-Kamenetsky, N. Yip Ka, R. Alvarez, W. Y. Sah Dinah, A. de Fougerolles, K. Fitzgerald, V. Koteliansky, A. Akinc, R. Langer and G. Anderson Daniel, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1864–1869 CrossRef CAS PubMed.
- Y. Dong, K. T. Love, J. R. Dorkin, S. Sirirungruang, Y. Zhang, D. Chen, R. L. Bogorad, H. Yin, Y. Chen, A. J. Vegas, C. A. Alabi, G. Sahay, K. T. Olejnik, W. Wang, A. Schroeder, A. K. Lytton-Jean, D. J. Siegwart, A. Akinc, C. Barnes, S. A. Barros, M. Carioto, K. Fitzgerald, J. Hettinger, V. Kumar, T. I. Novobrantseva, J. Qin, W. Querbes, V. Koteliansky, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3955–3960 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and 1H NMR structure determination. See DOI: 10.1039/c6bm00683c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |