Biophysical insight into the interaction of the bioflavonoid kaempferol with triple and double helical RNA and the dual fluorescence behaviour of kaempferol†
Abstract
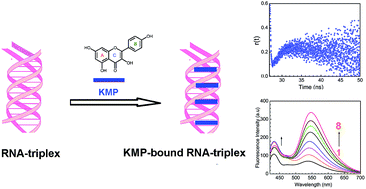
The interaction of the naturally occurring flavonoid, kaempferol (KMP) with single, double and triple helical forms of RNA has been investigated by different spectroscopic and viscometric techniques. It was found that KMP binds with triple helical [poly(U).poly(A)*poly(U), hereafter U.A*U, the dot represents the Watson–Crick and asterisk represents Hoogsteen base pairing respectively] and double helical [poly(A).poly(U), hereafter A.U] forms of RNA, whereas no interaction was observed with single stranded polyuridylic acid [poly-U] under identical experimental conditions. The binding of KMP was found to be stronger with U.A*U (20 × 104 M−1) compared to that of the parent duplex A.U (9.5 × 104 M−1). From the Stern–Volmer quenching constant, viscosity measurement and perturbation of CD spectra of RNA revealed that KMP binds to the U.A*U structure by intercalation while partial intercalation has been proposed for the binding to the duplex RNA structure. Thermodynamic data obtained from the temperature dependence study showed that the association was favoured by negative enthalpy and positive entropy changes. Experimental observations indicated that KMP binds and stabilizes the RNA-triplex more than its parent duplex counterpart.

 Please wait while we load your content...
Please wait while we load your content...