A remarkable sensitivity enhancement in a gold nanoparticle-based lateral flow immunoassay for the detection of Escherichia coli O157:H7†
Xi Cuiab,
Youju Huang*b,
Jingyun Wangab,
Lei Zhangb,
Yun Rongb,
Weihua Lai*a and
Tao Chen*b
aState Key Laboratory of Food Science and Technology, Nanchang University, Nanchang 330047, China. E-mail: talktolaiwh@163.com
bDivision of Polymer and Composite Materials, Ningbo Institute of Material Technology and Engineering, Chinese Academy of Science, No. 1219 Zhongguan West Road, Zhenhai District, Ningbo 315201, China. E-mail: yjhuang@nimte.ac.cn; tao.chen@nimte.ac.cn
First published on 8th May 2015
Abstract
Because of the distinctive features of ease-of-use, low cost and portable detection, a gold nanoparticle (AuNP) based lateral flow immunoassay (LFA) is an effective and currently used method for the detection of Escherichia coli O157:H7; however, its low sensitivity limits its practical use. In the present study, the size and uniformity of AuNPs have been systematically optimized to maximally amplify both the visual inspection signals (the color of test line) and the quantitative data (light intensity) recorded using a bioassay reader. The remarkable enhancement of detection sensitivity can be increased to 102 colony forming units per mL by taking advantage of the optimized AuNPs and the separated incubation of the AuNPs/antibody/E. coli O157:H7 complex. Quantitative detection of E. coli O157:H7 was partially obtained in a wide concentration range with good repeatability. The new, optimized AuNPs-based LFA is well suited to fast quantitative and qualitative food analysis.
1. Introduction
Since its discovery in 1982, Escherichia coli O157:H7 has become a public health concern worldwide because of its capability of contaminating food and water, causing severe intestinal infection in humans such as gastrointestinal illness, hemolytic uremic syndrome, and occasionally kidney failure. The great demand for a rapid, accurate sensor that can be used widely is an urgent and challenging task for food safety. In the past few decades, numerous efforts have been devoted to various methods for the detection of E. coli O157:H7, including a molecular biological method (polymerase chain reaction-based analysis),1 DNA and RNA probes,2 surface plasmon resonance,3 immunomagnetic separation analysis,4 electrochemical biosensor,5 and quartz crystal microbalance techniques.6 Although each approach has its own advantages, most reports are usually of a time consuming method, and/or one that requires expensive equipment and skilled workers.From the standpoint of ease/broadness of use, low cost and portable detection, lateral flow immunoassay (LFA), particularly, gold nanoparticle (AuNPs) based LFA, is an effective and currently used method,7,8 which is ideally suitable for on-site testing. Wang et al.9 have reported a visually inspected LFA using AuNPs for simultaneous detection of three pesticides. Parolo et al.10 have reported an enhanced LFA using AuNPs loaded with enzymes capable of detecting human immunoglobulin G (IgG). This paper was the inspiration establishing a special AuNP-based LFA to detect E. coli O157:H7,8 but the sensitivity (104 colony forming units (CFU) mL−1) was not very high and limits its practical use as a commercial product. The objective of the present study was to develop a high performance AuNP-based LFA for E. coli O157:H7 detection that would achieve high sensitivity and stability.
Generally, there are three ways to improve the detection sensitivity of an AuNP-based LFA such as designing a sensitive bioassay reader, optimizing the test procedure and improving the properties of the probe materials.11 Compared with the other two ways, exploring the optimized label probes for LFA is a simple and effective means to improve the detection sensitivity. It is well known that AuNPs play a critical role in LFA, because both visual inspection signals (the color of the test line) and quantitative data (light intensity) recorded by the ‘Assay Reader’ originate from AuNPs, which are mainly dependent on their size, shapes, uniformity and aggregation behaviours,12–27 as well as the wavelength coupling between the light source and AuNPs.
In this present study, the size and uniformity of AuNPs were systematically optimized to maximally amplify the detection signals to improve the sensitivity. In addition, the separated incubation of AuNPs/antibody/E. coli O157:H7 complex was used to modify the conventional AuNP-based LFA to improve the stability. The new optimized AuNPs-based LFA allowed us to detect E. coli O157:H7 with an estimated detection limit of 102 CFU mL−1, which is much lower (two orders in concentration) than that of a conventional AuNP-based LFA.
2. Experimental section
2.1. Reagents
Hydrogen tetrachloroaurate(III) trihydrate (HAuCl4·3H2O), sodium citrate (C6H5Na3O7·2H2O), and bovine serum albumin (BSA) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). The sample pad, nitrocellulose (NC) membrane, conjugate release pad, and absorbent pad were obtained from Schleicher and Schuell GmbH (Dassel, Germany). The antibody pairs, namely, murine anti-E. coli O157:H7 monoclonal antibody and goat anti-E. coli O157:H7 polyclonal antibody and donkey anti-mouse IgG were purchased from Meridian Life Science, Inc (Memphis, TN, USA). Other reagents were of analytical grade and purchased from Sinopharm Chemical Corp. (Shanghai, China).2.2. Instruments
A SkanFlexi BioAssay reader system for reading the AuNP-based LFA strip was supplied by the Skannex Biotech Co., Ltd. (Oslo, Norway). The BioDot XYZ platform combined with a motioncontroller, BioJet Quanti3000k dispenser and an AirJetQuanti3000k dispenser for solution dispensing were purchased from BioDot (Irvine, CA USA). The structures of the nanoparticles were observed using transmission electron microscopy (TEM) on a Jeol JEM-2010 electron microscope. Ultraviolet-visible (UV-Vis) absorption spectra were recorded using a TU-1810 UV-Vis spectrophotometer obtained from the Purkinje General Instrument Co. Ltd. (Beijing, China).2.3. Bacterial strains and growth condition
Bacterial strains were prepared according to a method reported by Xie et al.8 E. coli O157:H7 strain ATCC 43888 was cultured in Luria–Bertani medium (Oxoid, Basingstoke, UK) at 37 °C for 20 h before use. Serial dilutions of cultures in phosphate buffered saline (PBS, Sigma Chemical Company, St. Louis, MO, USA, 0.01 M, pH 7.4) were made and plated onto trypticase soy agar (Becton, Dickinson and Company, Sparks Glencoe, MD, USA) to determine the number of viable E. coli O157:H7 cells. The plates were then incubated at 37 °C for 24 h. The number of cells was counted when the bacterial colonies could be clearly seen on the plates.2.4. Preparation of AuNPs of different sizes
AuNPs were prepared according to a method given in a previous report.26,28 Briefly, 100 mL of 2.5 × 10−4 M HAuCl4 solution was heated to 120 °C in an oil bath under vigorous stirring for 30 min. Then, a predetermined volume of 1% sodium citrate solution was added into the previous solution with continued boiling. After 20 min, the color of the boiled solution changed to ruby red, indicating the formation of AuNPs. The solution was cooled to room temperature before use in further experiments. Different predetermined volumes of sodium citrate solution would give different sized AuNPs.2.5. Preparation of AuNPs with different polydispersity indices (PDIs)
AuNP solutions with different PDI were prepared by mixing two AuNP samples with the pre-widened PDI. The AuNPs with a PDI of 0.20 were prepared by the previously mentioned Frens method.24 The other sample with a PDI of 0.65 was prepared as follows: 1 mL of 1% sodium citrate solution was added into 100 mL of 2.5 × 10−4 M boiled HAuCl4 solution heated with an electric heating jacket. A series of AuNPs with the same plasmonic wavelength peak and different PDI such as 0.26, 0.39, 0.47, 0.52, and 0.61 were prepared by adjusting the ratio of the amount of the two AuNPs samples with pre-widened PDI.2.6. Preparation of AuNP–antibody complex
The anti-E. coli O157:H7 monoclonal antibody (300 μL) with a concentration of 166 μg mL−1 was added into 3 mL of a pH adjusted AuNP solution and was stirred for 60 min. Then, 0.15 mL of 3% (w/v) BSA solution was added and the solution was stirred for another 30 min. Later, 0.15 mL of 10% (w/v) PEG20000 solution was added and the mixture was stirred for another 30 min. The mixture was centrifuged at 9000g for 30 min and the precipitate was then dissolved in 300 μL of 50 mM Tris/HCl buffer. The AuNP-labeled monoclonal antibody complex was dried for 2 h at 30 °C before further use.2.7. Preparation of AuNP test strips
A sample pad was pre-treated with 50 mM of borate buffer (pH = 7.4) containing 1% BSA, 0.5% Tween 20, and 0.05% sodium azide, and further dried at 60 °C for 2 h. The goat anti-E. coli O157:H7 polyclonal antibody (1.0 mg mL−1) and donkey anti-mouse IgG (1.0 mg mL−1) were sprayed onto the test (T) and control (C) lines on the NC membrane, respectively, and then dried at 35 °C. The NC membrane was blocked with 5% BSA in 0.02 M PBS (pH = 7.2) to avoid the background of the strip. The absorption pad and glass fibre membrane were used without further treatment. The NC membrane, absorption pad, glass fibre membrane, and pre-treated sample pad were assembled on a poly(vinylchloride) plate to form the final test strip.2.8. Detection of E. coli O157:H7
The E. coli O157:H7 culture was diluted into different concentrations from 2.8 × 107 to 2.8 × 102 CFU mL−1 in sterile PBS, which were then mixed with the AuNP–antibody complex. Then, the mixtures were placed onto the test strip. The SkanFlexiBioAssay Reader was used to quantitatively detect the E. coli O157:H7 after 10 min. All the experiments were performed in triplicate.3. Results and discussion
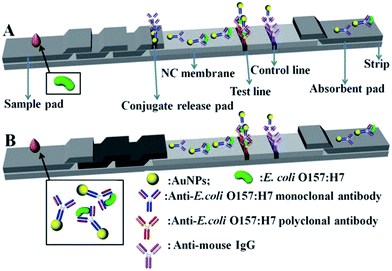
Fig. 1 shows the schematic of conventional and modified AuNP-based LFA strips. Generally, the strip contains mainly three parts such as sample pad, NC membrane, and absorbent pad (AuNP–antibody conjugate pad, T and C line with modified corresponding antibodies). In the conventional strip (Fig. 1A), the target analyte (E. coli O157:H7 culture) flow laterally through the test strip. Firstly they interact with the antibody-labelled AuNPs in the conjugate pad, and aggregate subsequently at the T line because of the specific interaction between the analyte and the antibody, which leads to a darkening color of the T line which is then detected optically. The non-E. coli O157:H7 cannot interact with the antibody-labelled AuNPs and no red line develops at the test line. The colored antibody-labelled AuNPs should bind to the C line and form a red-colored band regardless of the presence of E. coli O157:H7.Unlike the conventional strip, a separated incubation of the target analysts was proposed for the modified strip (Fig. 1B), which gives LFA three distinctive advantages. Firstly, it eliminates the AuNP–antibody conjugate pad, making the fabrication of the strip easier. Secondly, the separated incubation of E. coli O157:H7 allows easier control of the reaction time (about 5 min), resulting in the complete interaction with antibody labelled AuNPs, rather than the instantaneous reaction at the AuNP–antibody conjugate pad. This effective incubation of E. coli O157:H7 with antibody labelled AuNPs gives a high detection performance. Lastly, the amount of the separated incubation of E. coli O157:H7 can be randomly increased, and used many times for a large quantity of strips, which significantly improves the stability and repeatability of the test.
It is well known that the size of the AuNPs has an important effect on the performance of AuNP-based LFA.28 Large sized AuNPs usually have a high steric hindrance, which affects the interaction with the labelled antibody/target analytes and impedes the fluidity in the strip, while AuNPs that are too small are not the best option for showing visual inspection signals. In addition, different labelled antibodies/target analytes favour the interaction with different size AuNPs. For the detection of E. coli O157:H7 on a strip, people usually randomly choose AuNPs in the range from 20 to 60 nm; there is not much reported in the literature about the precise investigation of the small window of size effect on the detection performance.
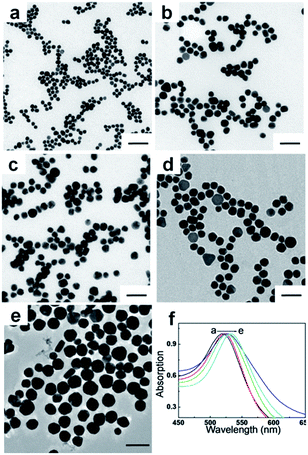
In order to explore the cut-off size where both visual inspection signals and quantitative data (light intensity) could be optimal, AuNPs (gold spheres) were successfully synthesized in a finely tuned size range from 20 to 54 nm. Fig. 2 shows the TEM images of AuNPs of various sizes. It was demonstrated that the smallest AuNPs have a relatively good uniform shape, whereas larger AuNPs are mixed with non-spherical AuNPs, and have a relatively broad size distribution. The average sizes that were obtained from the TEM results were 20 ± 1 nm, 28 ± 1 nm, 35 ± 1 nm, 43 ± 2 nm, and 54 ± 3 nm. The UV-Vis absorption spectra (Fig. 2f) correspond to samples shown in Fig. 2a–e. As the size of the AuNPs increases from 20 to 54 nm, the absorption peaks are noticeably red-shifted from 518 to 533 nm, showing a good agreement with results from previous reports. The different sized AuNPs obtained were used as labelled probes for testing the signal source to investigate the detection efficiency and sensitivity of E. coli O157:H7 in LFA.
The different sized AuNPs were first labelled with a monoclonal antibody, which interacts with E. coli O157:H7 and forms an AuNP/antibody/E. coli O157:H7 complex via the separated incubation. The complex flowed in the strip and was captured by the goat anti-E. coli O157:H7 polyclonal antibody at the T line. The visually inspected color and optical intensity at the T line can be used to evaluate the detection performances quantitatively and qualitatively, respectively.
Fig. 3 shows a typical batch of AuNP-based LFA strips used to investigate the detection performances of E. coli O157:H7 with 35 nm AuNP-based LFA. The detection procedure was repeated at least three times. In the present study, the visual inspection limit was defined as the minimum analyte concentration required for no obvious visual color on the T line. Following this definition, the visual qualitative inspection limit achieved with 35 nm AuNPs was around 103 CFU mL−1. The other sizes of AuNPs labelled LFA samples showed a visual qualitative inspection limit of 103, 104, 105 and 106 CFU mL−1, corresponding to AuNPs sizes of 20, 28, 43 and 54 nm, respectively.
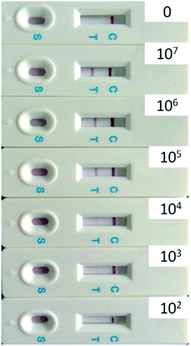 | ||
| Fig. 3 Specificity of strip for the detection of E. coli O157:H7 (from 0 to 107 CFU mL−1) using 35 nm AuNPs. | ||
In order to precisely and quantitatively study the detection performance of different sized AuNP probes, the test strips were further subjected to optical density analysis. The light scattering signal of the AuNP aggregates on the T line was monitored using a specific reader. The optical densities of the T line under different concentrations of E. coli O157:H7 from different sized AuNP labelled LFA T lines were recorded and are shown in Table 1. Generally, the optical densities decreased with the decrease of the concentration of E. coli O157:H7. Usually, a threshold value was set to determine the detection limits based on the optical density from the instrument, which can be roughly calculated from average signal intensity and standard deviation (SD) of the negative control (PBS) on the T line. The threshold value was the total contribution of the average signal intensity and three times the SD (average intensity + 3SD). The corresponding concentration of E. coli O157:H7 can be seen as the detection limit, when the detected optical value is bigger than the threshold value. The values highlighted in red in Table 1 were used to show the detection limit from different sized AuNP labelled LFA T lines. The optical density (64 au) on the T line from a 35 nm AuNPs labelled probe was strong enough to be detected even though the concentration of E. coli O157:H7 was as low as 102 CFU mL−1. The same detection process was repeated three times and similar detection limits of 102 CFU mL−1 were obtained, which indicated good reproducibility.
| Concentration (CFU mL−1) | Signal intensity of different sized AuNPs based LFA in T line | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 20 nm | 28 nm | 35 nm | 43 nm | 54 nm | ||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| PBS | 28 | 1 | 63 | 5 | 37 | 2 | 44 | 5 | 82 | 2 |
| 2.8 × 107 | 238 | 10 | 208 | 2 | 746 | 32 | 377 | 21 | 202 | 19 |
| 2.8 × 106 | 413 | 15 | 712 | 23 | 722 | 28 | 865 | 50 | 443 | 39 |
| 2.8 × 105 | 284 | 8 | 287 | 8 | 872 | 26 | 782 | 31 | 392 | 32 |
| 2.8 × 104 | 125 | 6 | 132 | 9 | 324 | 31 | 209 | 18 | 110 | 15 |
| 5.6 × 103 | 101 | 6 | 101 | 5 | 108 | 12 | 126 | 15 | 109 | 12 |
| 2.8 × 103 | 58 | 5 | 97 | 10 | 121 | 15 | 95 | 11 | 65 | 7 |
| 5.6 × 102 | 7 | 2 | 90 | 3 | 100 | 7 | 8 | 2 | 32 | 3 |
| 2.8 × 102 | 10 | 3 | 56 | 6 | 64 | 10 | 32 | 5 | 30 | 5 |
Fig. 4 shows the clear relationship between the size of the AuNPs and the detection performances of E. coli O157:H7 in LFA T lines. It is observed that the detection limit from the optical density at the T line firstly decreases as the size of the AuNPs becomes larger up to 35 nm, and then suddenly increases as the size of the AuNPs continues to increase. This finding implies that neither a small nor a big size of AuNPs benefits the detection limits of E. coli O157:H7. The cut-off size is optimized to 35 nm for the best detection performances of E. coli O157:H7 in AuNP-based LFA. As far as steric hindrance and the optical properties of AuNPs are concerned, smaller sized AuNPs generally have smaller steric hindrance, which provides a strong absorption with antibodies and E. coli O157:H7, and also facilitates their fluidity in the strip, while the larger sized AuNPs in a small size window would offer better optical signals for detection performances. Another possible reason is that the optimized size (35 nm) of the AuNPs has a plasmonic resonance wavelength of 525 nm, which is the same as the wavelength of the light source in the bioassay reader. This coupling effect of light wavelength29,30 would maximally amplify the optical properties of AuNPs for the detection of E. coli O157:H7.
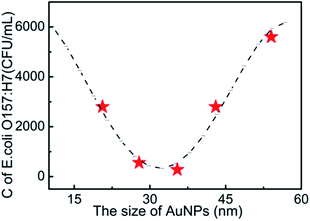 | ||
| Fig. 4 The relationship between the size of AuNPs and the detection limits of E. coli O157:H7. C is the concentration of E. coli O157:H7 at the detection limit. | ||
The size uniformity of AuNPs was further investigated to evaluate the detection performances of E. coli O157:H7 in AuNP-based LFA. In order to eliminate the light wavelength coupling effect, all the samples with different PDI (ESI Fig. 1f†) have the same plasmonic resonance wavelength at 525 nm, which were prepared by artificially mixing the different sized AuNP solutions together. The PDI of AuNPs (ESI Fig. 1a–e†) was tuned into 0.26, 0.39, 0.47, 0.52, and 0.61. The optical densities of T line under different concentrations of E. coli O157:H7 with AuNP labelled LFA with different PDIs are shown in Table 2. It is very clear to see that the higher PDIs lead to the higher detection limit (Fig. 5). It is speculated that the possible reason is follows: all samples have the same plasmonic resonance wavelength at 525 nm, which corresponds to AuNPs of 35 nm, which is the optimized size for the best detection. A sample with a lower PDI should contain more AuNPs with 35 nm, which offer bigger contributions to the detection performance; whereas the fewer AuNPs with a size of 35 nm in samples with a higher PDI would lead to the worst detection limits. A sample with a lower PDI was of much more higher uniformity than that with a higher PDI, which provided the optimized optical signals and smaller steric hindrance for absorption with the antibody and E. coli O157:H7. These conditions would lead to the best detection performance.
| Concentration (CFU mL−1) | Signal intensity of AuNPs with different PDI based LFA in T line | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.26 | 0.39 | 0.47 | 0.52 | 0.61 | ||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| PBS | 37 | 2 | 55 | 3 | 99 | 2 | 53 | 1 | 60 | 2 |
| 2.8 × 107 | 746 | 23 | 237 | 15 | 312 | 10 | 148 | 12 | 171 | 12 |
| 2.8 × 106 | 722 | 26 | 645 | 50 | 566 | 32 | 413 | 22 | 351 | 23 |
| 2.8 × 105 | 872 | 31 | 242 | 16 | 467 | 41 | 587 | 31 | 127 | 11 |
| 2.8 × 104 | 324 | 27 | 168 | 20 | 143 | 15 | 128 | 8 | 58 | 10 |
| 5.6 × 103 | 108 | 12 | 99 | 11 | 73 | 6 | 52 | 3 | 33 | 5 |
| 2.8 × 103 | 121 | 15 | 69 | 9 | 42 | 5 | 50 | 2 | 34 | 2 |
| 5.6 × 102 | 100 | 8 | 42 | 10 | 40 | 5 | 16 | 3 | 30 | 3 |
| 2.8 × 102 | 64 | 9 | 14 | 2 | 33 | 6 | 12 | 3 | 28 | 3 |
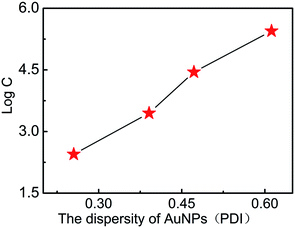 | ||
| Fig. 5 The relationship between the PDI of AuNPs and the detection limits of E. coli O157:H7 (C is the concentration of E. coli O157:H7). | ||
4. Conclusions
In this research it has been shown that a new modified AuNP-based LFA can remarkably improve the detection performance of E. coli O157:H7. Firstly, the separated incubation of E. coli O157:H7 and the antibody labelled AuNPs significantly improve the stability of the strip and the repeatability of the experimental data. Secondly, a cut-off size of AuNPs in a small size window was found to maximally amplify both visual inspection and optical signals. The newly optimized AuNP-based LFA will provide the possibility of fast quantitative and qualitative food analysis.Acknowledgements
We thank the Chinese Academy of Science for Hundred Talents Program, Chinese Central Government for Thousand Young Talents Program, the Natural Science Foundation of China (21404110, 51473179, 51303195, 21304105), Excellent Youth Foundation of Zhejiang Province of China (LR14B040001), and Ningbo Science and Technology Bureau (Grant 2014B82010). Earmarked Fund for Jiangxi Agriculture Research System (JXARS-03), Jiangxi Education Bureau Technology Put into Use project (KJLD13009), and the Nanchang Technological Program (2012-CYH-DW-SP-001).Notes and references
- D. Hardegger, D. Nadal, W. Bossart, M. Altwegg and F. Dutly, J. Microbiol. Methods, 2000, 41, 45–51 CrossRef CAS.
- X.-T. Mo, Y.-P. Zhou, H. Lei and L. Deng, Enzyme Microb. Technol., 2002, 30, 583–589 CrossRef CAS.
- H. Uchida, K. Fujitani, Y. Kawai, H. Kitazawa, A. Horii, K. Shiiba, K. Saito and T. Saito, Biosci., Biotechnol., Biochem., 2004, 68, 1004–1010 CrossRef CAS PubMed.
- X. Cui, Q.-R. Xiong, Y.-H. Xiong, S. Shan and W.-H. Lai, Chin. J. Anal. Chem., 2013, 41, 1812–1816 CrossRef CAS.
- C. M. Pandey, I. Tiwari and G. Sumana, RSC Adv., 2014, 4, 31047–31055 RSC.
- X. Su, S. Low, J. Kwang, V. H. T. Chew and S. F. Y. Li, Sens. Actuators, B, 2001, 75, 29–35 CrossRef CAS.
- E.-H. Lee, Y. A. Kim, Y. T. Lee, B. D. Hammock and H.-S. Lee, Food Agric. Immunol., 2012, 24, 129–138 CrossRef PubMed.
- Q.-Y. Xie, Y.-H. Wu, Q.-R. Xiong, H.-Y. Xu, Y.-H. Xiong, K. Liu, Y. Jin and W.-H. Lai, Biosens. Bioelectron., 2014, 54, 262–265 CrossRef CAS PubMed.
- L. M. Wang, J. Cai, Y. L. Wang, Q. K. Fang, S. Y. Wang, Q. Cheng, D. Du, Y. H. Lin and F. Q. Liu, Microchim. Acta, 2014, 181, 1565–1572 CrossRef CAS.
- C. Parolo, A. de la Escosura-Muniz and A. Merkoci, Biosens. Bioelectron., 2013, 40, 412–416 CrossRef CAS PubMed.
- L. Zhang, Y. Huang, J. Wang, Y. Rong, W. Lai, J. Zhang and T. Chen, Langmuir, 2015 DOI:10.1021/acs.langmuir.5b00592 , in press.
- H. Chen, L. Shao, Q. Li and J. Wang, Chem. Soc. Rev., 2013, 42, 2679–2724 RSC.
- L. M. Liz-Marzan, C. J. Murphy and J. Wang, Chem. Soc. Rev., 2014, 43, 3820–3822 RSC.
- Y. Huang, A. R. Ferhan, Y. Gao, A. Dandapat and D.-H. Kim, Nanoscale, 2014, 6, 6496–6500 RSC.
- J. Allouche, S. Soule, J.-C. Dupin, S. Masse, T. Coradin and H. Martinez, RSC Adv., 2014, 4, 63234–63237 RSC.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, Chem. Soc. Rev., 2012, 41, 2740–2779 RSC.
- Y. Huang, A. R. Ferhan and D.-H. Kim, Nanoscale, 2013, 5, 7772–7775 RSC.
- C. P. Rao, D. S. Yarramala and S. Doshi, RSC Adv., 2015, 5, 32761–32767 RSC.
- Y. Huang and D.-H. Kim, Nanoscale, 2011, 3, 3228–3232 RSC.
- J. Xin, R. Zhang and W. Hou, RSC Adv., 2014, 4, 5834–5837 RSC.
- S. Eustis and M. A. El-Sayed, Chem. Soc. Rev., 2006, 35, 209–217 RSC.
- Y. Huang, L. Wu, X. Chen, P. Bai and D.-H. Kim, Chem. Mater., 2013, 25, 2470–2475 CrossRef CAS.
- H. Yuan, K. P. F. Janssen, T. Franklin, G. Lu, L. Su, X. Gu, H. Uji-i, M. B. J. Roeffaers and J. Hofkens, RSC Adv., 2015, 5, 6829–6833 RSC.
- G. Frens, Nature (London), Phys. Sci., 1973, 241, 20–22 CrossRef CAS PubMed.
- L. Dykman and N. Khlebtsov, Chem. Soc. Rev., 2012, 41, 2256–2282 RSC.
- Y. Huang and D.-H. Kim, Langmuir, 2011, 27, 13861–13867 CrossRef CAS PubMed.
- N. Khlebtsov and L. Dykman, Chem. Soc. Rev., 2011, 40, 1647–1671 RSC.
- L. Zhang, L. Dai, Y. Rong, Z. Liu, D. Tong, Y. Huang and T. Chen, Langmuir, 2015, 31, 1164–1171 CrossRef CAS PubMed.
- H. Su, Y. Zhong, T. Ming, J. Wang and K. S. Wong, J. Phys. Chem. C, 2012, 116, 9259–9264 CAS.
- W. Ni, T. Ambjörnsson, S. P. Apell, H. Chen and J. Wang, Nano Lett., 2010, 10, 77–84 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra06237c |
| This journal is © The Royal Society of Chemistry 2015 |