 Open Access Article
Open Access ArticleEmerging nanosensor platforms and machine learning strategies toward rapid, point-of-need small-molecule metabolite detection and monitoring
Shi Xuan
Leong
a,
Yong Xiang
Leong
a,
Charlynn Sher Lin
Koh
a,
Emily Xi
Tan
 a,
Lam Bang Thanh
Nguyen
a,
Jaslyn Ru Ting
Chen
a,
Carice
Chong
a,
Desmond Wei Cheng
Pang
a,
Howard Yi Fan
Sim
a,
Xiaochen
Liang
a,
Nguan Soon
Tan
a,
Lam Bang Thanh
Nguyen
a,
Jaslyn Ru Ting
Chen
a,
Carice
Chong
a,
Desmond Wei Cheng
Pang
a,
Howard Yi Fan
Sim
a,
Xiaochen
Liang
a,
Nguan Soon
Tan
 bc and
Xing Yi
Ling
bc and
Xing Yi
Ling
 *ab
*ab
aDivision of Chemistry and Biological Chemistry, School of Chemistry, Chemical Engineering and Biotechnology, Nanyang Technological University, Singapore. E-mail: xyling@ntu.edu.sg
bLee Kong Chian School of Medicine, Nanyang Technological University, Singapore
cSchool of Biological Sciences, Nanyang Technological University, Singapore
First published on 13th September 2022
Abstract
Speedy, point-of-need detection and monitoring of small-molecule metabolites are vital across diverse applications ranging from biomedicine to agri-food and environmental surveillance. Nanomaterial-based sensor (nanosensor) platforms are rapidly emerging as excellent candidates for versatile and ultrasensitive detection owing to their highly configurable optical, electrical and electrochemical properties, fast readout, as well as portability and ease of use. To translate nanosensor technologies for real-world applications, key challenges to overcome include ultralow analyte concentration down to ppb or nM levels, complex sample matrices with numerous interfering species, difficulty in differentiating isomers and structural analogues, as well as complex, multidimensional datasets of high sample variability. In this Perspective, we focus on contemporary and emerging strategies to address the aforementioned challenges and enhance nanosensor detection performance in terms of sensitivity, selectivity and multiplexing capability. We outline 3 main concepts: (1) customization of designer nanosensor platform configurations via chemical- and physical-based modification strategies, (2) development of hybrid techniques including multimodal and hyphenated techniques, and (3) synergistic use of machine learning such as clustering, classification and regression algorithms for data exploration and predictions. These concepts can be further integrated as multifaceted strategies to further boost nanosensor performances. Finally, we present a critical outlook that explores future opportunities toward the design of next-generation nanosensor platforms for rapid, point-of-need detection of various small-molecule metabolites.
1. Introduction
Small-molecule metabolites are promising and reliable biomarkers for diverse clinical uses including early disease detection, drug identification, toxicological screening of new drugs, and drug pharmacology studies to advance personalized medicine.1–3 They also play central roles in agricultural and food sectors as key indicators for environmental stress detection in plants, food and wine authentication, and identification of microbial-related off-flavors in food.4–11 Defined as low molecular-weight (<1500 Da) intermediates or products of metabolic pathways, small-molecule metabolites include organic compounds such as amino acids, toxins, volatile organic compounds (VOCs) and adenosine triphosphate (ATP), as well as inorganic compounds like carbon dioxide, hydrogen and nitrous oxide. To detect and monitor these metabolites in various biological and agri-food samples, nanomaterial-based sensors (nanosensors) are rapidly emerging as excellent candidates for versatile and ultrasensitive detection owing to their highly configurable properties.12–15 Moreover, due to their fast readout, multiplexing capability, portability and ease of use, nanosensors are ideally suited for rapid, point-of-need applications. Common nanomaterials employed in nanosensors range from semiconductor quantum and carbon dots to metallic and metal oxide nanostructures, upconversion nanoparticles as well as carbon nanotubes.16,17 Their unique nanoscale optical, electrical and electrochemical properties dictate the signal transduction mechanisms and modes of the as-fabricated nanosensors. Notably, most nanomaterials have multimodal properties and can be used for more than one type of nanosensor, depending on the analytical technique used.One emerging optical-based method is surface-enhanced Raman scattering (SERS), which leverages intense electromagnetic (EM) fields from localized surface plasmon resonances (LSPR) in plasmonic nanomaterials to enhance the metabolites' inherent weak Raman scattering modes by up to 1010-fold.18–22 Besides SERS, other popular optical-based nanosensors include colorimetric and fluorescence nanosensors, which measure metabolite-triggered absorbance and fluorescence changes.23–26 For the former, nanomaterials with size- and/or shape-dependent absorbance peak shifts (i.e. color changes) are used to detect metabolites via nanoparticle aggregation/cross-linking or nanomaterial growth/etching. For the latter, intrinsically fluorescent nanomaterials such as quantum dots and upconversion nanoparticles with high quantum yields and photostability are popular candidates as fluorescent probes.27–29 The surface plasmon fields of metallic nanomaterials such as Au and Ag can also enhance the luminescence of adjacent/conjugated fluorophores by interacting with their dipole moments. On the other hand, electrical-based nanosensors such as chemiresistors and electrochemical sensors record changes in electrical resistance as a result of analyte interactions and changes in current upon electrochemical oxidation or reduction of redox-active metabolites, respectively.30,31 Besides the abovementioned emerging techniques, nanosensors can also synergize with conventional, de facto technologies including mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy.32–37 Such synergy provides orthogonal, complementary (bio)molecular information to boost detection sensitivity and specificity. Importantly, the plethora of analytical techniques available introduces greater flexibility in methodology and clinical designs by empowering the user with a versatile toolkit to tailor fit to the specific application in mind.
Nonetheless, we have identified four main prerequisites that are essential to translate the rich biochemical information offered by small-molecule metabolite analysis to real-life screening, therapeutics, and detection, namely high nanosensor sensitivity and selectivity, excellent multiplexing capability, as well as more sophisticated data analysis. First, many target metabolites are typically found in low physiological concentrations at ppb or nM levels, which demand high detection sensitivity.38,39 Second, highly selective nanosensors are desirable to circumvent interfering signals from other interfering species in real-life biological sample matrices such as urine, cerebrospinal fluids and sputum while enabling unambiguous sample analysis. Such increased analyte selectivity will also be useful to differentiate structurally similar metabolites whose relative concentrations reveal underlying metabolic changes, as they often generate highly similar signals that are difficult to distinguish.40–43 Next, it is essential to improve the nanosensors' multiplexing capabilities for concurrent detection of a panel of metabolites and/or differentiation of metabolite mixtures since there is usually no single unique biomarker that is specific to a particular disease or microorganism. Finally, more sophisticated data analysis is necessary to better accommodate the multivariate nature of acquired data and elucidate underlying interrelationships and subtle signal variances for more accurate predictions. It is thus imperative to review contemporary and emerging strategies developed to tackle the aforementioned challenges, targeted at improving nanosensors' capabilities as next-generation toolkits for screening and monitoring purposes.
Herein, we offer a critical outlook of the current research status on nanosensors for small-molecule metabolite detection and monitoring, highlighting the latest advancements from 2016 to 2022 in addressing the aforementioned challenges. We broadly categorize these strategies into three concepts: (1) customization of designer nanosensor platform configurations, (2) development of hybrid techniques and (3) use of machine learning tools (Fig. 1). First, we will examine designer nanosensor platform configurations, including chemical- and physical-based modifications of nanosensors to increase metabolite's surface affinity, thereby achieving higher detection sensitivity and chemoselectivity. We will also discuss array-based configurations that combine various tailored nanosensors for selective, pattern recognition-based detection to discriminate chemically diverse analyte mixtures without identifying individual components. Next, we will highlight multimodal techniques, where the coupling of two or more techniques such as SERS and electrochemistry achieves the ‘best of both worlds’ and generates multidimensional information for more comprehensive metabolite identification. We then evaluate how machine learning algorithms can transform the assimilation and interpretation of complex data by discerning more patterns hidden within the data, thereby achieving higher sensitivity and selectivity. We have also consolidated a non-exhaustive list of recent studies exemplifying these strategies in Tables 1–3 to provide an easily accessible overview on current research in this area. Finally, we conclude with our perspective on potential research directions in this area. It is noteworthy to mention that although some of the analytes mentioned in subsequent sections are not strictly of metabolic origin (e.g. methylbenzenethiol and atmospheric CO2), the strategies employed to detect these small molecules can be readily extrapolated for metabolite detection. All in all, we envisage that these insights can stimulate the development of innovative and hybrid detection methods across the entire analytical discipline to resolve longstanding challenges in small-molecule metabolite sensing, especially in real-life sample matrices.
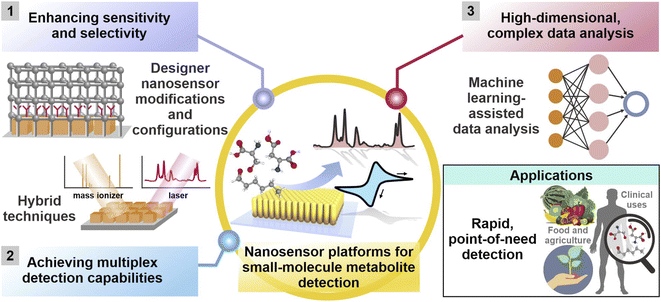 | ||
| Fig. 1 Current and emerging research strategies to enhance sensitivity and selectivity, achieve multiplex detection capabilities and facilitate analysis of complex, high-dimensional datasets for small-molecule metabolite detection using various nanosensor platforms across diverse applications. They are broadly categorized into (1) customization of platform modifications and designer platform configurations, (2) development of hybrid techniques involving two or more analytical techniques, and (3) complementary use of machine learning algorithms. Adapted and reprinted with permission from ref. 49. | ||
| Nanosensor modification strategy | Nanomaterials | Transduction mode(s) | Additional description | Analyte(s) | LOD/Selectivity | Ref. |
|---|---|---|---|---|---|---|
| Chemical-based modifications using molecular receptors | Reduced graphene oxide/Au NPs | Electrochemical | 4-Mercaptophenylboronic acid | Natural glycoside toxins including α-solanine and α-chaconine | Chemoselective over minerals (K+, Ca2+) and vitamins (lutein); <3.4 μM LOD | 31 |
| Ag nanocubes | SERS | 3-Mercaptobenzoic acid | Adenosine; cytosine | — | 44 | |
| Single-walled carbon nanotubes | Fluorescence | Polyethylene glycol | 7 plant polyphenols including tannic and caffeic acid | LOD in low μg mL−1 range | 45 | |
| Yb/Tm@NaGdF4 core–shell upconversion nanoparticles | Fluorescence | Photocleavable linker-modified DNA aptamers | Adenosine triphosphate (ATP) | Chemoselective over GTP, CTP and UTP | 48 | |
| Au nanorods | SERS | 4-Mercaptophenylboronic acid and n-hexanethiol | 11 aromatic enantiomer pairs including mandelic acid and phenylalanine | — | 49 | |
| Au nanospheres and Au nanorods | SERS | Arg-Gly-Asp (RGD) peptides and nuclear localization signal (NLS) peptide | Phenylalanine and its derivatives | — | 50 | |
| Ag NPs | SERS | 2,2′-disulfanediylbis(N-(2-aminophenyl)acetamide) | Nitric oxide | Live bacterium as SERS platform; high selectivity over other reactive oxygen species (ClO−, H2O2, O2−); <100 nM LOD | 51 | |
| Physical-based modifications | Ag nanocubes | Fluorescence | ZIF-8 encapsulation | Cu2+ ions | 4 × 10−4 M LOD | 57 |
| Ag NPs | LSPR sensing | HKUST-1 coating | CO2 gas | 14-Fold increase in signal responses with MOF coating | 58 | |
| Ag nanocubes | SERS | ZIF-8 coating | Gaseous 4-methylbenzenthiol (proof-of-concept) | 2.5-Fold increase in signal responses as MOF thickness increases from 8–146 nm | 59 | |
| Pd NPs@ZnO nanowires | Electrical resistivity | ZIF-8 coating | H2 gas | 10 ppm LOD; selectivity over interfering gases (e.g. benzene, ethanol, acetone) | 61 | |
| PtPd NPs | Electrochemical | ZIF-67 encapsulation | Aqueous phenylketonuria biomarkers e.g. phenylpyruvic and phenylacetic acid | Selectivity over common amino acids with similar structural backbones | 62 | |
| Combined chemical- and physical-based modifications | Ag nanocubes/octahedra | SERS | SPHB platform + 4-mercaptophenylboronic acid | Pregnane and tetrahydrocortisone in urine samples | LOD at ppt levels | 54 |
| Au superparticles (GSP) | SERS | ZIF-8 coating + 4-aminothiophenol | Gaseous aldehyde lung cancer biomarkers | Selectivity over other functional groups e.g. alcohols, esters and amines | 69 | |
| Array-based configurations | Ag nanocubes | SERS | 3 different molecular receptors i.e. 4-mercaptobenzoic acid, 4-aminothiophenol and 4-mercaptopyridine | BVOC profiles of COVID-19 patients | Increased sensitivity from 80% using a single-probe platform to 96.2% with the multiprobe array | 70 |
| Au NPs | Chemiresistance | 8 different hydrocarbon thiol ligands e.g. dodecanethiol, butanethiol | Breath volatile organic compounds (BVOC) profiles of COVID-19 patients | 76–95% accuracy | 73 | |
| Au and Ag NPs | Colorimetric | 8 different chemical species e.g. chitosan, glucose, glutathione | 45 gaseous VOCs from 9 chemical families | <10 ppb LOD | 79 |
| Nanosensor modification strategy | Nanomaterials | Transduction mode(s) | Additional description | Analyte(s) | LOD/Selectivity | Ref. |
|---|---|---|---|---|---|---|
| Multimodal techniques | N,P-codoped carbon dots/Au NPs | Colorimetric + fluorescence | Serves as complementary cross-validation | Aqueous uric acid | Colorimetric quantification: 0.1–10 μM | 82 |
| Fluorescence quantification: 0.5–10 μM | ||||||
| Chitosan-functionalized MoS2–Au@Pt and Au NP-supported MnO2 nanoflowers labelled with aptamers | Electrochemical + colorimetric | Serves as complementary cross-validation | Ochratoxin A | Colorimetric quantification: 0.1–200 ng mL−1 | 83 | |
| Electrochemical LOD: 1 × 10−4 ng mL−1 | ||||||
| SiO2/TiO2 core/shell (T-rex) beads | SERS + SALDI MS | Increased range of detectable metabolites | Structurally analogous (1R, 2S)-(−)-ephedrine and amphetamine; regioisomers theobromine and theophylline | — | 84 | |
| Au NPs | Paper spray ionization MS (PSI-MS) + SERS | Increased differentiating ability, where JWH-018 isomers could be distinguished while MS alone cannot differentiate the samples | 5 illicit drugs including 2C–B, cocaine, fentanyl, hydrocodone, and JWH-018 | Quantification of 0–100 ng; 99.8% accuracy in a blind study | 85 | |
| Hyphenated techniques | Ag NP-coated screen-printed electrodes | Electrochemical-SERS | Strongest SERS intensity with −0.8V potential | Aqueous uric acid in NaF synthetic urine | 0.2 mM LOD | 86 |
| Multi-layered Au/Ag-coated screen-printed electrodes | Electrochemical-SERS | Quantitative detection in clinically useful range (0.1–1.0 mM) | Aqueous uric acid in NaF synthetic urine | 0.19 mM LOD | 87 | |
| Au nanospheres-decorated nanocone array polycarbonate substrate | Electrochemical-SERS | 35-Fold increase in SERS intensity upon applying −1.0V potential | Aqueous uric acid | 8.7 × 10−8 M LOD | 88 | |
| Ag NP-coated screen-printed electrodes | Electrochemical-SERS | Highest SERS intensity with −1.0V potential | 6-Thiouric acid in synthetic urine | 1 μM LOD | 89 | |
| Ag NP-coated screen-printed electrodes | Electrochemical-SERS | Negligible SERS responses with applied potential | Cannabinoids tetrahydrocannabinol and carboxy-tetrahydrocannabinol | — | 90 | |
| Au nanoporous gold nanobowls | Electrochemical-SERS | Enantiospecific differentiation only observed upon applying −0.6V potential | Aqueous L/D-tryptophan; R/S-propranolol | — | 91 | |
| Negatively and positively charged AuNPs@SiO2NPs nanoconjugates | Nanosensor-assisted NMR | Partial chemoselectivity based on electrostatic interactions | Serotonin; l-serine; homovanillic acid; l-phenylalanine; dopamine | 10 μM LOD | 92 | |
| Ag colloidal nanoparticles | Reverse-phased liquid chromatography (RP-LC)-SERS | Sequential metabolite separation and detection to reduce cross-interferences | Drug molecule methotrexate (MTX) and its metabolites, 7-hydroxy methotrexate (7-OH MTX) and 4-diamino-N(10)-methylpteroic acid (DAMPA) in urine | μM LOD | 93 | |
| Hyphenated techniques | Ag NPs | Paper chromatography-SERS | Sequential metabolite separation and detection to reduce cross-interferences | Aqueous β-carotene and lycopene | Low-cost; disposable | 95 |
| Ag nanostructured SERS substrate | LC-SERS with sheath flow confinement | Comparable metabolite detection capability to LC-MS | 2-Amino-3-hydroxypyridine (AHP) in tumor sample lysates | — | 96 | |
| MMTV-Wnt1, MMTV-Neu tumor samples and healthy mammary gland sample from their SERS metabolic fingerprint |
| Machine learning algorithms | Nanomaterials | Transduction mode(s) | Additional description | Analyte(s) | LOD/Selectivity | Ref. |
|---|---|---|---|---|---|---|
| Unsupervised clustering algorithms to uncover data interrelationships | Highly doped 3D nanoprobes | SERS | PCA for metastasis onset prediction | Invasive metastatic, less-invasive progeny and non-stem cells based on metabolite cues | — | 108 |
| Cysteamine-grafted Au NPs | SERS | PCA to establish correlations between cancerous exosomes and protein biomarkers | Nonsmall-cell lung cancer (NSCLC) exosomes and normal cell-derived exosomes | — | 109 | |
| Dual nanosensor arrays consisting of diversely modified Au NPs and SWCNTs | Chemiresistance | HC analysis to establish similarities/differences in BVOC profiles of different diseases | BVOC profiles from patients with 17 different disease conditions | — | 30 | |
| Supervised classification and regression algorithms for predictions of new, unknown samples | 4-Mercaptophenylboronic acid-functionalized Ag nanocubes on SPHB surface | SERS | PLS for multiplex quantification | Pregnane and tetrahydrocortisone in urine samples | High R2 cross-validation linear coefficient of 0.99; low absolute deviations between predicted and actual pregnane% by 0.0–3.1% | 54 |
| 7-Electrode array | Electrochemical (voltammetric e-tongues) | PLSDA for prostate cancer detection | Urine metabolic profiles | 91% sensitivity and 73% specificity | 116 | |
| Array-based sensor with 73 different indicators | Colorimetric | Random forest for tuberculosis detection | Urine metabolic profiles | 85.5% sensitivity and 79.5% specificity | 117 | |
| Au nanoraspberries-coated nanopipettes | SERS | CNN for multiplex monitoring of 8 metabolite concentration gradients | Pyruvate, lactate, ATP, ADP, glucose, glutamine, urea, and CO2 | >86.8% sorting accuracy | 118 |
2. Designer nanosensor platform configurations
In general, nanosensors are designed to utilize the unique nanoscale properties of various nanomaterials for sensitive metabolite detection via different transduction mechanisms, including optical-, electrical- and electrochemical-based modes. To improve a nanosensor's detection sensitivity and selectivity, diverse chemical analyte capturing and physical analyte confinement strategies can be adopted to enhance nanosensor–metabolite interactions (Table 1). The former capitalizes on chemical interactions with different surface-grafted moieties to capture and drive metabolites to the nanosensor surface, while the latter physically confines and encapsulates metabolites close to the nanosensor surface.12–15,44 These tailored nanosensors in turn serve as important building blocks for further assembly into various user-defined, highly customizable arrays depending on the choice of target analytical method and application.2.1 Chemical analyte capturing strategies
Chemical analyte capturing strategies utilize chemical interactions between metabolites and various receptor molecules grafted on the nanosensors to facilitate electron transfer and enable the metabolites to better access the nanomaterial's EM field and/or catalytic active sites, thus enhancing signal responses. Commonly employed receptors range from small molecular probes including sulfonated aromatic compounds to nucleic acids and other macromolecules.45–48 Key advantages of these chemical analyte capturing strategies include (1) increased signal sensitivity, (2) selective metabolite isolation and detection in complex sample matrices and (3) accurate differentiation of structurally similar analogs such as enantiomers.To bring the metabolites close to the nanosensor surface and increase the effective metabolite concentrations for higher signal responses, the chosen receptors should possess functional groups which interact favorably with specific chemical moieties on the target metabolite(s) (Fig. 2Ai). Commonly employed interactions range from noncovalent interactions such as electrostatic interactions, hydrogen bonding and van der Waals' forces to covalent and coordination bonds. For instance, abundant ether groups (–O–) on single-wall carbon nanotubes modified with polyethylene glycol (PEG)-phospholipids (PEG-PL-SWCNT) form numerous hydrogen bonds with the hydroxyl groups (–OH) on target plant polyphenols, including genistein and trihydroxypterocarpan, to attract these chemical defense metabolites close to the SWCNTs (Fig. 2Aii).45 These interactions render the modified SWCNTs highly sensitive to plant polyphenol detection via near-infrared fluorescence quenching in a low μg mL−1 range, even in plant tissue extracts and culture media (Fig. 2Aiii).
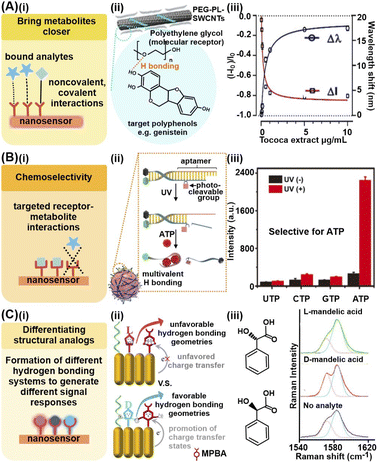 | ||
| Fig. 2 Chemical analyte capturing strategies. (A) (i) Use of chemical interactions to bring metabolites close to nanosensor for enhanced signals. (ii) Depiction of effective capture and detection of pathogen-induced polyphenol secretion by soybean (Glycine max) culture using polyethylene glycol-phospholipid single-walled carbon nanotube (PEG-PL-SWCNT)-based fluorescent sensors via hydrogen bonding. (iii) Nanosensor response against purified polyphenol extract from Tococa spp., showing a decrease in NIR fluorescence and concurrent redshift of emission wavelengths (mean ± SD, n = 3, colored line = hyperbolic fit). (B) (i) Achieving chemoselectivity via targeted receptor-metabolite chemical interactions. (ii) Schematic illustration of UV light-activatable ATP sensing mechanism of the nanosensor, whereby ATP selectively hybridizes to the aptamer, likely via multivalent H bonding. (iii) Response of aptamer-modified upconversion nanoparticles to 5 mM of different nucleoside triphosphates with and without 365 nm light irradiation, showing selective fluorescence only in the presence of ATP. (C) (i) Differentiation of structural analogs via formation of different hydrogen bonding systems. (ii) Pictorial representation of proposed differentiation mechanisms. (iii) Different 4-mercaptophenylboronic acid (MPBA) SERS spectra in the 1540–1620 cm−1 region in the presence of D- and L-mandelic acid. Reprinted and adapted with permission from (A) ref. 45, (B) ref. 48 and (C) ref. 49. Copyright 2020 Wiley-VCH. Copyright 2021 R. Niβler, A.T. Müller, F. Dohrman. Angew. Chem., Int. Ed. Published by Wiley-VCH GmbH. Copyright 2017 American Chemical Society. | ||
To achieve chemoselectivity, the strategic employment of receptors which form covalent bonds via specific chemical reactions with target metabolites can help to suppress interferences from other species present in the sample matrices (Fig. 2Bi). For instance, the cyclic esterification of boronic acid-based receptors is a useful chemical reaction for selective capture and detection of diol-containing metabolites, with good functional group selectivity.46,47 In one study, this esterification reaction was utilized for selective electrochemical detection of natural glycoside toxins containing numerous diol groups such as α-solanine and α-chaconine, by modifying a reduced graphene oxide/Au nanosensor with 4-mercaptophenylboronic acid.31 In comparison, the nanosensor exhibited negligible responses to minerals such as potassium and calcium, as well as vitamins such as lutein, thus demonstrating good anti-interference ability. Apart from specific covalent bonds, multivalent hydrogen bonding can also be leveraged for specific capture of only those metabolites which can form these interactions at the corresponding sites with the receptors, akin to a ‘lock-and-key’ mechanism. For example, highly specific complementary base pairing between upconversion nanoparticles conjugated with photocleavable linker-modified DNA aptamers and adenosine triphosphate (ATP) is utilized for selective fluorescence imaging of ATP in live HeLa cells and mice over other nucleotides, namely UTP, GTP and CTP (Fig. 2Bii and iii).48
Finally, it has been demonstrated that certain receptors such as mercaptophenylboronic acid and aminothiophenol form different hydrogen bonding systems with structurally identical enantiomers to generate different charge transfer states for facile enantiomeric differentiation (Fig. 2Ci and ii). For instance, in one work, the authors exploit this phenomenon to differentiate 11 aromatic enantiomeric pairs via SERS, including mandelic acid and phenylalanine, using Au nanorod arrays modified with 4-mercaptophenylboronic acid and n-hexanethiol, where each enantiomer forms differential MPBA-enantiomer complex (Fig. 2Ciii).49 However, this strategy is currently only reported for receptors capable of forming hydrogen bonds. More in-depth understanding of the underlying working principle is thus essential to investigate if the phenomenon can be extended to other chemical interactions.
Other than analyte capture, these chemical modifications also facilitate targeted endocytosis and translocation of these nanosensors to specific cellular sites for subsequent in vivo metabolite monitoring, which is useful to elucidate cellular pathways and mechanisms. For instance, Au nanospheres were functionalized with Arg-Gly-Asp (RGD) peptides to mediate their internalization by cancer cells via RGD-integrin interactions, as well as with nuclear localization signal (NLS) peptides for translocation near the nucleus via NLS-importin interactions.50 This enabled subsequent monitoring of distinct increases in the SERS signals of phenylalanine and its derivatives at 1000, 1207 and 1580 cm−1 after plasmonic photothermal therapy, consistent with phenylalanine-induced apoptosis pathways. Interestingly, the chemical analyte capturing techniques can even be applied directly to live organisms for in vivo small-molecule metabolite monitoring. In one study, 2,2′-disulfanediylbis(N-(2-aminophenyl)acetamide) (OPD-dTGA)-functionalized Ag nanocubes were modified on the surface of a live methicillin-resistant Staphylococcus aureus bacterium to monitor the release of nitric oxide during different antibiotic treatments.51 This is observed through the decrease in Raman intensities of characteristic OPD-dTGA peaks at 1233, 1263, 1322, 1349 and 1446 cm−1, attributed to NO-induced cleavage of aromatic o-phenylenediamine groups. Overall, chemical analyte capturing strategies afford better metabolite affinities and can also be readily tailored to detect different metabolites based on the intended application, be it to differentiate enantiomers or for partially selective detection in complex media. The tailored nanosensors can also be deposited on diverse substrates from conventional wafer chips to flexible PDMS substrates and even live organisms, thereby expanding their potential applications for in situ and in vivo investigations. Nonetheless, most platforms rely on passive analyte diffusion toward functionalized nanosensors for interactions, which is not ideal for the detection of metabolites at ultratrace concentrations. Hence, there is a need to develop strategies that enable a more active accumulation of target molecules near the nanosensor surfaces through physical confinement.
2.2 Physical confinement and encapsulation strategies
Physical confinement and encapsulation strategies aim to concentrate metabolites near the nanosensor surface to enhance intermolecular interactions, thereby attaining stronger signal responses and higher sensitivity. For liquid-phase metabolite detection at trace concentrations, an analyte enrichment effect can be achieved through the reduction of analyte spread, which minimizes the analyte-sensor contact area and localizes analyte concentration. Alternatively, for both liquid and gaseous samples, nanosensors can be integrated with secondary sorbent materials with high molecular sorption capabilities to accumulate metabolites within the pores close to the nanosensor surface via multifold host-guest chemistry. These physical-based strategies are pertinent in detecting a wide range of small-molecule metabolites because they do not rely on the chemical nature or surface affinity of the analyte, function well even at ultratrace concentrations and require minimal amounts of sample.One prominent physical molecular enrichment strategy to reduce analyte spread for aqueous samples is to form superhydrophobic surfaces by modulating the nanosensors' surface wetting properties (Fig. 3Ai). Superhydrophobicity is typically imbued by (1) modification with hydrophobic chemical functionalities or (2) enhancement of nano/microscale surface roughness with roughened template supports such as nanopillars or via nanoparticle deposition.52,53 The superhydrophobic surface reduces the analyte spread and confines the drying spot, thus concentrating the analytes within an area that is up to 104-fold smaller. In one study, the electrostatic self-assembly of Ag nanocubes and octahedra into a rough metallic array, followed by chemical surface modification with perfluorodecanethiol, was employed to render superhydrophobic SERS substrates (contact angle ∼158 ± 8°). When compared to a hydrophilic substrate, the superhydrophobic nanosensor concentrated pregnane and tetrahydrocortisone, which are key urinary biomarkers for threatened miscarriage, into a ∼185-fold smaller area to enable ultrasensitive SERS detection at sub-nanomolar (ppt) levels (Fig. 3Aii).54
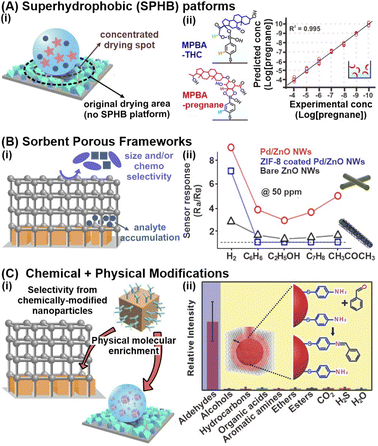 | ||
Fig. 3 Physical confinement strategies and multifunctional platforms synergizing both chemical and physical-based strategies. (A) (i) Schematic illustration of the analyte concentrating effect on superhydrophobic (SPHB) platforms. (ii) Sensitive detection of pregnane at sub-nanomolar concentrations (ppt levels) on SPHB SERS substrate using 4-mercaptophenylboronic acid (MPBA)-functionalized Ag nanocubes. Corresponding structures for MPBA-pregnane and MPBA-tetrahydrocortisone, another urine biomarker, are included. (B) (i) Schematic illustration of the key advantages in physically modifying nanosensors with sorbent porous frameworks. (ii) Resistance changes (where Ra and Rg are the resistances in the absence and presence of the target gas) to 50 ppm of H2, C6H6, C7H8, C2H5OH and CH3COCH3 gases at 200 °C using bare ZnO, Pd/ZnO and ZIF-8 coated Pd/ZnO nanowires. (C) (i) Illustration of selective confinement and enrichment by synergizing chemical and physical modification strategies. (ii) Relative intensities of 1623 cm−1 peak indexed to imine C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching of the cross-linked product when exposed to different functional groups, illustrating the selectivity of gold superparticles coated with ZIF-8 (GSP@ZIF-8). The schematic representation of GSP@ZIF-8 and the selective Schiff-base reaction between ATP and aldehydes is included as inset. Reprinted and adapted with permission from (A) ref. 54, (B) ref. 61. (C) ref. 69. Copyright 2017 Wiley-VCH. Copyright 2018, 2020 American Chemical Society. N stretching of the cross-linked product when exposed to different functional groups, illustrating the selectivity of gold superparticles coated with ZIF-8 (GSP@ZIF-8). The schematic representation of GSP@ZIF-8 and the selective Schiff-base reaction between ATP and aldehydes is included as inset. Reprinted and adapted with permission from (A) ref. 54, (B) ref. 61. (C) ref. 69. Copyright 2017 Wiley-VCH. Copyright 2018, 2020 American Chemical Society. | ||
However, superhydrophobic nanosensors are less effective at concentrating analytes in less polar/organic media of low surface tensions, which tend to spread more readily and further dilute analytes. Thus, one key research direction is the fabrication of superoleophobic nanosensors with strong organic liquid repellency. This is crucial to expand the range of detectable metabolites because many metabolites in biofluids, food, and environmental samples are oil-soluble, including free fatty acids, lipid-soluble vitamins (E, A, D and K), and various oral drugs. Although current works on superoleophobic platforms are largely still limited to the detection of test dye molecules with strong surface affinities, we anticipate an increase in its application for the detection of a wider range of biologically relevant metabolites for real-life detection.52,55 Improvements in superoleophobicity of nanosensor platforms can potentially be achieved with strategic designs of re-entrant and doubly re-entrant nano/microstructures on top of increasing surface roughness and modulating surface chemistry. Future work could also be directed to the development of superomniphobic platforms that exhibit both superhydrophobicity and superoleophobicity to achieve universal, two-in-one detection of both lipid- and aqueous-soluble analytes.
Next, for the enrichment of both liquid/aqueous and gaseous metabolites, one versatile strategy is to integrate nanosensors with sorbent porous materials, such as metal–organic frameworks (MOFs), formed by extensive coordination of metal ions/clusters with organic linkers (Fig. 3Bi). Owing to their ultrahigh specific areas and high porosities, these sorbent porous frameworks are excellent molecular sponges for efficient metabolite sorption and preconcentration, even for intrinsically dilute and highly mobile gaseous compounds.56 For instance, ZIF-8 encapsulation around fluorescent BSA-coated Ag nanocubes (Ag NCs) increased the fluorescence quenching responses of bare Ag NCs and BSA-coated Ag NCs in the presence of Cu2+ ions by 7-fold and 3-fold respectively, enabling sensitive Cu2+ detection in blood down to 4 × 10−4 M.57 The increased fluorescence response is attributed to ZIF-8's porous network, which facilitates efficient adsorption and accumulation of Cu2+ ions. Furthermore, the imidazole moieties within ZIF-8 serve as specific recognition elements to capture Cu2+ from the sample media. In another study, Ag NPs coated with 30 HKUST-1 layers exhibited a 14-fold increase in LSPR spectral shifts upon gaseous CO2 exposure compared to the uncoated sensor, effectively amplifying the signal responses.58 Regardless of the type of metabolites and detection method, it is of note that systematic optimization of the MOF thickness and surface coverage is essential to maximize efficient molecular capture near the nanosensor surface. For instance, an increase in ZIF-8 thickness over a Ag nanocube array from 8 to 146 nm results in a 2.5-fold rise in SERS signals of gaseous 4-methylbenzenethiol (MBT), due to greater gas accumulation along the z-depth of the array for enhanced EM interactions.59 Beyond the optimal thickness, there are negligible changes to the SERS activity because the MBT molecules are too far away from the nanosensor surface to experience the EM field. Similar thickness and coverage-dependent signal amplifications are also observed for other transduction modes.60
In addition to molecular enrichment, these porous networks also enhance size selectivity by serving as molecular sieves to prevent larger species from accessing the nanosensor surface, controlled by the porous networks' tunable pore and cavity sizes (Fig. 3Bi). In one study, ZIF-8-coated Pd NPs@ZnO nanowires exhibited excellent sensitivity and selectivity for gaseous H2 sensing down to 10 ppm, with negligible resistance changes from interfering gases, including benzene, ethanol and acetone (Fig. 3Bii).61 Certain MOFs also demonstrate chemoselectivity. Such selectivities (both analyte size and chemoselectivity) are crucial to facilitate selective detection in complex biomatrices. For instance, a PtPd@ZIF-67 electrochemical sensor displayed high current responses in the presence of aqueous phenylketonuria (PKU) biomarkers such as phenylpyruvic and phenylacetic acid, but negligible changes for common amino acids with similar structural backbones.62 The chemoselectivity is permitted by selective acylation between the imidazole ring of ZIF-67 and carboxyl groups of selected PKU biomarkers only, controlled by the electron-withdrawing functional group on the carbonyl α-carbon. For additional in-depth understanding of the fabrication and applications of nanoparticle-MOF nanohybrid sensors, interested readers can refer to the relevant reviews cited here.15,63,64
Due to the numerous benefits of these physical molecular enrichment strategies, we expect sustained, strong interest in their developments for small-molecule metabolite detection. Some up-and-coming research directions include the design and fabrication of chiral MOFs using chiral ligands, and the integration of chiral nanosensors with MOFs for breakthroughs in enantiomeric sensing. Through rational design of the recognition sites, chiral MOFs demonstrate huge potential for chiral sensing with greater stereoselective adsorptions due to enhanced, selective metabolite confinement from both the MOF scaffold and the conformational rigidity of specific recognition sites.65–67 It is also important to increase the types of MOFs selected (beyond the conventional ZIFs and HKUSTs), as well as explore emerging porous materials such as porous organic polymers (POPs) as potential host matrix candidates, where their integration may enable new performance breakthroughs.63,68 For instance, besides the water stability of the host porous matrix material, other important considerations include its stability in organic solvents to enable the detection of oil-soluble metabolites.
2.3 Combining chemical and physical-based strategies for multifunctional nanomaterials
The combination of both chemical capture and physical molecular enrichment strategies is also a promising strategy to achieve both sensitivity and specificity by capitalizing on their individual advantages (Fig. 3Ci). For instance, lung cancer biomarkers in exhaled breath, such as 4-ethylbenzaldehyde, can be detected down to 10 ppb using 4-aminothiophenol (ATP)-grafted Au superparticles (GSPs) encapsulated with ZIF-8 (Fig. 3Cii).69 The two-pronged strategy first leverages highly porous ZIF-8 to facilitate the sorption and concentration of aldehyde biomarkers near the GSP surface. The ATP receptors then selectively react with these aldehydes via a Schiff base reaction, where the product can be readily detected using SERS through the appearance of the 1623 cm−1 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching mode. Due to the specific reaction between the aldehyde and amino groups, the GSPs exhibited high anti-interference ability with negligible SERS changes in the presence of other functional groups, such as alcohols, esters, organic acids, and amines.
N stretching mode. Due to the specific reaction between the aldehyde and amino groups, the GSPs exhibited high anti-interference ability with negligible SERS changes in the presence of other functional groups, such as alcohols, esters, organic acids, and amines.
In addition to utilizing sorbent frameworks to physically confine molecules, superhydrophobic surfaces can also be combined with chemoselective nanoparticles to concentrate trace molecules in a small area. For example, urine metabolites correlating with miscarriage can be detected in urine samples down to 10−10 M by first mixing with 4-mercaptophenylboronic acid (MPBA)-grafted Ag nanocubes, followed by dropcasting on a superhydrophobic Ag nanocube/octahedra surface.54 MPBA selectively captures the target urine metabolites pregnane and tetrahydrocortisone through boronate ester bond formation, exhibiting distinct SERS spectral changes between 1140 and 1380 cm−1, which are in good agreement with the simulated results. The superhydrophobic surface with a high surface roughness of 158 ± 8° further concentrates these metabolites to a small surface area for ultrasensitive SERS detection.
Collectively, these examples highlight the synergistic importance of coupling chemoselective capturing with physical molecular trapping to enhance the selective detection of trace metabolites in complex sample matrices. Such platform designs also create immense opportunities in diverse fields ranging from biomedical applications to food and environmental analyses as they require small analysis volumes (μL), have fast readouts, and can be readily miniaturized for point-of-need applications. Hence, we foresee similar synergistic platform designs for other analytical techniques, such as infrared spectroscopy and chemiresistors.
2.4 Array-based strategies
Thus far, our discussion of various analyte capturing and enrichment strategies have revolved around the specific detection of individual analytes and/or analyte classes. However, disease screening and monitoring often demands multiplex metabolite detection or differentiation of complex, multicomponent mixtures without identifying individual components. One powerful strategy to directly differentiate complex metabolite mixtures is to combine multiple individually modified nanosensor platforms for pattern-based recognition (Fig. 4A). These array-based configurations typically leverage various chemical interactions between multiple receptors and different metabolites to generate high-dimensional signal patterns specific to different analyte mixtures. By measuring cumulative responses from multiple receptors, such pattern-based recognition mimics human sensory systems, which comprise sophisticated networks of diverse sensory receptors to convey large amounts of chemical information to the brain.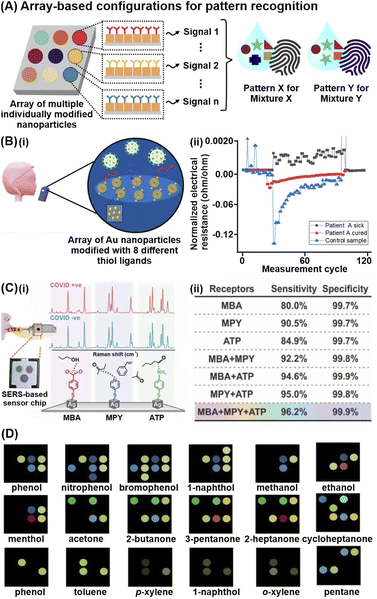 | ||
| Fig. 4 Array-based techniques for differentiation of complex metabolites mixtures. (A) Schematic depiction of high-dimensional pattern fingerprints generated from array-based configurations. (B) (i) Illustration of chemiresistive nanoarray for COVID-19 detection comprising of 8 sensor elements. (ii) Representative response of one sensor in a chemiresistive array to three different breath samples: infected COVID-19 patient A; recovered COVID-19 patient A; and a healthy control. Each unit represents one sensor cycle. (C)(i) Representative SERS spectra of each probe (MBA: 4-mercaptobenzoic acid, MPY: 4-mercaptopyridine, ATP: 4-aminothiophenol) in the presence of COVID-positive and COVID-negative breath samples. A total of 74 COVID-positive (31 asymptomatic) and 427 COVID-negative samples are measured. Schematic of multiprobe SERS-based sensor is included as inset. (ii) Multiprobe Ag nanocube platform demonstrates enhanced classification sensitivity and specificity for COVID-19 infection status. (D) Color difference maps of 18 representative volatile organic compound vapors using paper-based optoelectronic noses (OENs) fabricated from gold and nanoparticles modified with 8 different capping agents. Reprinted and adapted with permission from (B) ref. 73, (C) ref. 70 and (D) ref. 79. Copyright 2019, 2020, 2022 American Chemical Society. | ||
The immense potential of array-based recognition is evidenced by the burgeoning number of studies that utilize them for the detection of small-molecule metabolites, particularly volatile organic compounds (VOCs), in complex breath and aqueous biological matrices.70–75 One notable technique that best exemplifies this strategy is chemiresistive sensing. For instance, a chemiresistor nanoarray comprising Au NPs functionalized with 8 different hydrocarbon thiol ligands successfully enables COVID-19 detection via exhaled breath in 130 participants, achieving 76–95% classification accuracy (Fig. 4Bi).73 The 8-sensor configuration increases the data dimensionality to generate a wide range of signal patterns for better differentiation, where each sensor emits different electrical resistance signals of varying intensities and signs due to nanomaterial swelling/aggregation and/or changes in permittivity of the organic layer upon different VOC adsorption (Fig. 4Bii).
Such pattern-based recognition using array configurations is also increasingly applied for other analytical techniques, including SERS, fluorescence, and colorimetric assays.76–78 In one study involving 501 participants, a multiprobe Ag nanocube array modified with 4-mercaptobenzoic acid, 4-aminothiophenol and 4-mercaptopyridine recorded distinct SERS changes upon multiplex adsorption of breath VOCs, enabling COVID-19 detection even in asymptomatic patients (Fig. 4Ci).70 Importantly, the breath-induced spectral changes are consistent with experimental and in silico spectral changes triggered by pure VOC vapors of potential COVID-19 biomarkers, including alcohols, aldehydes and ketones, affirming that the multiprobe nanocube array chemically interacts with the diverse BVOCs to elicit disease-specific spectral profiles. The additive effect of array-based configurations is highlighted by a notable improvement in sensitivity from 80% using a single-probe platform to 96.2% using the multiprobe platform, which emphasizes the importance of combining multiple receptors to provide a more complete description of complex matrices for better differentiation (Fig. 4Cii). Another notable study used Au and Ag NPs modified with 8 different chemical species, including chitosan, glucose, and cysteine, to generate unique color-difference maps for univocal gaseous VOC recognition, where different VOCs induce aggregation of each modified NP to various extents due to differential interactions.79 For instance, cysteine-modified and glutathione-modified AgNPs preferentially interact with esters and carboxylic acids respectively. Notably, this metabolite-induced nanoparticle aggregation is observed even with gaseous analytes, successfully differentiating 45 target gaseous VOCs in 9 chemical families including phenols, amines, arenes, hydrocarbons, and esters, at detection limits <10 ppb (Fig. 4D).
Collectively, these examples emphasize the universal nature of array-based techniques to detect a wide range of VOCs of diverse chemical functionalities, owing to their capability to cumulatively encode various chemical interactions as distinct signal fingerprint patterns specific to each analyte or mixture. They also highlight the versatility and broad applicability of array-based techniques to probe both gaseous and aqueous biomatrices without prior separation/pretreatment. It is of note that array-based platforms are often combined with multivariate chemometric tools and machine learning algorithms to facilitate pattern analysis of more complex array-based sensing data.75,80,81 In this aspect, a key understanding and careful selection of suitable algorithms are thus necessary to advance the development of array-based platforms for diverse applications, which will be discussed in Section 4 below. Finally, while array-based nanosensors are well-poised to play increasingly important roles in biology and medicine, a key scientific challenge remains in reconciling the data-rich pattern outputs with metabolomic advances to better understand what the arrays are responding to.
3. Hybrid techniques
In addition to strategies that isolate target metabolites and/or attract metabolites closer to nanosensors, hybrid techniques couple two or more analytical methods to further bolster detection sensitivity and increase the range of detectable metabolites by synergizing the strengths of individual techniques (Table 2). These techniques can be multimodal, such as colorimetric/fluorescence-SERS and SERS-MS, or hyphenated, such as electrochemical-SERS (EC-SERS) and nanosensor-enhanced NMR and MS (Fig. 5A). The former integrates multiple types of transduction signals to generate multi-tiered information for complementary cross-validation and higher data dimensionality. In contrast, the latter typically couples additional separation or molecule adsorption enhancement techniques to complement a primary analyte detection method, which provides the resultant signal readout. Regardless of the type of hybrid technique, the combinatorial use of two or more analytical techniques aims to achieve the ‘best of both worlds’ to enable breakthrough performance through synergistic effects.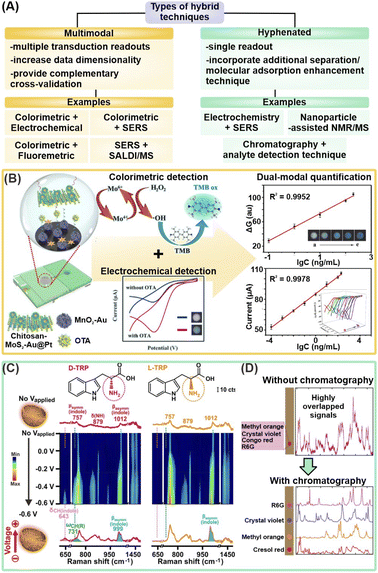 | ||
| Fig. 5 Hybrid techniques combining two or more analytical methods. (A) Schematic summary of different types of hybrid techniques, including multimodal and hyphenated techniques. SERS: surface-enhanced Raman scattering; SALDI/MS: surface-assisted laser desorption/ionization-mass spectrometry; NMR: nuclear magnetic resonance. (B) Schematic illustration of dual-modal colorimetric-electrochemical nanosensor platform for detection of ochratoxin A (OTA) using a sandwiched complex of chitosan-functionalized MoS2–Au@Pt and Au NP-supported MnO2 nanoflowers respectively labelled with aptamers (left), and corresponding quantification models for cross-validation (right). (C) Electrochemical-SERS differentiation of L- and D-tryptophan (TRP), where the enantiomers exhibited identical spectra without Vapplied and showcased differential spectral changes at Vapplied = −0.6 V. (D) Schematic illustration of coupling paper chromatography for analyte separation prior to SERS measurements to obtain distinct SERS fingerprints, using a mixture of 4 organic dyes as a proof-of-concept. Reprinted and adapted with permission from (B) ref. 83, (C) ref. 91 and (D) ref. 95. Copyright 2021 Elsevier B.V. Copyright 2018, 2021 American Chemical Society. | ||
3.1 Emerging multimodal techniques
By integrating multiple types of transduction signals, the multidimensional information obtained from multimodal platforms serve as important cross-validation to increase the overall reliability of each detection mode, which is imperative for future translations to practical workflows. Such multifunctional platforms are achieved by capitalizing on the multimodal nature of most nanomaterials, which allows them to be measured by more than one analytical technique. For instance, a dual-modal colorimetric and fluorometric uric acid sensor was developed using N,P-codoped carbon dots (CDs) coupled with Au nanoparticles.82 Upon the addition of uric acid and Ag+ ions, uric acid reduces Ag+ onto the Au nanoparticles to induce a red-to-orange color change due to blueshifts of the surface plasmon resonance (SPR), which permits UV-vis quantification from 0.1 to 10 μM. At the same time, fluorescence quenching arising from the SPR overlap of the Ag@Au nanoparticles with the emission peaks of N,P-codoped CDs can also be used for quantification from 0.5 to 10 μM to offer an additional layer of verification. In another example, an electrochemical-colorimetric sensor was developed for the detection of ochratoxin A (OTA), a carcinogenic contaminant produced by fungi (Fig. 5B).83 In brief, OTA triggers the formation of a sandwich complex with chitosan functionalized MoS2–Au@Pt and Au nanoparticles-supported MnO2 nanoflowers modified with the label and capture aptamers, respectively, catalyzing H2O2 reduction to generate an EC response. Simultaneously, the generated ·OH radicals induce 3,3′,5,5′-tetramethylbenzidine (TMB) to change color. Visual detection enables quantification in the range of 0.1–200 ng mL−1, while EC detection permits further quantification down to 1 × 10−4 ng mL−1. Importantly, they act as complementary validation methods. Overall, generating multiple readouts aids in the cross-validation of acquired results, thereby alerting users to potential experimental errors in the form of contradictory results. Such two-fold result verification is essential to increase the nanosensor reliability for the translation of these emerging platforms into robust technologies for real-life clinical applications.In addition to cross-validation, a key advanatage of multimodal platforms is the ability of the selected methods to compensate for the limitations of each method, and enable unambiguous molecule recognition over a wider analyte scope. In one example, SiO2/TiO2 core/shell (T-rex) beads are used as a multifunctional platform for plasmon-free SERS and SALDI mass spectrometry (RaMassays).84 By controlling the thickness of the TiO2 shell, the optical properties of the T-rex beads can be tuned to achieve both efficient visible light trapping for SERS detection and UV absorption for laser desorption/ionization process in SALDI-MS. The same dual-modal RAMassay is used to differentiate (1R, 2S)-(−)-ephedrine and amphetamine, two important alkaloids related by the same phenethylamine skeletal structure, by MS, and regioisomers theobromine and theophylline by SERS. Notably, employing only SERS does not allow for the differentiation of the alkaloids as they give similar Raman fingerprints while employing only MS will fail to differentiate the regioisomers of similar molecular weights. Hence, the combination of two molecule-specific analytical methods provides complementary molecular information which effectively expands the list of target analytes that the sensor can detect.
Owing to their aforementioned advantages, multimodal platforms offer promising synergy for robust and sensitive detection of a wide range of small-molecule metabolites. In addition, compared to conventional workflows that analyze metabolite samples on separate instruments using their respective sensor platforms, the same multimodal nanosensor platform can be used to obtain complementary molecular information, which is more convenient and requires lower sample volumes. Nonetheless, there are several important considerations for their future developments. First, it is essential to develop two-in-one portable instrumental platforms for practical on-site usage without the need to load the nanosensor platform onto two separate instruments. For instance, a recent coupling of paper spray ionization mass spectrometry (PSI-MS) with SERS permits dual signal readouts on a single instrumental platform by using a custom movable holder that can be shifted forward/backward for detection via portable Raman spectrometer or ambient ionization (AI)-MS system.85 It is also noteworthy to emphasize that the nanosensor platform needs to be compatible with the chosen detection techniques to achieve such multimodal analysis.
3.2 Emerging hyphenated techniques
Hyphenated techniques that couple analyte detection methods with additional molecule adsorption enhancement techniques offer enhanced detection capabilities to address current challenges of complex matrix interferences, growing demand for lower detection limits, and better differentiation of structural analogs for biological and biomedical applications. A notable methodology in this category is electrochemical (EC)-assisted SERS, whereby SERS signals are recorded during an applied potential. The applied potential (Vapplied) is critical to increase adsorption of metabolites and/or desorption of interferents from the plasmonic surface via surface charge manipulation. Such close analyte-nanosensor proximity is vital to boost overall SERS signal intensity for increased detection sensitivity by enhancing the metabolites' interactions with the nanosensor's EM field and facilitating charge transfer for larger chemical SERS enhancement. Several notable works have demonstrated the potential of EC-SERS for rapid, sensitive, and on-site detection of uric acid for preeclampsia diagnosis.86–88 For instance, a −1.0 V applied potential increased the SERS fingerprint of uric acid by ∼35-fold on a Au nanospheres-decorated nanocone array polycarbonate substrate, likely attributed to the cumulative effect of the aforementioned phenomena.88 EC-assisted SERS enhancement subsequently enabled sensitive uric acid detection in synthetic urine at a detection limit of 8.7 × 10−8 M, using its characteristic 1586 cm−1 peak. This has led to a sustained interest in EC-SERS over the past few years, in particular with expanding its analyte scope. Other target metabolites investigated include 6-thiouric acid for chemotherapy monitoring and tetrahydrocannabinol for cannabis control.89,90Next, the close analyte-nanosensor proximity also enables metabolites to better access asymmetric adsorption sites on designer nanoparticles, thereby triggering the effective formation of molecular-level metabolite–nanosensor interactions for better differentiation of structural analogs and enantiomers. In one work, EC-SERS is synergized with nanoporous gold nanobowls (NPGs), which are designed to comprise numerous asymmetric surface atomic defects, to induce enantiospecific interactions with target L/D-tryptophan and R/S-propranolol for SERS differentiation.91 Notably, each enantiomer pair displayed enantiospecific SERS fingerprints that are readily differentiated at Vapplied = −0.6 V. In contrast, they exhibited identical, indistinguishable SERS spectra without any applied potential, which emphasizes the importance of the applied potential for maximal interactions between the metabolites and stereospecific NPG adsorption sites (Fig. 5C). In overall, these studies thus highlight the prospects of ultrasensitive small-molecule metabolite detection by incorporating additional molecule adsorption enhancement techniques, even in complex (bio)sample matrices.
Another hyphenated strategy is to leverage separation and/or isolation of target metabolites, such as chromatography or chemoselective nanosensors, prior to detection to improve resolution and suppress interfering signals.92–96 In one study, glass microfiber filter paper coated with Ag nanoparticles achieves sequential paper-chromatographic separation and SERS detection of two carotenoids, β-carotene and lycopene, in tomato, carrot, and commercially available fruit and vegetable juice (Fig. 5D).95 Distinct, isolated SERS fingerprint of each analyte is obtained via SERS mapping of the entire paper at regular intervals. In another example, chemoselective AuNPs@SiO2NPs nanoconjugates are integrated with NMR spectroscopy for selective extraction of NMR spectra of specific metabolites to reduce spectral overcrowding by interfering species in complex mixtures.92,94 Electrostatic interactions form preferentially between the negatively charged nanoconjugates and positively charged serotonin but not with negatively charged L-phenylalanine, thus only serotonin's NMR signals are recorded, even in a complex serotonin/L-phenylalanine mixture.92
Although many novel hyphenated techniques appear to provide exciting new opportunities, most of them are still in their infancy with relatively few studies and a narrow focus on potential technique combinations. Hence, there remains significant room for future research and development. For example, the hyphenation of chromatographic techniques with SERS is of great interest at the moment as the former helps to reduce matrix complexity and remove interferences. We foresee similar potential in hyphenating chromatographic techniques with other detection techniques such as surface-enhanced infrared absorption (SEIRA), electrochemical, and fluorescence/colorimetric methods for reduced signal overlap, especially for techniques that easily suffer from cross-interferences due to non-molecule-specific signals. Another valuable research direction is the miniaturization of these hyphenated platforms to realize point-of-need applications since the hyphenation of two techniques often results in bulky instrumentation which is not operationally feasible.
All in all, the key to enable breakthrough detection capabilities using multimodal/hyphenated platforms lies in the prudent choice of nanomaterials and careful tuning of their physicochemical properties to fulfill the requirements of the chosen analytical methods. For instance, plasmonic nanoparticles such as Ag and Au exhibit strong visible light adsorption for excellent SERS activities, while semiconductor-based nanoparticles demonstrate high optical tunability in the UV range and are thus more suitable for SALDI ionization. Similarly, the nanoparticle size is also an important consideration to achieve good colorimetric and SERS performances concurrently. Next, for hybrid techniques that involve sequential measurements, it is crucial to ensure that the sample is not damaged after each process. Finally, the datasets obtained from individual methods are still analyzed separately with individual detection limits, which does not fully utilize the high-dimensional nature of the sample's overall dataset. We expect that higher sensitivities can be achieved through the parallel combination of the individual datasets to leverage the molecular information available for a more comprehensive analysis. In this aspect, we hypothesize that the use of machine learning algorithms will be critical to assimilating and analyzing these complex, high-dimensional data.
4. Machine learning-assisted data analysis
With continued advancement in sensing strategies at the nanobio interface, we have witnessed an expansion in the range of detectable analytes. Together with higher data dimensionality arising from multimodal techniques and/or array-based strategies that range from fluorescence and colorimetric images to spectroscopic data and electrical readouts, the resultant datasets have become increasingly complex. In addition, as we shift toward detection in real-life samples, the data complexity is further compounded by increased sample variability, such as high heterogeneity within patient populations. Consequently, the inability to identify minor differences or complex interrelations in signal readouts through conventional manual data analyses offers strong motivation to utilize more sophisticated tools in data analyticswhen translating complex raw signals into meaningful and comprehensible results.In recent years, machine learning (ML) is increasingly favored for the robust interrogation of these complex datasets due to their advanced pattern recognition capabilities (Table 3).97–99 These algorithms can elucidate underlying trends and interrelationships within large quantities of complex input data, which cannot be performed readily via traditional data analytical approaches which rely on single-variable changes or perform simple correlations. There are numerous ML algorithms available including unsupervised clustering algorithms such as principal component analysis and hierarchical clustering, as well as supervised classification and regression algorithms such as partial least-squares, support vector machines, tree-based algorithms and neural networks.100,101 For detailed conceptual understanding on the working principles of these ML algorithms and workflows on ML implementation for data analysis of various data types such as vibrational and mass spectra, interested readers can refer to the relevant reviews cited.102–104 Here, we highlight two broad areas in which ML has achieved exponential progress—(1) data exploration to uncover new, hidden interrelationships and (2) construction of prediction models for metabolite identification and/or quantification.99,100,105,106
4.1 Machine learning for data exploration
First, ML can be leveraged to elucidate new, unknown patterns and interrelationships within the study population of interest in a given feature/variable space without considering any pre-identified class labels and associated metadata. This is typically accomplished using unsupervised clustering algorithms such as principal component analysis (PCA) and hierarchical clustering (HC) (Fig. 6A).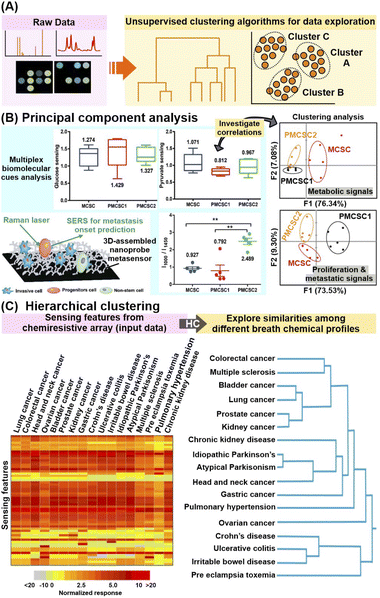 | ||
| Fig. 6 Application of unsupervised machine learning algorithms. (A) Workflow schematic which describes the use of unsupervised machine learning (ML) algorithms to uncover hidden interrelationships or construct prediction models for metabolite identification and/or quantification. (B) Application of principal component analysis (PCA) in conjunction with multiplex metabolite and proliferation-metastatic biomolecular cue signal analysis e.g. glucose, pyruvate and phenylalanine to investigate the prediction of metastasis onset using SERS. SERS spectra of metastatic (MCSC), premetastatic (PMCSC) and nonmetastatic (NMCSC) cancer stem-like cells from a highly metastatic cancer phenotype obtained on a 3D-assembled nanoprobe metasensor are used as input data. (C) Application of hierarchical clustering to explore the similarities in sensor response profiles (and thus breath profiles) among subjects suffering from 17 different diseases. 59 numerical sensing features obtained from multi-sensor nanoarray exposed to different breath samples, namely relative change of sensor's resistance at the beginning, middle and end of breath exposure, and the area under the curve for each sensor element, are used as input data. Reprinted and adapted with permission from (B) ref. 108 and (C) ref. 30. Copyright 2016, 2021 American Chemical Society. | ||
In brief, PCA extracts dominant patterns from the input datasets by converting the data into a complementary, orthogonal set of PC scores and loadings, whereby more similar data will have similar PC scores and be clustered closer together.107 In addition, the corresponding loadings plot of each PC reflects the contribution of each feature (e.g. spectra datapoint) to the PC, thus facilitating the identification of key diagnostic features which contribute to the interclass variances. For example, PCA was employed to investigate the biomolecular cues of metastatic cancer stem-like cells for metastasis onset prediction from SERS profiles of invasive metastatic, less-invasive premetastatic and non-stem cells (Fig. 6B).108 By focusing on specific Raman regions corresponding to metabolite (800–1450 cm−1) and proliferative-metastatic (1000–1650 cm−1) signals, PCA reveals distinct clustering of metastatic cells from their progeny and non-stem types. This validates the presence of multiplex hallmark signals, which is attributed to concentration variations of metabolites such as glucose, pyruvate and phenylalanine, as inherent cues to predict the metastatic ability of a tumor. In another study, through in-depth analysis of the PC score plot and the corresponding loadings, 26 SERS bands were selected as potential candidates for differentiation of nonsmall-cell lung cancer (NSCLC) exosomes from normal cell-derived exosomes, of which 21 were verified with ratiometric analysis.109 Notably, these selected spectral signatures were correlated back to several potential protein biomarkers such as CD9, CD81 EGFR and EpCAM, highlighting the use of PCA for identification of relevant disease biomarkers.
Apart from PCA, HC is also a useful clustering algorithm to examine sample interrelationships within the dataset by building a hierarchy of clusters, whereby the degrees of similarity between samples are represented as dendrograms and/or heatmaps.110 Notably, HC analysis on chemiresistive signals of 1404 breath samples collected from patients with 17 different disease conditions revealed strong signal resemblances between disease subgroups with common pathophysiologies (Fig. 6C).30 For instance, a high breath VOC profile similarity was found among diseases associated with increased inflammatory activity, namely Crohn's disease, ulcerative colitis, and pre-eclampsia.
In general, these examples highlight the prospects of clustering algorithms for rapid and sensitive extraction of relevant, informative interrelations among different sample subgroups, even for complex signal outputs with multiplex, overlapping influences from multiple biomarkers. Nonetheless, it is important to enrich such ML-based data exploration with domain knowledge on various pathophysiological and cellular–level interactions, as well as molecular information to validate that the observed clustering has strong chemical/biological correlations.
4.2 Machine learning for robust prediction models
In addition to data exploration, supervised classification and regression ML algorithms can be trained using labelled datasets (e.g. known metabolite identity and concentration or disease status) to construct prediction models for new, unlabeled data (Fig. 7A). The prediction can be categorical, as in the case of a disease status, or along a continuous regression scale, as in the case of metabolite quantification or disease progression. Many algorithms such as partial least-squares, random forests and artificial neural networks have variants to tackle both problem types and can cater to both binary, as well as more complex multi-class classification.111–113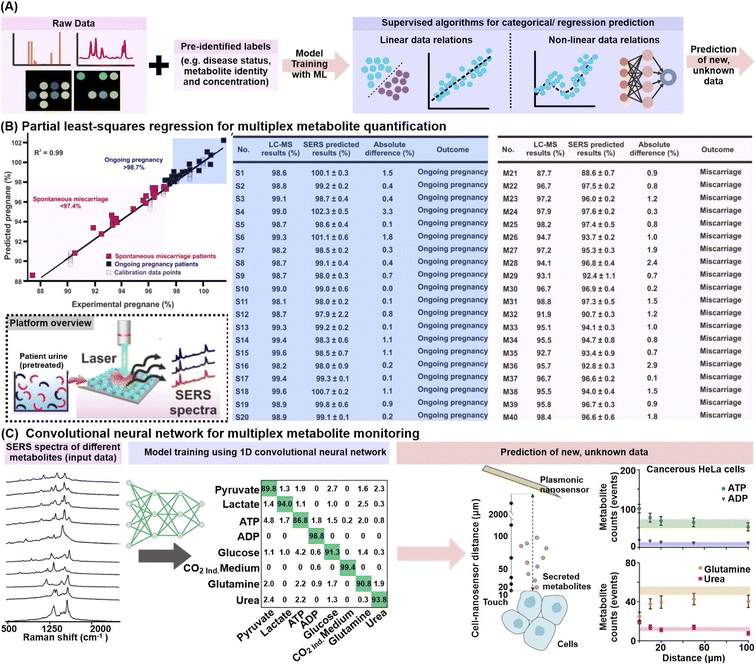 | ||
| Fig. 7 Application of supervised machine learning algorithms. (A) Workflow schematic which describes the use of supervised machine learning (ML) algorithms to construct prediction models for metabolite identification and/or quantification. (B) (left) Construction of a partial least-squares (PLS) regression model for multiplex quantification of pregnane % (relative pregnane/tetrahydrocortisone ratio) by mixing various pregnane % (at 10−10 M) using nonpregnant women's urine samples. Predicted pregnane % from 20 ongoing pregnancy (blue) and 20 spontaneous miscarriage (pink) urine samples are included. (right) Comparison of relative pregnane % measured from the surface-enhanced Raman scattering (SERS) nanosensor against liquid chromatography-mass spectrometry (LC-MS) analyses for the aforementioned ongoing pregnancy (S1–S20) and miscarriage (M1–M20) samples. The brief workflow from urine sample preparation to SERS measurement is included as an inset. (C) ML-driven SERS optophysiology to reveal multiplexed metabolite gradients near healthy and cancerous cells. SERS spectra acquired from 7 pure aqueous metabolite solutions, or the CO2-independent cell culture medium (background) were obtained, randomly separated into 60/20/20% train/test/validation sets for model training with a 1D convolutional neural network. Only representative SERS spectra are shown. The model was used to predict the metabolite counts near living cells, where those of ATP, ADP, glutamine and urea were shown for cancerous HeLa cells. Reprinted and adapted with permission from (B) ref. 54 and (C) ref. 118. Copyright 2019, 2020 American Chemical Society. | ||
In particular, partial least-squares (PLS) and its variant to tackle categorical classification, PLS-discriminant analysis (PLS-DA), are popular choices to analyze linearly correlated datasets. Briefly, the PLS algorithm aims to find a set of latent factors which best explains the maximum multidimensional covariance between the input data and given descriptors, e.g. known metabolite concentrations or disease status, before performing least-squares regression analyses to eventually construct regression or classification models.114 Notably, the PLS algorithms are well-suited to interrogate multidimensional datasets because they can accommodate multicollinearity among independent variables (e.g. SERS spectra datapoints) and are also designed to handle multiplex quantification of two or more biomarkers, unlike conventional statistical methods.115 For instance, PLS regression was applied to simultaneously quantify the amount of pregnane and tetrahydrocortisone in urine specimens of 40 women presenting symptoms of spontaneous miscarriage from their SERS profiles (Fig. 7B).54 The constructed regression curve shows an excellent fit, with a high cross-validation value R2 of 0.99, which allows the sensor to effectively monitor the pregnane/tetrahydrocortisone ratio and potentially prompt timely medical intervention. In another study, PLS-DA was employed for the classification of urine samples from 22 prostate cancer and 15 non-cancer patients using the multivariate electrochemical profiles measured on a 7-electrode electronic voltammetric tongue, achieving 91% sensitivity and 73% specificity.116 The aforementioned examples thus illustrate the ability of PLS algorithms to construct accurate classification or regression models from multivariate datasets, which portray distinct linear trends or separations.
To interrogate nonlinear or higher-order data relationships, ML algorithms such as support vector machine (SVM), tree-based algorithms including decision trees and random forests, as well as neural networks are preferred. For instance, a random forest (RF) classifier was trained on the colorimetric signatures of urine VOCs from tuberculosis and non-tuberculosis patients based on a sensor array comprising 73 different indicators responsive to chemically diverse metabolites including amines, sulfides, nitric oxides and carbonyl compounds.117 Among 22 tuberculosis cases and 41 symptomatic controls, the RF classifier attained 85.5% sensitivity and 79.5% specificity using repeated stratified 10 × 10-fold cross-validation. In another example, a convolutional neural network (CNN) model allowed multiplex SERS monitoring of 8 metabolite concentration gradients, namely pyruvate, lactate, ATP, ADP, glucose, glutamine, urea, and CO2 in the extracellular matrices of cancer, healthy and control cells (Fig. 7C).118 The CNN was first trained with a series of SERS spectra associated with different adsorption orientations for each metabolite, achieving high sorting accuracy of >86.8%. The model was then applied to classify SERS spectra measured near living cells, revealing an increase in ATP, ADP, lactate, and urea, with a concomitant decrease in glucose and glutamine near cancerous cells, notably in good agreement with expected metabolic pathways.
Given the plethora of ML algorithms available, careful design of the model architecture including algorithm selection and hyperparameter optimization based on the intended use and desired information is vital to construct robust and appropriate prediction models. For instance, tree-based algorithms such as random forests offer higher model explainability for scientists to better correlate the ML prediction with fundamental chemical/biochemical knowledge. This is because they can identify key features (e.g. spectral datapoints) that are important in model construction using methods such as Gini importance and mean decrease in accuracy. On the other hand, neural networks do not impose strict restrictions on the input data and variables, making them especially useful to model datasets with high inherent variability. Various dimension reduction and feature selection techniques such as recursive feature elimination (RFE) can also be incorporated prior to/during model training to further improve prediction accuracies by preserving only the key, relevant features while removing redundant features.119,120
Overall, ML algorithms are powerful tools that can be used to predict the identity and quantity of one or more biomarkers and uncover intricate relationships between samples. Together with the increasing availability of open-source code repositories, affordable high-performance computing infrastructure, and flexibility to apply to various data types, ML is thus well-poised to establish a new standard for scientific data processing.121–123 However, to achieve the ultimate end goal of practical ML implementation in clinical practices, there needs to be careful data curation to ensure the data is representative and of good quality (e.g. low spectral noise) to avoid false conclusions and construct prediction models with high accuracies. In addition, cross-validation techniques such as leave-one-out cross-validation or Venetian blinds are quintessential during the construction of ML models to avoid overfitting and increase model generalizability. Recognizing these caveats is critical in ensuring the constructive use of ML algorithms for small molecule analyses at the nanobio interface.
5 Conclusions
In this Perspective, we have provided an overview of the progress in various emerging strategies for enhanced small-molecule metabolite detection on nanosensor platforms. These strategies are crucial to overcoming some of the key roadblocks which hinder the real-life detection of metabolites, including ultratrace metabolite concentrations, the presence of interfering species in complex sample matrices, and signal ambiguity from structurally similar analogs.At the nanobio interface, we first highlight how analyte manipulation and enrichment strategies boost nanosensor–metabolite interactions, thereby enabling the metabolites to better access the nanosensor's EM field and/or catalytically active sites for more sensitive signal responses. Based on their underlying working principles, we identify two main categories which exploit (1) chemical interactions between surface-grafted receptors and target metabolites and (2) physical metabolite accumulation near the nanosensor surface. We also showcase the strategic combination of these individually modified nanosensors in array-based configurations to generate high-dimensional signal patterns useful for pattern-based differentiation of multiplex metabolite mixtures. Notably, these strategies can be facilely applied for detection via diverse signal transduction modes, ranging from optical to electrical and electrochemical, which is important to introduce high flexibility in methodology and clinical design based on the intended end-use. Next, from the detection mode perspective, we cover the burgeoning efforts to couple different analytical techniques in hyphenated or multimodal configurations, to ‘achieve the best of both worlds’ by complementing the limitations of each and serving as important cross-validation. Finally, with respect to data analysis, we emphasize the integral role of machine learning toolkits to assimilate and analyze increasingly complex and high-dimensional data arising from developments in the aforementioned strategies. Notable applications include the rapid construction of robust prediction models for metabolite identification and/or quantification, and the uncovering of new interrelationships. Given the plethora of strategies, we can synergize one or more strategies to tailor the nanosensor platform and select the appropriate analytical instrument(s) and machine learning algorithms for data analysis based on target metabolites, analytical medium, and intended end-use.
Although nanosensors for metabolite detection have achieved tremendous progress over the past decade with a highly prospective outlook, there are still several major considerations and potential research directions toward the practical translation of these technologies. First, real-life adoption of these nanosensor platforms requires highly reproducible and efficient nanosensor fabrication on a larger scale to ensure that the collected data is comparable and reliable. However, most nanosensor platforms involve multiple modifications whereby each step is susceptible to human errors, which can lead to high inter-batch performance inconsistencies. For instance, although array-based configurations can augment the overall sensitivity through the addition of individual signal responses, batch-to-batch signal deviations for each modified nanosensor will be similarly amplified, which may overwhelm actual but subtle metabolite-induced signal changes. In this aspect, we have seen the advent of robotic platforms for nanomaterial synthesis and characterization to reduce hands-on operation and minimize human-related errors.124–126 We postulate a growing interest in the development of similar automated platforms specializing in different nanosensor modifications to produce highly consistent nanosensors, such as surface functionalization with target receptor moieties and controlled MOF encapsulation.
Next, the integration of ML-assisted nanosensor platforms in real-life detection workflows is also hindered by a fundamental lack of trust in the prediction results due to poor model interpretability. As datasets become more high-dimensional, scientists need to turn towards more complex ML algorithms for better prediction performances. However, these ML systems are often ‘black boxes’ in nature and suffer from ambiguous and unclear decision-making processes, which affects their reliability. Hence, we have observed a prominent shift in focus towards ‘explainable ML’ to improve the model interpretability by enabling the algorithms to trace and rationalize their decision-making processes in a manner that can be comprehended by scientists. For example, methods such as SHapley Additive exPlanations (SHAP) help to characterize the relative importance of input features for a constructed model, which can be interpreted together with domain knowledge to better guide the selection of scientifically relevant input features.127,128 Recently, a novel model framework Model Understanding through Subspace Explanations (MUSE) is also proposed to allow scientists to incorporate additional constraints to output various interpretable explanations that include only selected features of interest.129 This is especially useful to generate more specific interpretations using different input feature subspaces that can explain either a single sample or a smaller sample subset. These explainable ML algorithms will be vital to building greater confidence in the robustness and scientific soundness of the ML models, facilitating their widespread integration into various healthcare, environmental, and agri-food setting and normalizing their use.
Third, the demand for rapid, point-of-need platforms requires further research on the miniaturization of analytical platforms. This is especially pertinent for hybrid techniques since the combination of two techniques often results in bulky instrumentation. Current advances in microelectronics have allowed the miniaturization of spectrometry instruments from entire rooms to benchtops and into the hand. In addition, advancements in multiplex-friendly microfluidic chips have also introduced additional advantages including minimizing contamination and manual handling as well as increasing overall platform stability and detection efficiency.130 Nevertheless, future research can focus on improving the sensitivity and stability of the signal readouts of such handheld instruments so that miniaturization will be more than just a space saver. Fundamental investigations to understand the physicochemical fluid dynamics in the microenvironments are also essential to truly realize the lab-on-a-chip concept for biodetection.
Finally, to future-proof the technology to detect and discover novel metabolites that we do not currently have profiles for, effective data analyses and mining to conduct similarity analysis within and between classes of small-molecule metabolites are pertinent.131 For instance, the construction of a ‘protein structure space’ employing pairwise structural similarity space demonstrates strong structure-to-property relationships, whereby proteins with similar structures and protein functions cluster closer together on the map.132 A similar clustering map performed on 3116 chemicals associated with skin sensitization enabled facile visualization of the molecular structure–property relationships of positive sensitizers and construction of models to predict skin sensitization of new chemicals.133 These studies emphasize the importance of leveraging molecular structure similarity analyses of metabolites to allow rapid identification, classification, and inference of their function, expression, and regulation. Importantly, such repositories will enable the prediction and assignment of functions to novel, uncharacterized homologues.
All in all, given the flexible customizability of the nanosensor platform, detection technique and advanced data analytic algorithms, we anticipate the continuous burgeoning research in the design of next-generation nanosensor platforms for rapid, point-of-need detection of various small-molecule metabolites. Such advancements will enable us to explore exciting new opportunities in fundamental nanobio investigations as well as real-world applications in key areas including biomedicine, pharmaceutics, agri-food and environmental surveillance.
Author contributions
Writing-original draft: S. X. L., Y. X. L., C. S. K. L., E. X. T., L. B. T. N., J. R. T. C., C. C., D. W. C. P., H. Y. F. S. and X. L. performed literature search and wrote the original manuscript draft, with the following section allocations: S. X. L. for Sections 1, 2.3 and Conclusion, D. W. C. P., H. Y. F. S. and X. L. for Sections 2.1–2.3, E. X. T. and J. R. T. C. for Section 2.4, C. S. K. L. and C. C. for Section 3, Y. X. L. and L. B. T. N. for Section 4. Visualization: S. X. L., Y. X. L., C. S. K. L., E. X. T., L. B. T. N., J. R. T. C. and C. C. prepared the tables and figures. Writing-review & editing: S. X. L., Y. X. L., N. S. T. and X. Y. L. reviewed and edited the manuscript drafts. Conceptualization and supervision: X. Y. L. provided the initial perspective direction and supervised the writing and literature consolidation process.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This research is supported by the Singapore National Medical Research Council COVID-19 Grant (MOH-000584) and A*STAR AME Individual Research Grant (A20E5c0082). S. X. L. and L. B. T. N. acknowledge Nanyang President's Graduate Scholarship support from Nanyang Technological University, Singapore.References
- S. Lamichhane, P. Sen, A. M. Dickens, T. Hyötyläinen and M. Orešič, in Comprehensive Analytical Chemistry, ed. J. Jaumot, C. Bedia and R. Tauler, Elsevier, 2018, vol. 82, pp. 387–413 Search PubMed.
- T. Ramirez, M. Daneshian, H. Kamp, F. Y. Bois, M. R. Clench, M. Coen, B. Donley, S. M. Fischer, D. R. Ekman, E. Fabian, C. Guillou, J. Heuer, H. T. Hogberg, H. Jungnickel, H. C. Keun, G. Krennrich, E. Krupp, A. Luch, F. Noor, E. Peter, B. Riefke, M. Seymour, N. Skinner, L. Smirnova, E. Verheij, S. Wagner, T. Hartung, B. van Ravenzwaay and M. Leist, ALTEX, 2013, 30, 209–225 CrossRef PubMed.
- R. D. Beger, M. A. Schmidt and R. Kaddurah-Daouk, Metabolites, 2020, 10, 129 CrossRef CAS.
- Z. Jia, A. Patra, V. K. Kutty and T. Venkatesan, Metabolites, 2019, 9, 52 CrossRef CAS PubMed.
- L. Deng, K. Ismond, Z. Liu, J. Constable, H. Wang, O. I. Alatise, M. R. Weiser, T. P. Kingham and D. Chang, Cancer Epidemiol., Biomarkers Prev., 2019, 28, 1283–1291 CrossRef PubMed.
- Y. Cho, Y. Park, B. Sim, J. Kim, H. Lee, S.-N. Cho, Y. A. Kang and S.-G. Lee, Sci. Rep., 2020, 10, 3825 CrossRef CAS PubMed.
- P. Pandey, V. Irulappan, M. V. Bagavathiannan and M. Senthil-Kumar, Front. Plant Sci., 2017, 8, 537 Search PubMed.
- R. Jónsdóttir, G. Ólafsdóttir, E. Chanie and J.-E. Haugen, Food Chem., 2008, 109, 184–195 CrossRef PubMed.
- E. Cubero-Leon, R. Peñalver and A. Maquet, Food Res. Int., 2014, 60, 95–107 CrossRef CAS.
- A. Erban, I. Fehrle, F. Martinez-Seidel, F. Brigante, A. L. Más, V. Baroni, D. Wunderlin and J. Kopka, Sci. Rep., 2019, 9, 9697 CrossRef PubMed.
- S. Pavagadhi and S. Swarup, Metabolites, 2020, 10, 197 CrossRef CAS.
- K. Kobayashi, J. Wei, R. Iida, K. Ijiro and K. Niikura, Polym. J., 2014, 46, 460–468 CrossRef CAS.
- S. X. Leong, Y. X. Leong, C. S. L. Koh, J. R. T. Chen and X. Y. Ling, in Principles and Clinical Diagnostic Applications of Surface-Enhanced Raman Spectroscopy, ed. Y. Wang, Elsevier, 2022, pp. 33–79, DOI: DOI:10.1016/B978-0-12-821121-2.00005-6.
- Y.-Q. Chen and C.-J. Lu, Sens. Actuators, B, 2009, 135, 492–498 CrossRef CAS.
- C. S. L. Koh, H. Y. F. Sim, S. X. Leong, S. K. Boong, C. Chong and X. Y. Ling, ACS Mater. Lett., 2021, 3, 557–573 CrossRef CAS.
- M. Holzinger, A. Le Goff and S. Cosnier, Front. Chem., 2014, 2, 63 Search PubMed.
- H. Kumar, K. Kuča, S. K. Bhatia, K. Saini, A. Kaushal, R. Verma, T. C. Bhalla and D. Kumar, Sensors (Basel, Switzerland), 2020, 20, 1966 CrossRef CAS.
- K. Ataka and J. Heberle, Anal. Bioanal. Chem., 2007, 388, 47–54 CrossRef CAS PubMed.
- F. Neubrech, C. Huck, K. Weber, A. Pucci and H. Giessen, Chem. Rev., 2017, 117, 5110–5145 CrossRef CAS PubMed.
- X. X. Han, R. S. Rodriguez, C. L. Haynes, Y. Ozaki and B. Zhao, Nat. Rev. Methods Primers, 2022, 1, 87 CrossRef.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- W. R. Premasiri, J. C. Lee, A. Sauer-Budge, R. Théberge, C. E. Costello and L. D. Ziegler, Anal. Bioanal. Chem., 2016, 408, 4631–4647 CrossRef CAS PubMed.
- C.-Y. Chang, H.-T. Lin, M.-S. Lai, T.-Y. Shieh, C.-C. Peng, M.-H. Shih and Y.-C. Tung, Sci. Rep., 2018, 8, 11812 CrossRef PubMed.
- J.-S. Chen, P.-F. Chen, H. T.-H. Lin and N.-T. Huang, Analyst, 2020, 145, 7654–7661 RSC.
- B. Liu, J. Zhuang and G. Wei, Environ. Sci.: Nano, 2020, 7, 2195–2213 RSC.
- Z. Li, J. R. Askim and K. S. Suslick, Chem. Rev., 2019, 119, 231–292 CrossRef CAS PubMed.
- Y. Zhang and T. H. Wang, Theranostics, 2012, 2, 631–654 CrossRef CAS PubMed.
- M. Li, T. Chen, J. J. Gooding and J. Liu, ACS Sens., 2019, 4, 1732–1748 CrossRef CAS PubMed.
- Z. Zhang, S. Shikha, J. Liu, J. Zhang, Q. Mei and Y. Zhang, Anal. Chem., 2019, 91, 548–568 CrossRef CAS PubMed.
- M. K. Nakhleh, H. Amal, R. Jeries, Y. Y. Broza, M. Aboud, A. Gharra, H. Ivgi, S. Khatib, S. Badarneh, L. Har-Shai, L. Glass-Marmor, I. Lejbkowicz, A. Miller, S. Badarny, R. Winer, J. Finberg, S. Cohen-Kaminsky, F. Perros, D. Montani, B. Girerd, G. Garcia, G. Simonneau, F. Nakhoul, S. Baram, R. Salim, M. Hakim, M. Gruber, O. Ronen, T. Marshak, I. Doweck, O. Nativ, Z. Bahouth, D.-y. Shi, W. Zhang, Q.-l. Hua, Y.-y. Pan, L. Tao, H. Liu, A. Karban, E. Koifman, T. Rainis, R. Skapars, A. Sivins, G. Ancans, I. Liepniece-Karele, I. Kikuste, I. Lasina, I. Tolmanis, D. Johnson, S. Z. Millstone, J. Fulton, J. W. Wells, L. H. Wilf, M. Humbert, M. Leja, N. Peled and H. Haick, ACS Nano, 2017, 11, 112–125 CrossRef CAS PubMed.
- Z. Shi, Y. Lu, Z. Chen, C. Cheng, J. Xu, Q. Zhang, Z. Yan, Z. Luo and Q. Liu, Sens. Actuators, B, 2021, 329, 129197 CrossRef CAS.
- G. A. Gowda, S. Zhang, H. Gu, V. Asiago, N. Shanaiah and D. Raftery, Expert Rev. Mol. Diagn., 2008, 8, 617–633 CrossRef CAS PubMed.
- A. H. Emwas, R. Roy, R. T. McKay, L. Tenori, E. Saccenti, G. A. N. Gowda, D. Raftery, F. Alahmari, L. Jaremko, M. Jaremko and D. S. Wishart, Metabolites, 2019, 9, 123 CrossRef CAS PubMed.
- J.-L. Ren, A.-H. Zhang, L. Kong and X.-J. Wang, RSC Adv., 2018, 8, 22335–22350 RSC.
- I. Aretz and D. Meierhofer, Int. J. Mol. Sci., 2016, 17, 632 CrossRef PubMed.
- C. B. Clish, Cold Spring Harbor Mol. Case Stud., 2015, 1, a000588 CrossRef PubMed.
- C. H. Johnson, J. Ivanisevic and G. Siuzdak, Nat. Rev. Mol. Cell Biol., 2016, 17, 451–459 CrossRef CAS PubMed.
- S. Grassin-Delyle, C. Roquencourt, P. Moine, G. Saffroy, S. Carn, N. Heming, J. Fleuriet, H. Salvator, E. Naline, L.-J. Couderc, P. Devillier, E. A. Thévenot and D. Annane, EBioMedicine, 2021, 63, 103154 CrossRef CAS PubMed.
- S. O. Kelley, ACS Sens., 2017, 2, 193–197 CrossRef CAS PubMed.
- Y. Kiriyama and H. Nochi, Scientifica (Cairo), 2016, 2016, 6494621 Search PubMed.
- Z. Wei Poh, C. Heng Gan, E. J. Lee, S. Guo, G. W. Yip and Y. Lam, Sci. Rep., 2015, 5, 14355 CrossRef PubMed.
- J. Iida, J. Dorchak, R. Clancy, J. Slavik, R. Ellsworth, Y. Katagiri, E. N. Pugacheva, T. H. van Kuppevelt, R. J. Mural, M. L. Cutler and C. D. Shriver, Exp. Cell Res., 2015, 330, 358–370 CrossRef CAS PubMed.
- G. Vessella, J. A. Vázquez, J. Valcárcel, L. Lagartera, D. T. Monterrey, A. Bastida, E. García-Junceda, E. Bedini, A. Fernández-Mayoralas and J. Revuelta, Polymers, 2021, 13, 313 CrossRef CAS PubMed.
- S. X. Leong, L. K. Koh, C. S. L. Koh, G. C. Phan-Quang, H. K. Lee and X. Y. Ling, ACS Appl. Mater. Interfaces, 2020, 12, 33421–33427 CrossRef CAS.
- R. Nißler, A. T. Müller, F. Dohrman, L. Kurth, H. Li, E. G. Cosio, B. S. Flavel, J. P. Giraldo, A. Mithöfer and S. Kruss, Angew. Chem., Int. Ed., 2022, 61, e202108373 Search PubMed.
- R. Ma, J. Hu, Z. Cai and H. Ju, Nanoscale, 2014, 6, 3150–3156 RSC.
- K. V. Kong, Z. Lam, W. K. O. Lau, W. K. Leong and M. Olivo, J. Am. Chem. Soc., 2013, 135, 18028–18031 CrossRef CAS PubMed.
- J. Zhao, J. Gao, W. Xue, Z. Di, H. Xing, Y. Lu and L. Li, J. Am. Chem. Soc., 2018, 140, 578–581 CrossRef CAS PubMed.
- Y. Wang, X. Zhao, Z. Yu, Z. Xu, B. Zhao and Y. Ozaki, Angew. Chem., Int. Ed., 2020, 59, 19079 CrossRef CAS PubMed.
- M. R. K. Ali, Y. Wu, T. Han, X. Zang, H. Xiao, Y. Tang, R. Wu, F. M. Fernández and M. A. El-Sayed, J. Am. Chem. Soc., 2016, 138, 15434–15442 CrossRef CAS PubMed.
- Z. Zhang, X. Han, Z. Wang, Z. Yang, W. Zhang, J. Li, H. Yang, X. Y. Ling and B. Xing, Chem. Commun., 2018, 54, 7022–7025 RSC.
- X. Li, H. K. Lee, I. Y. Phang, C. K. Lee and X. Y. Ling, Anal. Chem., 2014, 86, 10437–10444 CrossRef CAS.
- H. K. Lee, Y. H. Lee, Q. Zhang, I. Y. Phang, J. M. R. Tan, Y. Cui and X. Y. Ling, ACS Appl. Mater. Interfaces, 2013, 5, 11409–11418 CrossRef CAS.
- Y.-C. Kao, X. Han, Y. H. Lee, H. K. Lee, G. C. Phan-Quang, C. L. Lay, H. Y. F. Sim, V. J. X. Phua, L. S. Ng, C. W. Ku, T. C. Tan, I. Y. Phang, N. S. Tan and X. Y. Ling, ACS Nano, 2020, 14, 2542–2552 CrossRef CAS PubMed.
- X. Huang, B. Li, L. Wang, X. Lai, H. Xue and J. Gao, ACS Appl. Mater. Interfaces, 2019, 11, 24533–24543 CrossRef CAS PubMed.
- H. K. Lee, Y. H. Lee, J. V. Morabito, Y. Liu, C. S. L. Koh, I. Y. Phang, S. Pedireddy, X. Han, L.-Y. Chou, C.-K. Tsung and X. Y. Ling, J. Am. Chem. Soc., 2017, 139, 11513–11518 CrossRef CAS.
- C. Fan, X. Lv, F. Liu, L. Feng, M. Liu, Y. Cai, H. Liu, J. Wang, Y. Yang and H. Wang, ACS Sens., 2018, 3, 441–450 CrossRef CAS PubMed.
- L. E. Kreno, J. T. Hupp and R. P. Van Duyne, Anal. Chem., 2010, 82, 8042–8046 CrossRef CAS PubMed.
- C. S. L. Koh, H. K. Lee, X. Han, H. Y. F. Sim and X. Y. Ling, Chem. Commun., 2018, 54, 2546–2549 RSC.
- L. Liu, C. He, S. P. Morgan, R. Correia and S. Korposh, Proc. SPIE, Seventh European Workshop on Optical Fibre Sensors, 2019, vol. 11199, pp. 111990Z Search PubMed.
- M. Weber, J.-H. Kim, J.-H. Lee, J.-Y. Kim, I. Iatsunskyi, E. Coy, M. Drobek, A. Julbe, M. Bechelany and S. S. Kim, ACS Appl. Mater. Interfaces, 2018, 10, 34765–34773 CrossRef CAS PubMed.
- X. Xu, D. Ji, Y. Zhang, X. Gao, P. Xu, X. Li, C.-C. Liu and W. Wen, ACS Appl. Mater. Interfaces, 2019, 11, 20734–20742 CrossRef CAS PubMed.
- W.-T. Koo, J.-S. Jang and I.-D. Kim, Chem, 2019, 5, 1938–1963 CAS.
- L. Liu, Y. Zhou, S. Liu and M. Xu, ChemElectroChem, 2018, 5, 6–19 CrossRef CAS.
- Z. Han, K. Wang, Y. Guo, W. Chen, J. Zhang, X. Zhang, G. Siligardi, S. Yang, Z. Zhou, P. Sun, W. Shi and P. Cheng, Nat. Commun., 2019, 10, 5117 CrossRef PubMed.
- S. Okur, P. Qin, A. Chandresh, C. Li, Z. Zhang, U. Lemmer and L. Heinke, Angew. Chem., Int. Ed., 2021, 60, 3566–3571 CrossRef CAS PubMed.
- Y. Zhu, Y. Zhou, X. Zhang, Z. Sun and C. Jiao, Adv. Opt. Mater., 2021, 9, 2001889 CrossRef CAS.
- S. Wang, H. Li, H. Huang, X. Cao, X. Chen and D. Cao, Chem. Soc. Rev., 2022, 51, 2031–2080 RSC.
- X. Qiao, B. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. Wang, Adv. Mater., 2018, 30, 1702275 CrossRef.
- S. X. Leong, Y. X. Leong, E. X. Tan, H. Y. F. Sim, C. S. L. Koh, Y. H. Lee, C. Chong, L. S. Ng, J. R. T. Chen, D. W. C. Pang, L. B. T. Nguyen, S. K. Boong, X. Han, Y.-C. Kao, Y. H. Chua, G. C. Phan-Quang, I. Y. Phang, H. K. Lee, M. Y. Abdad, N. S. Tan and X. Y. Ling, ACS Nano, 2022, 16, 2629–2639 CrossRef CAS PubMed.
- Y. Geng, W. J. Peveler and V. M. Rotello, Angew. Chem., Int. Ed., 2019, 58, 5190–5200 CrossRef CAS PubMed.
- L. Shang, C. Liu, B. Chen and K. Hayashi, ACS Sens., 2018, 3, 1531–1538 CrossRef CAS PubMed.
- B. Shan, Y. Y. Broza, W. Li, Y. Wang, S. Wu, Z. Liu, J. Wang, S. Gui, L. Wang, Z. Zhang, W. Liu, S. Zhou, W. Jin, Q. Zhang, D. Hu, L. Lin, Q. Zhang, W. Li, J. Wang, H. Liu, Y. Pan and H. Haick, ACS Nano, 2020, 14, 12125–12132 CrossRef CAS PubMed.
- A. D. Gill, B. L. Hickey, W. Zhong and R. J. Hooley, Chem. Commun., 2020, 56, 4352–4355 RSC.
- N. Kim, M. R. Thomas, M. S. Bergholt, I. J. Pence, H. Seong, P. Charchar, N. Todorova, A. Nagelkerke, A. Belessiotis-Richards, D. J. Payne, A. Gelmi, I. Yarovsky and M. M. Stevens, Nat. Commun., 2020, 11, 207 CrossRef CAS PubMed.
- A. Star, V. Joshi, S. Skarupo, D. Thomas and J.-C. P. Gabriel, J. Phys. Chem. B, 2006, 110, 21014–21020 CrossRef CAS PubMed.
- S. Pandit, T. Banerjee, I. Srivastava, S. Nie and D. Pan, ACS Sens., 2019, 4, 2730–2737 CrossRef CAS PubMed.
- B. Li, X. Li, Y. Dong, B. Wang, D. Li, Y. Shi and Y. Wu, Anal. Chem., 2017, 89, 10639–10643 CrossRef CAS PubMed.
- M. M. Bordbar, J. Tashkhourian and B. Hemmateenejad, ACS Sens., 2019, 4, 1442–1451 CrossRef CAS PubMed.
- W. Cuypers and P. A. Lieberzeit, Front. Chem., 2018, 6, 268 CrossRef.
- Y. Geng, W. J. Peveler and V. M. Rotello, Angew. Chem., Int. Ed., 2019, 58, 5190–5200 CrossRef CAS PubMed.
- T. Hallaj, N. Azizi and M. Amjadi, Microchem. J., 2021, 162, 105865 CrossRef CAS.
- X. Zhang, H. Zhi, M. Zhu, F. Wang, H. Meng and L. Feng, Biosens. Bioelectron., 2021, 180, 113146 CrossRef CAS PubMed.
- I. Alessandri, I. Vassalini, M. Bertuzzi, N. Bontempi, M. Memo and A. Gianoncelli, Sci. Rep., 2016, 6, 34521 CrossRef CAS.
- D. S. Burr, W. L. Fatigante, J. A. Lartey, W. Jang, A. R. Stelmack, N. W. McClurg, J. M. Standard, J. R. Wieland, J.-H. Kim, C. C. Mulligan and J. D. Driskell, Anal. Chem., 2020, 92, 6676–6683 CrossRef CAS.
- B. L. Goodall, A. M. Robinson and C. L. Brosseau, Phys. Chem. Chem. Phys., 2013, 15, 1382–1388 RSC.
- L. Zhao, J. Blackburn and C. L. Brosseau, Anal. Chem., 2015, 87, 441–447 CrossRef CAS PubMed.
- C.-Y. Huang and H.-C. Hsiao, Sensors, 2020, 20, 7066 CrossRef CAS PubMed.
- B. H. C. Greene, D. S. Alhatab, C. C. Pye and C. L. Brosseau, J. Phys. Chem. C, 2017, 121, 8084–8090 CrossRef.
- S. D. Bindesri, R. Jebailey, N. Albarghouthi, C. C. Pye and C. L. Brosseau, Analyst, 2020, 145, 1849–1857 RSC.
- S. X. Leong, C. S. L. Koh, H. Y. F. Sim, Y. H. Lee, X. Han, G. C. Phan-Quang and X. Y. Ling, ACS Nano, 2021, 15, 1817–1825 CrossRef CAS PubMed.
- F. De Biasi, D. Rosa-Gastaldo, F. Mancin and F. Rastrelli, Chem. Commun., 2021, 57, 3002–3005 RSC.
- A. Subaihi, D. K. Trivedi, K. A. Hollywood, J. Bluett, Y. Xu, H. Muhamadali, D. I. Ellis and R. Goodacre, Anal. Chem., 2017, 89, 6702–6709 CrossRef CAS.
- F. De Biasi, F. Mancin and F. Rastrelli, Prog. Nucl. Magn. Reson. Spectrosc., 2020, 117, 70–88 CrossRef CAS PubMed.
- J. D. Weatherston, R. K. O. Seguban, D. Hunt and H.-J. Wu, ACS Sens., 2018, 3, 852–857 CrossRef CAS.
- L. Xiao, C. Wang, C. Dai, L. E. Littlepage, J. Li and Z. D. Schultz, Angew. Chem., Int. Ed., 2020, 59, 3439–3443 CrossRef CAS PubMed.
- M. I. Jordan and T. M. Mitchell, Science, 2015, 349, 255–260 CrossRef CAS PubMed.
- J. Burrell, Big Data & Society, 2016, 3, 2053951715622512 Search PubMed.
- D. M. Camacho, K. M. Collins, R. K. Powers, J. C. Costello and J. J. Collins, Cell, 2018, 173, 1581–1592 CrossRef CAS.
- I. H. Witten, E. Frank and M. A. Hall, in Data Mining: Practical Machine Learning Tools and Techniques, ed. I. H. Witten, E. Frank and M. A. Hall, Morgan Kaufmann, Boston, 3rd edn, 2011, pp. 85–145, DOI: DOI:10.1016/B978-0-12-374856-0.00004-3.
- J. Vlasblom and S. J. Wodak, BMC Bioinf., 2009, 10, 99 CrossRef PubMed.
- F. Cui, Y. Yue, Y. Zhang, Z. Zhang and H. S. Zhou, ACS Sens., 2020, 5, 3346–3364 CrossRef CAS PubMed.
- U. W. Liebal, A. N. T. Phan, M. Sudhakar, K. Raman and L. M. Blank, Metabolites, 2020, 10, 243 CrossRef CAS PubMed.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, TrAC, Trends Anal. Chem., 2020, 124, 115796 CrossRef CAS.
- T. O. Ayodele, New Adv. Mach. Learn., 2010, 3, 19–48 Search PubMed.
- M. Fatima and M. Pasha, J. Intell. Learn Syst. Appl., 2017, 9, 1 Search PubMed.
- S. Wold, K. Esbensen and P. Geladi, Chemom. Intell. Lab. Syst., 1987, 2, 37–52 CrossRef CAS.
- P. Dharmalingam, K. Venkatakrishnan and B. Tan, ACS Nano, 2021, 15, 9967–9986 CrossRef PubMed.
- H. Shin, H. Jeong, J. Park, S. Hong and Y. Choi, ACS Sens., 2018, 3, 2637–2643 CrossRef CAS.
- F. Murtagh and P. Legendre, J. Classif., 2014, 31, 274–295 CrossRef.
- M. Barker and W. Rayens, J. Chemom., 2003, 17, 166–173 CrossRef CAS.
- E. Szymańska, E. Saccenti, A. K. Smilde and J. A. Westerhuis, Metabolomics, 2012, 8, 3–16 CrossRef.
- Z. R. Yang, R. Thomson, P. McNeil and R. M. Esnouf, Bioinformatics, 2005, 21, 3369–3376 CrossRef CAS PubMed.
- S. Wold, M. Sjöström and L. Eriksson, Chemom. Intell. Lab. Syst., 2001, 58, 109–130 CrossRef CAS.
- H. A. Farahani, A. Rahiminezhad, L. Same and K. immannezhad, Procedia Soc. Behav. Sci., 2010, 5, 1459–1463 CrossRef.
- L. Pascual, I. Campos, J.-L. Vivancos, G. Quintás, A. Loras, M. C. Martínez-Bisbal, R. Martínez-Máñez, F. Boronat and J. L. Ruiz-Cerdà, Analyst, 2016, 141, 4562–4567 RSC.
- S. H. Lim, R. Martino, V. Anikst, Z. Xu, S. Mix, R. Benjamin, H. Schub, M. Eiden, P. A. Rhodes and N. Banaei, ACS Sens., 2016, 1, 852–856 CrossRef CAS.
- F. Lussier, D. Missirlis, J. P. Spatz and J.-F. Masson, ACS Nano, 2019, 13, 1403–1411 CAS.
- H. Shin, S. Oh, S. Hong, M. Kang, D. Kang, Y.-g. Ji, B. H. Choi, K.-W. Kang, H. Jeong, Y. Park, S. Hong, H. K. Kim and Y. Choi, ACS Nano, 2020, 14, 5435–5444 CrossRef CAS.
- Y. Takahashi, M. Ueki, M. Yamada, G. Tamiya, I. N. Motoike, D. Saigusa, M. Sakurai, F. Nagami, S. Ogishima, S. Koshiba, K. Kinoshita, M. Yamamoto and H. Tomita, Transl. Psychiatry, 2020, 10, 157 CrossRef PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- J. Demšar, T. Curk, A. Erjavec, Č. Gorup, T. Hočevar, M. Milutinovič, M. Možina, M. Polajnar, M. Toplak and A. Starič, J. Mach. Learn. Res., 2013, 14, 2349–2353 Search PubMed.
- M. Abadi, P. Barham, J. Chen, Z. Chen, A. Davis, J. Dean, M. Devin, S. Ghemawat, G. Irving and M. Isard, arXiV: 1605.08695[cs], 2016.
- C. D. Ahrberg, J. Wook Choi and B. Geun Chung, Sci. Rep., 2020, 10, 1737 CrossRef CAS PubMed.
- H. Tao, T. Wu, M. Aldeghi, T. C. Wu, A. Aspuru-Guzik and E. Kumacheva, Nat. Rev. Mater., 2021, 6, 701–716 CrossRef.
- Y. Li, L. Xia, Y. Fan, Q. Wang and M. Hu, ChemPhysMater, 2022, 1, 77–85 CrossRef.
- R. Rodríguez-Pérez and J. Bajorath, J. Comput.-Aided Mol. Des., 2020, 34, 1013–1026 CrossRef PubMed.
- A. K. Chew, J. A. Pedersen and R. C. Van Lehn, ACS Nano, 2022, 16, 6282–6292 CrossRef CAS.
- H. Lakkaraju, E. Kamar, R. Caruana and J. Leskovec, presented in part at the Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society, Honolulu, HI, USA, 2019 Search PubMed.
- A. Jagannath, H. Cong, J. Hassan, G. Gonzalez, M. D. Gilchrist and N. Zhang, Biosens. Bioelectron.: X, 2022, 10, 100134 CAS.
- C.-S. Lee, Front. Sens., 2020, 1, 583035 CrossRef.
- J. Hou, S.-R. Jun, C. Zhang and S.-H. Kim, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 3651–3656 CrossRef CAS.
- T. Luechtefeld, A. Maertens, D. P. Russo, C. Rovida, H. Zhu and T. Hartung, ALTEX, 2016, 33, 135–148 Search PubMed.
| This journal is © The Royal Society of Chemistry 2022 |
