In situ formation of peptidic nanofibers can fundamentally optimize the quality of immune responses against HIV vaccine†
Ye
Liu
a,
Huaimin
Wang
b,
Dan
Li
c,
Yue
Tian
a,
Wenwen
Liu
a,
Lingmin
Zhang
a,
Wenshu
Zheng
a,
Yanling
Hao
c,
Jiandong
Liu
c,
Zhimou
Yang
b,
Yiming
Shao
*cde and
Xingyu
Jiang
*a
aBeijing Engineering Research Center for BioNanotechnology and CAS Key Laboratory for Biological Effects of Nanomaterials and Nanosafety, National Center for NanoScience and Technology, No., 11 Zhongguancun Beiyitiao, Beijing 100190, China. E-mail: xingyujiang@nanoctr.cn; Tel: +86 10 82545558
bState Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Bioactive Materials, Ministry of Education, and College of Life Sciences, and Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Nankai University, Tianjin 300071, China
cState Key Laboratory of Infectious Disease Prevention and Control, National Center for AIDS/STD Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China. E-mail: yshao08@gmail.com; Tel: +86 10 58900981
dCenter of Infectious Diseases, Peking University, Beijing, China and Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China
eSchool of Medicine, Nankai University, Tianjin, China
First published on 14th December 2015
Abstract
Herein, we report that the in situ formed peptidic nanofibers facilitate the induction of multiple crucial immunities against HIV DNA vaccine, including polyfunctional T cell response, broad IgG subclasses response, and V1/V2 loop-specific antibody response, all of which can hardly be triggered by HIV DNA vaccine alone. Such novel in situ formation fundamentally overcomes the big hurdle for the applications of such nanofibers, which previously can only trigger these crucial immune responses via adding exogenous alkaline phosphatase. Such robustness of peptidic nanofibers for inducing crucial immune responses may allow better inhibition against HIV than reported materials.
Conceptual insightsThis work employs peptidic nanofibers to significantly improve the efficacy of vaccines for HIV, a virus against which human beings currently have no effective vaccine. As adjuvant, such nanofibers can both improve the quality of immune responses against HIV and provide convenience for administration. These two improvements allow a significant advancement in the field of HIV vaccine, which is far limited because of the immense difficulties in obtaining a high-quality immune response and complex protocols for administration. This work provides an exciting way to overcome such difficulties via safe and convenient adjuvants, the in situ formed peptidic nanofibers. The superior performance of nanofibers as adjuvants on fundamentally optimizing HIV vaccine is a disruptive advancement in the field of adjuvant research, and will further promote the development of next-generation vaccines against AIDS and other diseases. |
Since the first case of human immunodeficiency virus/HIV discovered in 1983, this deadly virus has caused great harm to human health worldwide. Vaccination is recognized as one of the most effective and low-cost ways to control HIV spread worldwide.1 Up to now, a major goal for HIV vaccines is to induce good-quality, not only high-quantity, immune responses to achieve high protection efficacy of HIV vaccines.2 For example, a strong, mono-functional T cell response (a single T cell simultaneously secretes only one or two cytokines) is limited for the clearance and control of the virus in vivo.3 Likewise, a potent antibody titer against HIV alone does not necessarily translate into good neutralization (blocking the infection of virus).4 In contrast, some unique profiles of the immune response, such as both the polyfunctional T cell response5–8 and the broad IgG subclasses response,9–12 are featured in LTNPs, and the IgG response against the crucial epitope (V1/V2 loop of HIV Env) can effectively reduce HIV infection risk.13 We thus wonder if there is a way to optimize such profiles of immune responses of HIV vaccines.
Nanomaterials have obtained widespread attention because of their specific bio-effects in vivo, and also have been applied in the field of biomedicine.14,15 In particular, peptidic nanofibers may be useful for vaccination. They not only act as basic building blocks to comprise the complex cytoskeleton network to maintain cellular structure, but also construct the complex extracellular matrix to mediate the interactions between cells and their surrounding microenvironment.16 In recent years, numerous studies focused on investigating the bio-effects of peptidic nanofibers formed in vitro by some inducers, such as enzymes, heating, change of pH and so forth.17,18 However, such nanofibers still face many problems for practical applications, because they have to be constructed in vitro via adding inducers before injection into the body. A latest study from the Bing Xu's group reported that peptidic nanofibers can self-assemble on the surface of cancer cells in vitro, using the ALP secreted by cells themselves.19 It suggested that peptidic precursors might self-assemble in vivo dependent on an endogenous inducer, such as ALP. In one of our recent studies, a kind of peptidic precursor named NMe [it contains four amino acids backbone (glycine–phenylalanine–phenylalanine–tyrosine), in which the N-terminal and the C-terminal are modified by naphthalene acetic acid and the methylamino group respectively] can self-assemble into nanofibers in vitro after the hydrolysis by ALP. Such peptidic nanofibers as adjuvants can increase the strength of the T cell and the IgG response against the HIV DNA vaccine in mice, via compressing HIV DNA vaccines into more compact spheres to enter antigen-presenting cells more effectively.20 However, all above immunological effects were triggered by peptidic nanofibers which were assembled in vitro via addition of exogenous ALP. We thus wonder whether such peptidic nanofibers have similar effects when they are directly injected without exogenous ALP (a more convenient way for practical applications), considering endogenous ALP has an extensive contribution in vivo.
Herein, we investigated the effects of different assembly modes of peptidic nanofibers on regulating crucial immune responses induced by the HIV DNA vaccine, including the polyfunctional T cell response, the broad IgG subclasses response and the V1/V2-specific IgG response. Precursors of nanofibers that were directly injected in a body (a convenient route for application) had the same immunological effects as the nanofibers formed in vitro via addition of exogenous ALP, thus showing the robustness of such nanofibers. Our results not only explored multiple crucial immunological effects of peptidic nanofibers, but also cleared the hurdles for large-scale clinical applications by tremendously simplifying the route for administering such peptidic nanofibers.
Results and discussion
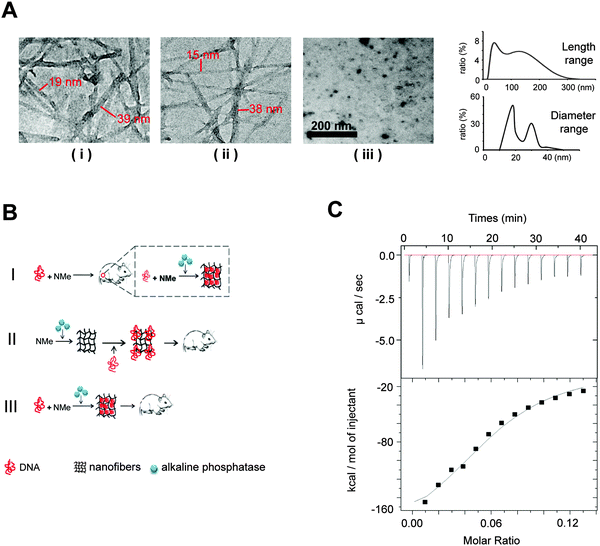
For enzyme-triggered peptidic nanofibers, the assembly mode directly determines their suitability as vaccine adjuvants. In most previous studies, enzyme-triggered nanofibers require exogenous enzymes to initiate the assembly process. Such operation procedure severely compromises the convenience, because users have to prepare the nanofibers every time before injection. Moreover, the safety of the vaccine will depend on the purity and safety of the enzymes, in addition to those of the nanofibers themselves. Having to include an enzyme in a vaccine will also significantly increase the cost for practical applications. We thus considered whether such nanofibers would successfully form in situ, employing only the endogenous enzymes in vivo.To verify the possibility that our nanofibers can be assembled in vivo using endogenous ALP, we first investigated the formation of such fibers in the cell culture system. We herein designed three experiment groups (Fig. 1A). (i) HeLa cells (a cancer cell line, 1 × 105 cells) cultured with the 0.1 wt% NMe precursor-added medium; (ii) medium supplemented by both ALP (60 U ml−1) and 0.1 wt% NMe precursors (positive control). HeLa cells lysis solution (1 × 105 cells) has similar catalytic capability to NMe precursors as 60 U ml−1 ALP (Fig. S1, ESI†); (iii) HeLa cells (1 × 105 cells) cultured with the medium alone (negative control). After 24 hours, the nanofiber network was observed in groups (i) and (ii), but not in group (iii) (observed via TEM). The range of length of nanofibers was between 15 nm and 300 nm (Fig. 1A). The range of diameter of nanofibers was between 15 nm and 40 nm (Fig. 1A). These results indicated that NMe can self-assemble into nanofibers in the cell system without exogenous ALP.
We wonder whether the in situ formed nanofibers (without exogenous ALP) have similar immunological effects to that formed in vitro via adding exogenous ALP before injection. To answer this question, we designed three modes of nanofibers (Fig. 1B). (i) Mode I (in situ formation, without exogenous ALP), the mixture of the DNA vaccine and NMe was injected directly into the mice body; (ii) Mode II, NMe assembled into nanofibers by adding exogenous ALP before combination with the HIV DNA vaccine, then were injected into mice together; (iii) Mode III, the DNA vaccine and NMe were mixed together in the presence of ALP before injection into mice. Mice injected with the DNA vaccine alone was used as a control.
We first investigated whether the precursors of NMe can bind to the HIV DNA vaccines to understand the probable working mechanism in above three models. Via an isothermal titration calorimetry (ITC) assay, we quantified the intermolecular interactions between the precursors of NMe and the HIV DNA vaccine (Fig. 1C). By titrating the precursors of NMe (250 μM) into the HIV DNA vaccine (153 nM), we obtained several critical thermodynamic parameters including binding affinity (K), enthalpy changes (ΔH) and binding stoichiometry (N). Approximately, one HIV DNA molecule will bind 13 precursors of NMe (N = 0.0806 ± 0.00517 sites) to form an intermediate. There is the binding (binding affinity is 2.68 ± 0.701 × 105 M−1) between the precursors of NMe and the HIV DNA vaccine (Fig. 1C). Such binding might help NMe to compress loose DNA into the compact structure along with the formation of the nanofiber network. The compact DNA will be more effective to enter the cells, finally causing enhanced immunity according to our previous report.20
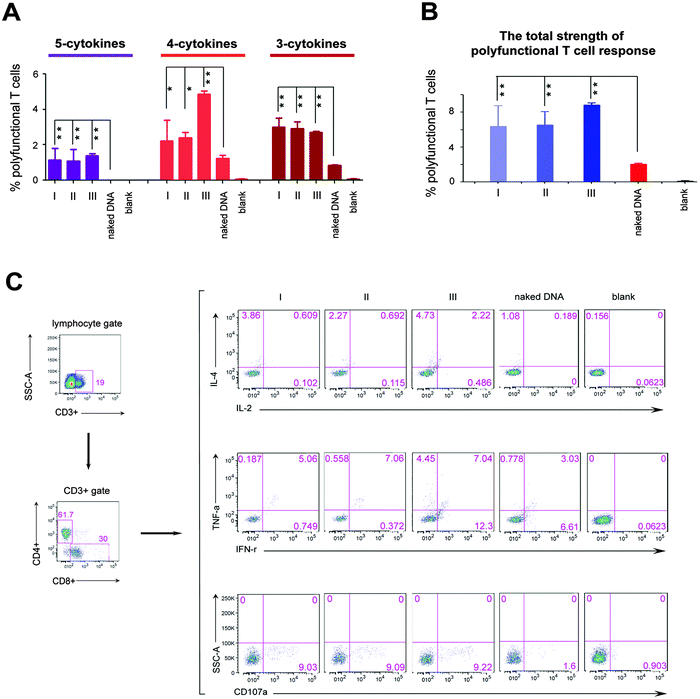
We next investigated the effects of three different formation modes of nanofibers on regulating the polyfunctional T cell response induced by the HIV DNA vaccine, because in the clinic, such immunity is inversely correlated with the disease progression in HIV infected individuals.6,8 In this study, the polyfunctionality of the T cell is defined as a single cell which can simultaneously secrete at least three cytokines among the following five cytokines, interleukin 2 (IL-2), interleukin 4 (IL-4), interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and cluster of differentiation molecule 107a (CD107a). All these cytokines are crucial parameters used to evaluate the polyfunctionality of T cells of the HIV vaccine.6,21 “5-Cytokine-secreting T cells” indicate cells that simultaneously secrete all these five cytokines. “4-Cytokine-secreting T cells” and “3-cytokine-secreting T cells” respectively represent the cells that secrete four and three cytokines among above five cytokines. The sum of the percentage of T cells secreting 3-, 4- and 5-cytokines represents the total strength of the polyfunctional T cell response. The 5-cytokine-secreting T cell response (1.05% for Mode I, 0.93% for Mode II and 1.37% for Mode III, Fig. 2A) was effectively induced in all three modes to a level not achievable by the DNA vaccine alone. Also, significantly increased ratios of T cells expressing 4-cytokines and 3-cytokines (Mode I: 2.39% and 3.31%; Mode II: 2.61% and 3.22%; Mode III: 4.86% and 2.7%, respectively) were triggered in three modes, compared with the DNA vaccine alone (1.24% and 0.83%) (Fig. 2A). The total strength of the polyfunctional T cell response (the sum of percentage of T cells secreting 3-, 4- and 5-cytokines) was improved markedly in three modes (P < 0.01; Fig. 2B). However, the total strength of the polyfunctional T cell response among three modes had no significant difference in statistics (Fig. 2B). In addition, peptidic nanofibers increased the numbers of combination ways of 3- and 4-cytokines, which respectively should be ten (C53 = 10) and five (C54 = 5) kinds of combination ways. i.e., four types of 4-cytokine combinations and seven types of 3-cytokine combinations were triggered in three modes, which respectively included (CD107a, IFN-γ, IL-2, IL-4), (CD107a, IFN-γ, IL-4, TNF-α), (CD107a, IL-2, IL-4, TNF-α), (CD107a, IFN-γ, IL-2, TNF-α) (Fig. S2, ESI†), as well as (CD107a, IFN-γ, IL-4), (CD107a, IL-2, TNF-α), (CD107a, IL-4, TNF-α), (IFN-γ, IL-2, IL-4), (IFN-γ, IL-4, TNF-α), (CD107a, IFN-γ, TNF-α), (CD107a, IL-2, IL-4) (Fig. S3, ESI†). The HIV DNA vaccine alone triggered only two combinations of 4-cytokines (Fig. S2, ESI†) and four combinations of 3-cytokines, respectively (Fig. S3, ESI†). All together, these results indicated that peptidic nanofibers can improve the HIV-specific polyfunctional T cell response. More importantly, in situ formed nanofibers had the same effect as that injected with exogenous ALP on immunoregulating the polyfunctional T cell response. It thus highlighted the advantages of such nanofibers for practical applications on improving the polyfunctional T cell response by the HIV DNA vaccine.
Such superior function of peptidic nanofibers on optimizing the polyfunctional T cell response also implies that they can serve as candidates to substitute existing ways for enhancing vaccine-specific polyfunctionality, e.g., some virus-derived vaccine vectors (vaccinia or yellow fever virus).22,23 Virus-derived vectors usually contain many undefined constituents that can be expressed in vivo and might incorrectly influence immune responses by some unknown and uncontrollable ways. For example, in the Merck phase II trials of the HIV vaccine, some unknown and in vivo expressed elements located in an adenoviral vector might be the factors that caused a higher infection ratio for vaccinated individuals.24 However, the components of peptidic nanofibers in this study have been identified clearly. Its safety is very good in vitro and in vivo.20 It suggests that such nanofibers might be a better option for inducing the polyfunctional T cell response, compared with the virus-derived vaccine vectors.
Moreover, the result that our in situ formed peptidic nanofibers increased IL-4 expression encouraged us to investigate the probable effect on regulating the maturation of B cells, since IL-4 is a crucial factor on promoting the maturation of B cells.25 We herein evaluated the maturation of B cells in four mice groups, which were respectively vaccinated with the HIV DNA vaccine alone and the HIV DNA vaccine aided by nanofibers via above three modes. An empty vaccine vector was used as a blank control. Three kinds of molecules on the B cell surface (CD40, CD69, CD86) were selected as markers for evaluating the maturation of B cells.26,27 The ratios of B cells expressing the above three molecules increased significantly in all three mice modes vaccinated with the HIV DNA vaccine aided by peptidic nanofibers, compared with those vaccinated with the HIV DNA vaccine alone (Fig. 3). Furthermore, the expression of the MHC molecule (the carrier of antigen epitope in cytoplasm) on the cell surface was significantly enhanced by peptidic nanofibers in three modes (Fig. 3). These results provided evidences for our above hypothesis that in situ formed peptidic nanofibers can promote B cell maturation, the same as that formed by adding exogenous ALP.
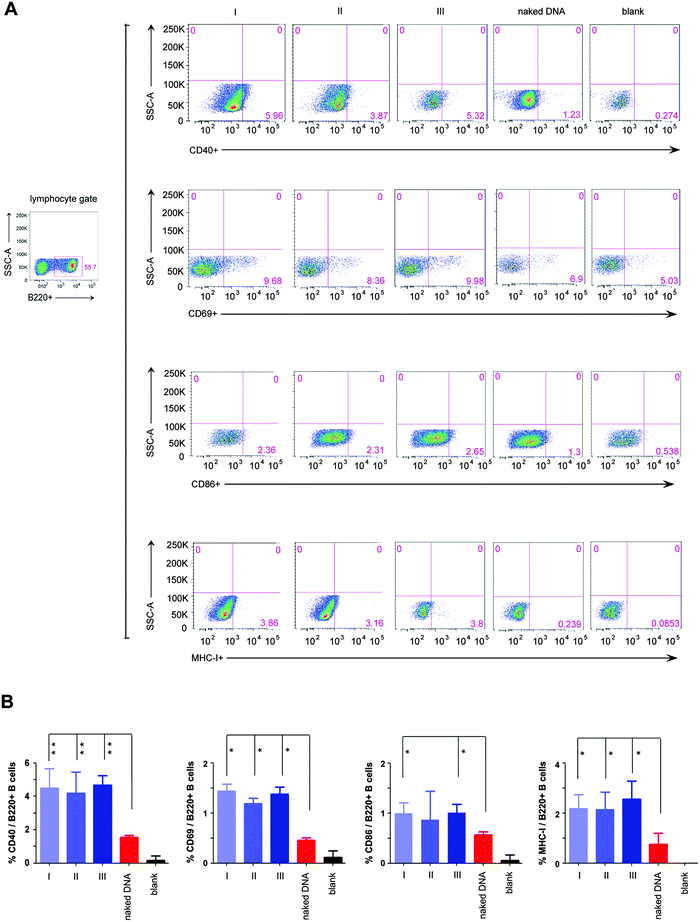 | ||
| Fig. 3 The maturation of B cells stimulated by peptidic nanofibers. Flow cytometric analysis for the expressions of four surface markers indicating B cell maturation, including CD40, CD69, CD86 and MHC I. These cells were gated from B220+ (one specific mark of B cells) lymphocytes derived from the mice spleen. (A) Flow cytometric analysis for above surface molecules in different mice groups. (B) Histograms (the percentage of B220+ B cells which respectively expressed CD40, CD69, CD86 and MHC I). ‘naked DNA’, ‘I’, ‘II’, ‘III’ and ‘blank’ are the same as Fig. 2. | ||
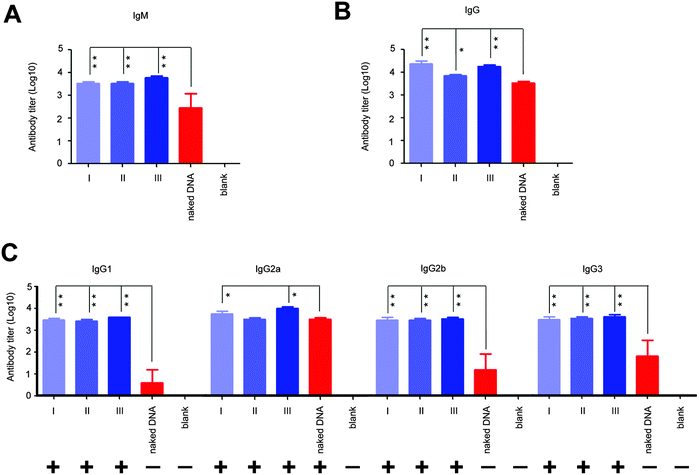
Next, we analyzed the effect of in situ formed peptidic nanofibers on the regulation of the humoral response via investigating antibody titers in the serum, because the facilitation of B cell maturation is consistent with the enhancement of the antibody response (B cells are in charge of secreting antibodies28). Compared with DNA alone, the DNA vaccine aided by peptidic nanofibers in all three modes induced significantly higher titers of IgM and IgG (Fig. 4A and B). We analyzed the influence of peptidic nanofibers on shaping the IgG subclass response, because the broad IgG subclass response is another important clinical feature for LTNPs.9–12 Herein, the positive IgG subclass response is defined as the OD value (difference value between 450 nm and 630 nm) being higher than 0.1 when serum samples are diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (log
100 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 value is 2 at least). Nanofibers in all three modes triggered positive responses against all four IgG subclasses (log
10 value is 2 at least). Nanofibers in all three modes triggered positive responses against all four IgG subclasses (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 values of IgG1, IgG2a, IgG2b, IgG3 were higher than 2), but DNA alone only induced positive IgG2a response (Fig. 4C). Therefore, in situ formed nanofibers, as effective as that formed via adding exogenous ALP, broadened the IgG subclasses response. Noteworthily, the function of such nanofibers on enhancing the IgG1 response by the HIV vaccine might further improve the value of such nanofibers. IgG1 is a main form of the neutralizing antibody in blood. Multiple broad-neutralizing monoclonal antibodies, such as F105, 2G12 and 2F5, belong to the IgG1 subclass.29 Moreover, high affinity of IgG1 with the Fc-γ receptor makes IgG1 become a crucial type of antibody on inhibiting HIV via antibody-dependent cell-mediated cytotoxicity (ADCC),11,30 which is a mechanism of immune defense where ADCC-effector cells (such as natural killer cells, macrophages or neutrophils) selectively attack pathogen-infected cells via the affinity-dependent recognition against antibodies binding on the surface of pathogen-infected cells (mediated by the Fc-γ receptor on the surface of ADCC-effector cells).31
10 values of IgG1, IgG2a, IgG2b, IgG3 were higher than 2), but DNA alone only induced positive IgG2a response (Fig. 4C). Therefore, in situ formed nanofibers, as effective as that formed via adding exogenous ALP, broadened the IgG subclasses response. Noteworthily, the function of such nanofibers on enhancing the IgG1 response by the HIV vaccine might further improve the value of such nanofibers. IgG1 is a main form of the neutralizing antibody in blood. Multiple broad-neutralizing monoclonal antibodies, such as F105, 2G12 and 2F5, belong to the IgG1 subclass.29 Moreover, high affinity of IgG1 with the Fc-γ receptor makes IgG1 become a crucial type of antibody on inhibiting HIV via antibody-dependent cell-mediated cytotoxicity (ADCC),11,30 which is a mechanism of immune defense where ADCC-effector cells (such as natural killer cells, macrophages or neutrophils) selectively attack pathogen-infected cells via the affinity-dependent recognition against antibodies binding on the surface of pathogen-infected cells (mediated by the Fc-γ receptor on the surface of ADCC-effector cells).31
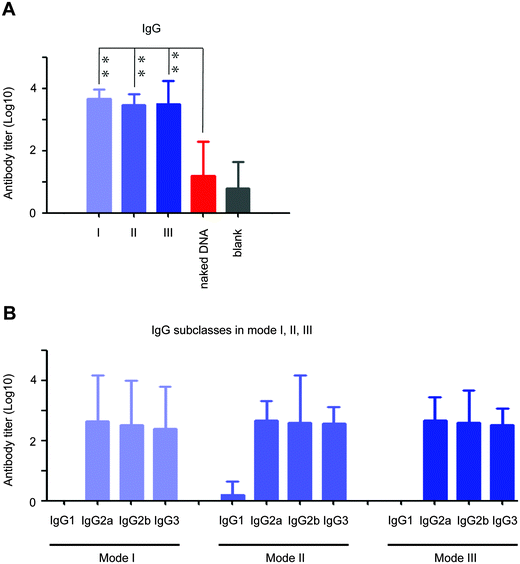
To our excitement, the peptidic nanofibers in all three modes improved the V1/V2 loop-specific IgG response (V1/V2-specific IgG2a, IgG2b and IgG3 responses; Fig. 5A and B), which was positively correlated with protection efficacy in an updated phase III HIV vaccine trial in Thailand.13 Such crucial immunity failed to be triggered by the HIV DNA vaccine alone (Fig. 5A). Recently, a NIH-led large-scale clinical trial (HVTN 505) was stopped, because the HIV vaccine regimen did not prevent HIV infection. One of major reasons might be that the crucial immune response, the V1/V2-specific IgG response, was not triggered.32 Considering our peptidic nanofibers can effectively optimize such immune response induced by the HIV vaccine, it thus might fundamentally optimize immunities of HIV vaccine regimens like HVTN 505. If such a clinical trial had incorporated the peptidic nanofibers, we would have predicted a better outcome.
Finally, we investigated whether NMe can still improve immunities by the HIV DNA vaccine when they are separately vaccinated. There is no binding between the HIV DNA vaccine and NMe in the separated vaccination. Such investigation is an important supplement for Mode I (injecting the HIV DNA vaccine and NMe together), and will further reveal the importance of the binding between the HIV DNA vaccine and NMe for enhancing immunities by the vaccine. One mouse group was injected with NMe precursors first, followed by the HIV DNA vaccine (the time interval is one hour). In contrast, another mouse group was injected with the HIV DNA vaccine first, followed by NMe precursors one hour later. Comparing with the mouse group vaccinated with the HIV DNA vaccine alone, neither the ‘HIV DNA vaccine first, followed by NMe’ nor ‘NMe first, followed by the HIV DNA vaccine’ significantly improve the immunities, including the polyfunctional T cell response, IgM, IgG, IgG subclass and V1/V2 loop-specific antibody responses (Fig. S4, ESI†). It suggested that a binding between NMe precursors and the HIV DNA vaccine is necessary to improve such immunities by the HIV DNA vaccine.
Conclusion
In summary, in situ formed peptidic nanofibers can improve the crucial immunities of the HIV DNA vaccine, including the polyfunctional T cell response, the broad IgG subclass response, and the V1/V2-specific IgG response, all of which can effectively delay the disease process or improve the protection from the infection of HIV. Such novel in situ formation (triggering the formation of peptidic nanofibers without exogenous ALP) thus highlights the robustness of the nanofibers as vaccine adjuvants for practical applications, since we can simplify the operation procedure when injecting the complex of nanofibers and the HIV DNA vaccine into the host body (without premixing with ALP). Also, such unique features will facilitate the peptidic nanofibers to be crucial components not only in next-generation HIV vaccines, but also for other infectious diseases and cancer immunotherapy vaccines, such as the yellow fever vaccine, and the lung cancer immunotherapy vaccine, considering that the importance of immunities improved by peptidic nanofibers on inhibiting the development of these diseases.Acknowledgements
The authors wish to acknowledge Prof. Bing Xu, Prof. Yuan Gao for their assistances in experiments and valuable suggestions in this study, and the financial supports from the Ministry of Science and Technology (2011CB933201, 2012AA022703, 2012AA030308), the Ministry of Health (2012ZX10001-008), the National Science Foundation of China (21025520, 51373043, 21105018 & 81361140345), the Chinese Academy of Science (XDA09030305) and Research Supported by the CAS/SAFEA International Partnership Program for Creative Research Teams.References
- D. H. Barouch and S. G. Deeks, Science, 2014, 345, 169–174 CrossRef CAS PubMed.
- J. McMichael, P. Borrow, G. D. Tomaras, N. Goonetilleke and B. F. Haynes, Nat. Rev. Immunol., 2010, 10, 11–23 CrossRef PubMed.
- P. Matthieu, L. Yves and P. Giuseppe, Curr. Opin. HIV AIDS, 2013, 8, 333–340 Search PubMed.
- S. W. Barnett, S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola and L. Stamatatos, J. Virol., 2001, 75, 5526–5540 CrossRef CAS PubMed.
- V. Velu, K. Titanji, B. Zhu, S. Husain, A. Pladevega, L. Lai, T. H. Vanderford, L. Chennareddi, G. Silvestri, G. J. Freeman, R. Ahmed and R. R. Amara, Nature, 2009, 458, 206–210 CrossRef CAS PubMed.
- M. R. Betts, M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer and R. A. Koup, Blood, 2006, 107, 4781–4789 CrossRef CAS PubMed.
- J. Liu, K. L. O’Brien, D. M. Lynch, N. L. Simmons, A. La Porte, A. M. Riggs, P. Abbink, R. T. Coffey, L. E. Grandpre, M. S. Seaman, G. Landucci, D. N. Forthal, D. C. Montefiori, A. Carville, K. G. Mansfield, M. J. Havenga, M. G. Pau, J. Goudsmit and D. H. Barouch, Nature, 2009, 457, 87–91 CrossRef CAS PubMed.
- J. R. Almeida, D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran and V. Appay, J. Exp. Med., 2007, 204, 2473–2485 CrossRef CAS PubMed.
- K. Banerjee, P. J. Klasse, R. W. Sanders, F. Pereyra, E. Michael, M. Lu, B. D. Walker and J. P. Moore, AIDS Res. Hum. Retroviruses, 2010, 26, 445–458 CrossRef CAS PubMed.
- G. D. Tomaras and B. F. Haynes, Curr. Opin. HIV AIDS, 2009, 4, 373–379 CrossRef PubMed.
- A. W. Chung, M. Ghebremichael, H. Robinson, E. Brown, I. Choi, S. Lane, A. S. Dugast, M. K. Schoen, M. Rolland, T. J. Suscovich, A. E. Mahan, L. Liao, H. Streeck, C. Andrews, S. Rerks-Ngarm, S. Nitayaphan, M. S. de Souza, J. Kaewkungwal, P. Pitisuttithum, D. Francis, N. L. Michael, J. H. Kim, C. Bailey-Kellogg, M. E. Ackerman and G. Alter, Sci. Transl. Med., 2014, 6, 228ra38 CrossRef PubMed.
- V. A. Sundqvist, A. Linde, R. Kurth, A. Werner, E. B. Helm, M. Popovic, R. C. Gallo and B. Wahren, J. Infect. Dis., 1986, 153, 970–973 CrossRef CAS PubMed.
- B. F. Haynes, P. B. Gilbert, M. Juliana McElrath, S. Zolla-Pazner, G. D. Tomaras, S. M. Alam, D. T. Evans, D. C. Montefiori, C. Karnasuta, R. Sutthent, H. X. Liao, A. L. DeVico, G. K. Lewis, C. Williams, A. Pinter, Y. Fong, H. Janes, A. DeCamp, Y. Huang, M. Rao, E. Billings, N. Karasavvas, M. L. Robb, V. Ngauy, M. S. de Souza, R. Paris, G. Ferrari, R. T. Bailer, K. A. Soderberg, C. Andrews, P. W. Berman, N. Frahm, S. C. De Rosa, M. D. Alpert, N. L. Yates, X. Shen, R. A. Koup, P. Pitisuttithum, J. Kaewkungwal and S. Nitayaphan, N. Engl. J. Med., 2012, 366, 1275–1286 CrossRef CAS PubMed.
- Z. Zhao, Z. Zhou, J. Bao, Z. Wang, J. Hu, X. Chi, K. Ni, R. Wang, X. Chen, Z. Chen and J. Gao, Nat. Commun., 2013, 4, 2266 Search PubMed.
- N. Gu and H. Xu, Sci. Rep., 2013, 3, 2655 Search PubMed.
- Y. Chen and G. Liang, Theranostics, 2012, 2, 139–147S CrossRef CAS PubMed.
- P. Huang, Y. Gao, J. Lin, H. Hu, H. Liao, X. Yan, Y. Tang, A. Jin, J. Song, G. Niu, G. Zhang, F. Horkay and X. Chen, ACS Nano, 2015, 9, 9517–9527 CrossRef CAS PubMed.
- Y. Gao, J. Shi, D. Yuan and B. Xu, Nat. Commun., 2012, 3, 1033 CrossRef PubMed.
- Y. Kuang, J. Shi, J. Li, D. Yuan, K. A. Alberti, Q. Xu and B. Xu, Angew. Chem., Int. Ed., 2014, 53, 8104–8107 CrossRef CAS PubMed.
- Y. Tian, H. Wang, Y. Liu, L. Mao, W. Chen, Z. Zhu, W. Liu, W. Zheng, Y. Zhao, D. Kong, Z. Yang, W. Zhang, Y. Shao and X. Jiang, Nano Lett., 2014, 14, 1439–1445 CrossRef CAS PubMed.
- U. Müller, D. Piehler, W. Stenzel, G. Köhler, O. Frey, J. Held, A. Grahnert, T. Richter, M. Eschke, T. Kamradt, F. Brombacher and G. Alber, Mucosal Immunol., 2012, 5, 299–310 CrossRef PubMed.
- M. L. Precopio, M. R. Betts, J. Parrino, D. A. Price, E. Gostick, D. R. Ambrozak, T. E. Asher, D. C. Douek, A. Harari, G. Pantaleo, R. Bailer, B. S. Graham, M. Roederer and R. A. Koup, J. Exp. Med., 2007, 204, 1405–1416 CrossRef CAS PubMed.
- D. Gaucher, R. Therrien, N. Kettaf, B. R. Angermann, G. Boucher, A. Filali-Mouhim, J. M. Moser, R. S. Mehta, D. R. Drake III, E. Castro, R. Akondy, A. Rinfret, B. Yassine-Diab, E. A. Said, Y. Chouikh, M. J. Cameron, R. Clum, D. Kelvin, R. Somogyi, L. D. Greller, R. S. Balderas, P. Wilkinson, G. Pantaleo, J. Tartaglia, E. K. Haddad and R. Sékaly, J. Exp. Med., 2008, 205, 3119–3131 CrossRef CAS PubMed.
- M. J. McElrath, S. C. De Rosa, Z. Moodie, S. Dubey, L. Kierstead, H. Janes, O. D. Defawe, D. K. Carter, J. Hural, R. Akondy, S. P. Buchbinder, M. N. Robertson, D. V. Mehrotra, S. G. Self, L. Corey, J. W. Shiver, D. R. Casimiro and the Step Study Protocol Team, Lancet, 2008, 372, 1894–1905 CrossRef CAS.
- B. Gao and T. L. Rothstein, J. Immunol., 2013, 191, 670–677 CrossRef PubMed.
- K. Attridge, R. Kenefeck, L. Wardzinski, O. S. Qureshi, C. J. Wang, C. Manzotti, K. Okkenhaug and L. S. K. Walker, J. Immunol., 2014, 192, 2195–2201 CrossRef CAS PubMed.
- P. J. Nadeau, A. Roy, C. Gervais-St-Amour, M. È. Marcotte, N. Dussault and S. Néron, Mol. Immunol., 2012, 49, 582–592 CrossRef CAS PubMed.
- Y. Bao and X. Cao, J. Autoimmun., 2014, 55, 10–23 CrossRef CAS PubMed.
- T. W. Baba, V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou and R. M. Ruprecht, Nat. Med., 2000, 6, 200–206 CrossRef CAS PubMed.
- P. Bruhns, B. Iannascoli, P. England, D. A. Mancardi, N. Fernandez, S. Jorieux and M. Daëron, Blood, 2009, 113, 3716–3725 CrossRef CAS PubMed.
- D. Rudnicka, A. Oszmiana, D. K. Finch, I. Strickland, D. J. Schofield, D. C. Lowe, M. A. Sleeman, D. M. Davis and HVTN 505 Study Team, Blood, 2013, 121, 4694–4702 CrossRef CAS PubMed.
- S. M. Hammer, M. E. Sobieszczyk, H. Janes, S. T. Karuna, M. J. Mulligan, D. Grove, B. A. Koblin, S. P. Buchbinder, M. C. Keefer, G. D. Tomaras, N. Frahm, J. Hural, C. Anude, B. S. Graham, M. E. Enama, E. Adams, E. DeJesus, R. M. Novak, I. Frank, C. Bentley, S. Ramirez, R. Fu, R. A. Koup, J. R. Mascola, G. J. Nabel, D. C. Montefiori, J. Kublin, M. J. McElrath, L. Corey and P. B. Gilbert, N. Engl. J. Med., 2013, 369, 2083–2092 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5nh00064e |
| This journal is © The Royal Society of Chemistry 2016 |