Compaction and binding properties of the intrinsically disordered C-terminal domain of Henipavirus nucleoprotein as unveiled by deletion studies†‡
David
Blocquel
,
Johnny
Habchi
,
Antoine
Gruet
,
Stéphanie
Blangy
and
Sonia
Longhi
*
CNRS, UMR 6098, 13288, Marseille, France. and Aix-Marseille Univ., Architecture et Fonction des Macromolécules Biologiques (AFMB), 13288, Marseille, France. E-mail: Sonia.Longhi@afmb.univ-mrs.fr; Fax: +33 4 91 26 67 20; Tel: +33 4 91 82 55 80
First published on 23rd November 2011
Abstract
Henipaviruses are recently emerged severe human pathogens within the Paramyxoviridae family. Their genome is encapsidated by the nucleoprotein (N) within a helical nucleocapsid that recruits the polymerase complex via the phosphoprotein (P). We have previously shown that in Henipaviruses the N protein possesses an intrinsically disordered C-terminal domain, NTAIL, which undergoes α-helical induced folding in the presence of the C-terminal domain (PXD) of the P protein. Using computational approaches, we previously identified within NTAIL four putative molecular recognition elements (MoREs) with different structural propensities, and proposed a structural model for the NTAIL–PXD complex where the MoRE encompassing residues 473–493 adopt an α-helical conformation at the PXD surface. In this work, for each NTAIL protein, we designed four deletion constructs bearing different combinations of the predicted MoREs. Following purification of the NTAIL truncated proteins from the soluble fraction of E. coli, we characterized them in terms of their conformational, spectroscopic and binding properties. These studies provided direct experimental evidence for the structural state of the four predicted MoREs, and showed that two of them have clear α-helical propensities, with the one spanning residues 473–493 being strictly required for binding to PXD. We also showed that Henipavirus NTAIL and PXD form heterologous complexes, indicating that the PXD binding regions are functionally interchangeable between the two viruses. By combining spectroscopic and conformational analyses, we showed that the content in regular secondary structure is not a major determinant of protein compaction.
Introduction
Henipaviruses are recently emerged human pathogens. Hendra virus (HeV) came to light in 1994 as the causative agent of a sudden outbreak of acute respiratory disease in horses in Brisbane (Australia) with a fatal human case. HeV infections currently pose a serious threat to livestock in Australia, where sporadic and lethal transmission to humans has also occurred. Nipah virus (NiV), the second known member of the Henipavirus genus, was recognized in 1998–1999 as the etiologic agent of an outbreak of disease in pigs and humans in Malaysia. 265 human cases of encephalitis with 105 deaths were reported and 1 million pigs were culled to stop the epidemic. Since 2001, and almost every year, including 2011,1 outbreaks of encephalitis caused by NiV occurred in Bangladesh with a case fatality rate approaching 75% (see ref. 2 and references therein cited). Fruit-eating bats are the natural reservoir of Henipaviruses (see ref. 3 and references therein cited). Occasional person-to-person NiV transmission is observed,4–7 thus further extending the potential of NiV to cause deadly outbreaks in humans. In addition to acute infection, these viruses cause asymptomatic infections in up to 60% of exposed people and may lead to late-onset of disease or relapse of encephalitis years after initial infection, as well as persistent or delayed neurological sequelae.8A few distinctive properties, including their much larger genome size, led to the classification of NiV and HeV within the Henipavirus genus of the Paramyxoviridae family.9 A number of Henipavirus strains have been isolated from humans, bats, horses and pigs over a wide geographic area in the last decade. Noteworthy, Henipaviruses are also found outside Australia and Asia, thus extending the endemic area of one of the most pathogenic virus genera known in humans.3 The susceptibility of humans, their high pathogenicity, the wide host range and interspecies transmission led to the classification of HeV and NiV as biosecurity level 4 (BSL-4) pathogens.10 To date, no therapeutic agents nor vaccines are available to fight against these severe pathogens.
As in all Mononegavirales members, the negative-strand, non-segmented RNA genome of Henipaviruses is encapsidated by the nucleoprotein (N) within a helical nucleocapsid. This helical N:RNA complex, and not naked RNA, is the substrate used by the polymerase complex during both transcription and replication. The helical nucleocapsid, made of genomic RNA encapsidated into a linear non-covalent polymer of the N protein, has the characteristic herringbone-like structure typically observed in other Paramyxoviridae members.11–17 When expressed in heterologous hosts, NiV N was found to form nucleocapsid-like particles that contain RNA.18–21
Minigenome replicon studies showed that in Henipaviruses, the N protein, the phosphoprotein (P) and the large protein (L) are necessary and sufficient to sustain replication of viral RNA.22 By analogy with other Paramyxoviridae members, the polymerase complex is thought to consist of the L protein and of the P protein, with the latter serving as a tether for the recruitment of L onto the nucleocapsid template. As in all Mononegavirales members, Henipavirus N and P proteins were shown to interact with each other, being able to form both homologous and heterologous N–P complexes.23,24
So far, high-resolution structural information is only available for the fusion (F) and attachment (G) glycoproteins.25–28 For the N and P proteins, the only molecular data come from our previous studies.21,29–31 Those studies showed that the N-terminal region of P (PNT, aa 1–404 or 1–406) and the C-terminal region of N (NTAIL, aa 400–532) belong to the family of intrinsically disordered proteins (IDPs), although they both contain short order-prone segments.29 The occurrence of some residual, transient secondary and/or tertiary structure within NTAIL and PNT led to their classification within the premolten globules (PMG) subfamily within the family of IDPs.32–37 IDPs are ubiquitous, functional proteins that lack highly populated secondary and tertiary structure under physiological conditions and in the absence of a partner/ligand (for recent reviews on IDPs, see ref. 38–43). In contrast, we showed that the C-terminal X domain of the Henipavirus P proteins (PXD, aa 660–709 of NiV P and aa 657–707 of HeV P) is folded, and adopts an α-helical conformation.21Henipavirus NTAIL and PXD domains were shown to form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric complex that is stable up to 1 M NaCl, and whose KD is within the μM range.21 Using bioinformatics approaches, we identified within both HeV and NiV NTAIL domains four putative molecular recognition elements (MoREs), where these latter are short order-prone regions within intrinsically disordered regions that have a propensity to undergo induced folding upon binding to partner(s).44–47 Among these predicted MoREs, at least two of them (aa 408–422 and 473–493) exhibit a clear α-helical nature, and spectroscopic studies carried out in the presence of trifluoroethanol (TFE) unveiled that both HeV and NiV NTAIL do possess α-helical propensities.29 In agreement, using far-UV circular dichroism (CD) and heteronuclear NMR we showed that binding to PXD triggers α-helical folding of NTAIL, with the resonance behavior suggesting a role for the predicted α-MoRE spanning residues 473–493 in binding to PXD.21 Using fluorescence spectroscopy, we showed that PXD has no impact on the chemical environment of a Trp residue introduced at position 527 of Henipavirus NTAIL, thus arguing for the lack of stable contacts between the C-terminus of NTAIL and PXD.21 Based on all these observations, and also by analogy with the related measles virus (MeV),48,49 we proposed a tentative structural model of the Henipavirus NTAIL–PXD interaction where the α-MoRE encompassing residues 473–493 adopt an α-helical conformation and are embedded between helices α2 and α3 of PXD, leading to a relatively small interface dominated by hydrophobic contacts.21
1 stoichiometric complex that is stable up to 1 M NaCl, and whose KD is within the μM range.21 Using bioinformatics approaches, we identified within both HeV and NiV NTAIL domains four putative molecular recognition elements (MoREs), where these latter are short order-prone regions within intrinsically disordered regions that have a propensity to undergo induced folding upon binding to partner(s).44–47 Among these predicted MoREs, at least two of them (aa 408–422 and 473–493) exhibit a clear α-helical nature, and spectroscopic studies carried out in the presence of trifluoroethanol (TFE) unveiled that both HeV and NiV NTAIL do possess α-helical propensities.29 In agreement, using far-UV circular dichroism (CD) and heteronuclear NMR we showed that binding to PXD triggers α-helical folding of NTAIL, with the resonance behavior suggesting a role for the predicted α-MoRE spanning residues 473–493 in binding to PXD.21 Using fluorescence spectroscopy, we showed that PXD has no impact on the chemical environment of a Trp residue introduced at position 527 of Henipavirus NTAIL, thus arguing for the lack of stable contacts between the C-terminus of NTAIL and PXD.21 Based on all these observations, and also by analogy with the related measles virus (MeV),48,49 we proposed a tentative structural model of the Henipavirus NTAIL–PXD interaction where the α-MoRE encompassing residues 473–493 adopt an α-helical conformation and are embedded between helices α2 and α3 of PXD, leading to a relatively small interface dominated by hydrophobic contacts.21
The objective of the present study was to directly assess the role of this α-MoRE in binding to PXD and to investigate the possible contribution of other short order-prone NTAIL segments to complex formation and folding. To this endeavor, for both HeV and NiV NTAIL proteins, we designed four constructs encoding truncated NTAIL proteins bearing various combinations of the four predicted MoREs. We report their bacterial expression, purification and characterization and show that removal of the α-MoRE spanning residues 473–493 does indeed abrogate binding to PXD. In contrast, removal of the other MoREs does not significantly affect the affinity of the binding reaction, although it impacts the folding propensities to various extents. Spectroscopic studies allowed us to shed light on the structural state of the various MoREs, showing that the MoRE encompassing residues 444–464 are unlikely to transiently populate a β structure. By combining spectroscopic and conformational analyses, we showed that the content in regular secondary structure is not a major determinant of protein compaction. Finally, we estimated the cross-reactivity between the HeV and NiV NTAIL–PXD interacting pairs and show that the PXD-binding regions are functionally interchangeable between the two viruses.
Results and discussion
Design, expression and purification of Henipavirus NTAIL deletion constructs
In order to provide direct experimental support for the ability of the α-MoRE encompassing residues 473–493 of Henipavirus NTAIL to bind to PXD and to assess the possible involvement of other short order-prone NTAIL fragments in PXD binding and in the formation of local transiently populated secondary structure elements, we have designed four deletion constructs bearing various combinations of the predicted MoREs29 (see Fig. 1A). For the sake of clarity, the four predicted MoREs are hereafter referred to as Box1, Box2, Box3 and Box4, with Box3 corresponding to the α-MoRE that has been modeled at the PXD surface.21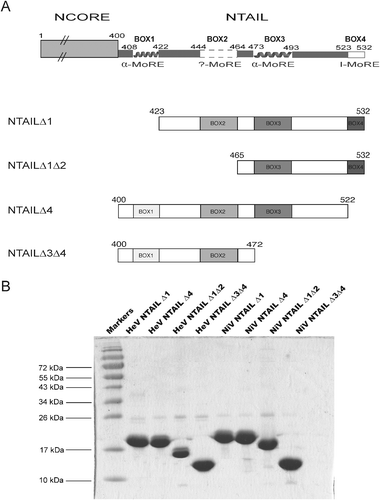 | ||
| Fig. 1 (A) Schematic representation of NTAIL deletion proteins. (Top) Structural organization of Henipavirus N proteins according to ref. 29 showing that they are composed of an N-terminal structured region (NCORE, aa 1–399) and a C-terminal disordered region (NTAIL, aa 400–532). The two putative α-MoREs (aa 408–422 and aa 473–493), with the second one corresponding to the region modeled at the PXD surface,21 are shown. A putative irregular MoRE (I-MoRE) (aa 523–532) and a putative MoRE of dubious state (aa 444–464, see region contoured by a dashed line)29 are also shown. The NTAILΔ1, NTAILΔ1Δ2, NTAILΔ4 and NTAILΔ3Δ4 deletion proteins are devoid of Box1, Box1 plus Box2, Box4 or Box3 plus Box4, respectively. They all possess an N-terminal hexahistidine tag. (B) 15% SDS-PAGE of purified deletion proteins followed by Coomassie blue staining. | ||
The gene fragments encoding the different NTAIL deletion proteins were cloned into the pDEST17OI vector to yield N-terminally histidine-tagged recombinant products, the expression of which is under the control of the T7 promoter being tightly controlled by IPTG. In all cases, the NTAIL proteins were readily expressed in E. coli and their solubility was high, thus allowing their recovery from the soluble fraction of the bacterial lysates. The proteins were purified to homogeneity (>95%) by immobilized metal affinity chromatography (IMAC) followed by preparative size exclusion chromatography (SEC) (Fig. 1B). The identity of the final purified proteins was confirmed by mass spectrometry analysis of the tryptic fragments obtained after digestion of the purified proteins excised from sodium dodecyl sulfate (SDS)-polyacrylamide gels: those analyses yielded for all NTAIL proteins a high sequence coverage attesting their identity and integrity (data not shown).
All the NTAIL proteins migrate in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with an apparent molecular mass higher than expected (see Fig. 1B and Table S1, ESI‡). Indeed the apparent molecular masses of the HeV and NiV NTAILΔ1 proteins are 18 and 19 kDa, respectively, while their expected molecular mass is 14 kDa (see Fig. 1B and Table S1, ESI‡). Likewise, the apparent molecular masses of HeV and NiV NTAILΔ4 proteins are 18 and 19 kDa, respectively, while their expected molecular mass is 15.5 kDa (see Fig. 1B and Table S1, ESI‡). The HeV and NiV NTAILΔ3Δ4 proteins both have an apparent molecular mass of approximately 12 kDa, while their expected mass is 10 kDa. As for the HeV and NiV NTAILΔ1Δ2 proteins, which have an expected molecular mass of 10 kDa, they display an even more abnormal electrophoretic mobility, with apparent molecular masses of approximately 14 kDa (HeV) and 17 kDa (NiV) (Fig. 1B and Table S1, ESI‡).
This abnormal migratory behavior has already been documented for both HeV and NiV full-length NTAIL proteins, where mass spectrometry analyses gave the expected results.29 This anomalous electrophoretic mobility is frequently observed in IDPs (for a few illustrative examples see ref. 12, 21, and 50–52) and has been ascribed to a rather high content of acidic residues as compared to globular proteins.53 The high negative charge is thought to lead to a larger Stokes radius of the protein–detergent complex together with a reduction in detergent binding.54 It should be noted however that the content in acidic residues of those NTAIL proteins that display the most aberrant migration, namely NiV and HeV NTAILΔ1Δ2 (see Fig. 1B and Table S1, ESI‡), is 11.1%, a value lower than the average acidic content (11.8%) of globular proteins53 (see Table S2, ESI‡). In addition, the content in acidic residues of the Henipavirus NTAILΔ1Δ2 proteins is also lower than that of the HeV and NiV NTAILΔ3Δ4 proteins (13.6% and 11.5%, respectively), where these latter have the same expected molecular mass as the NTAILΔ1Δ2 proteins and yet migrate less abnormally than these latter (see Tables S1 and S2, ESI‡). This observation suggests that other parameters, beyond the content in acidic residues, are responsible for the abnormal electrophoretic migration of IDPs. That the occurrence of PP and/or PXP motifs may be a possible determinant of the observed aberrant migration is unlikely. Indeed, while the HeV NTAILΔ1, NTAILΔ1Δ2 and NTAILΔ4 proteins possess a PP and a PKP motif, the NiV proteins, including the NiV NTAILΔ1Δ2 that displays the most aberrant migration, are all devoid of such motifs.
Sequence properties of NTAIL deletion proteins
Sequence analysis of NTAIL deletion proteins by the MeDor metaserver for the prediction of disorder55 and by the PONDR VL-XT predictor56,57 showed that they are all predicted to be mostly disordered (data not shown and Table S2, ESI‡). Consistent with these predictions, the mean hydrophobicity (〈H〉) value of NTAIL deletion proteins was found to be quite low and close to the value calculated for the full-length forms (see Table S2, ESI‡). Such a low hydrophobicity places the NTAIL proteins, including the full-length forms, in the IDP region of the mean net charge/mean hydrophobicity plot developed by Uversky and co-workers,58 with the only exception of NiV NTAILΔ3Δ4 that lies on the border between IDPs and folded proteins and is thus predicted to adopt a more compact conformation (Fig. 2). According to the distance from the boundary (referred to as CH-distance), HeV NTAILΔ1Δ2 is predicted to be the least ordered NTAIL protein, and HeV NTAIL proteins are predicted to be more disordered than NiV NTAIL proteins (Fig. 2 and Table S2, ESI‡). For both HeV and NiV NTAIL proteins, the extent of disorder follows the order NTAILΔ1Δ2 > NTAILΔ1 > NTAIL full-length > NTAILΔ4 > NTAILΔ3Δ4. Thus, for both HeV and NiV proteins, removal of Box1 plus Box2 and, to a minor extent, of Box1 alone leads to forms that are predicted to be less ordered (i.e. more extended) than the parental, full-length form. By contrast, removal of Box4 either alone or in conjunction with Box3 leads to forms that are predicted to be less extended. | ||
| Fig. 2 (A) Charge–hydropathy (CH) plot of Henipavirus NTAIL proteins. The mean net charge (R) is plotted against the mean hydropathy (H). In the left part of the CH plot, a protein is predicted to be intrinsically disordered, whereas it is predicted to be structured if it falls in the right part of it (see Experimental). | ||
Recent analyses of IDPs by the group of Pappu indicated sequence polarity as a determinant of IDP compaction: indeed polar IDPs were found to favor collapsed ensembles in water despite the absence of hydrophobic groups.59 To evaluate sequence polarity, we therefore calculated for each NTAIL protein the net charge per residue (NCR) as described by Pappu and co-workers59 (see Table S2, ESI‡). For all NTAIL proteins, the NCR is negative and well below the well-recognized threshold of 0.2, discriminating collapsed and extended IDPs.59 This suggests that the NTAIL proteins would preferentially populate collapsed states. According to Pappu and co-workers, these proteins would belong to the category of weak polyelectrolytes/polyampholytes, where |f+ − f−| ≤ 0.2 and each of f+ and f− are small.
Conformational properties of NTAIL deletion proteins
All the NTAIL proteins are monodisperse, as indicated by the elution profile observed in analytical SEC coupled to multiple angle laser light scattering (SEC-MALLS) and in preparative SEC (Fig. 3 and data not shown). The molecular masses inferred from SEC-MALLS were found to be in close agreement with the expected values (see Table 1), thus showing that the proteins are monomeric while confirming their integrity and ruling out possible proteolytic degradation. The Stokes radii (RS) of the NTAIL proteins, as derived by SEC-MALLS, are shown in Table 1. In all cases, the experimentally observed values were found to be higher than those expected for natively folded forms, as expected for proteins that are truncated forms of an IDP. Note that these elution profiles were found to be independent from ionic strength, as the same RS values were obtained irrespective of the NaCl concentration. For all NTAIL proteins, the observed RS values were found to be closer to the values expected for IDPs of the same molecular mass (as calculated based on the simple power-law model)60 than to those expected for fully unfolded proteins (in urea)61 (see Table 1). Interestingly, the experimentally observed Stokes radii were found to be in better agreement with the values expected for IDPs derived from the simple power-law model (see eqn (5) in Experimental) than with those derived from the sequence-based model (see eqn (6) in Experimental). The observed Stokes radii of HeV NTAILΔ1Δ2 and HeV NTAILΔ4 were found to be unexpectedly lower and higher, respectively, than those of the corresponding NiV proteins (see Table 1). Aside from these two exceptions, the other HeV NTAIL proteins were found to have RS values quite similar to those of the corresponding NiV NTAIL proteins. By comparing each experimentally determined RS with the theoretical values of the Stokes radius expected for various conformational states (RNFs: monomeric natively folded protein; RUs: fully unfolded state in urea; RPMGs: PMG conformation, see eqn (2)–(4) in Experimental), the HeV NTAILΔ1Δ2 and NTAILΔ3Δ4 proteins, and the NiV NTAILΔ4 and NTAILΔ3Δ4 proteins were found to have Stokes radii similar to the values expected for IDPs adopting a PMG-like conformation (see Table 1), as already observed in the case of both HeV and NiV full-length NTAIL proteins.29 Conversely, both HeV and NiV NTAILΔ1 proteins were found to have RS values closer to those expected for fully unfolded forms, while HeV NTAILΔ4 and NiV NTAILΔ1Δ2 have borderline RS values between PMG-like and random coil-like forms (see Table 1). In order to quantitatively estimate the degree of compaction of the various NTAIL proteins, we used the compaction index (CI) as described by Brocca and co-workers.62 This parameter (see eqn (7) in Experimental) does not mathematically depend on the residue number and hence allows comparison between proteins of different lengths. The CI value can vary between 0 and 1, with 0 indicating minimal compaction and 1 maximal compaction. For each NTAIL protein, we therefore calculated the CI using the reference RS values for unfolded or natively folded proteins as defined by either Uversky61 or Marsh and Forman-Kay.60 These two calculation methods yielded slightly different CI values. Irrespective of the calculation method used to derive the CI, for both NiV and HeV the least compact form was found to be NTAILΔ1 and the most compact form NTAILΔ3Δ4. Thus, this latter form has a CI that is even higher than that of the parental full-length protein, indicating that removal of Box3 plus Box4 leads to a more compact form. That removal of Box3 plus Box4 was to lead to a more compact form was rather unexpected, given the relative enrichment in hydrophobic clusters within these regions and the occurrence of a predicted α-helix within Box3 (see Fig. 1 in ref. 29). These latter features were indeed the basis for the speculation that those regions could correspond to MoREs, with the third MoRE (Box3) corresponding to a putative α-MoRE involved in binding to PXD.21 Notably, the unexpected high degree of compaction of NTAILΔ3Δ4 is in agreement with the sequence analyses shown in Fig. 2, which predict this protein to be the most compact. Strikingly, similar, although reciprocal, unexpected observations were done in the case of the intrinsically disordered Sic1 protein, where removal of a region predicted to be highly disordered led to a truncated form that was found to be less compact than the parental, full-length form.62 | ||
| Fig. 3 SEC-MALLS-RI analysis of HeV (A) and NiV (B) NTAIL proteins. The right y-axis represents the molecular mass and the left axis represents the differential refractive index. The horizontal traces show the molecular masses calculated from light-scattering intensity at different angles and differential refractive indexes as a function of the elution volume. The concentrations of the injected samples were as follows: HeV NTAILΔ1: 18.5 mg mL−1, HeV NTAILΔ4: 23.3 mg mL−1; HeV NTAILΔ1Δ2: 14.5 mg mL−1; HeV NTAILΔ3Δ4: 12 mg mL−1; NiV NTAILΔ1: 12.2 mg mL−1; NiV NTAILΔ4: 17 mg mL−1; NiV NTAILΔ1Δ2: 14.5 mg mL−1; NiV NTAILΔ3Δ4: 17.7 mg mL−1. | ||
| MMObs (kDa) | MMtheo (kDa) | R ObsS | R NFS | R IDPS | R PMGS | R US | R ObsS/RPMGS | R ObsS/RUS | CI1 | CI2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Values were derived by previously reported mass spectrometry studies.29 b Values were derived by previously reported SEC studies.29 | |||||||||||
| HeV | |||||||||||
| NTAIL | 15.3a | 15.2 | 28.0b ± 3 | 19.4 | 30.8 (26.6) | 28.0 | 34.0 | 1.001 | 0.826 | 0.410 | 0.475 |
| NTAILΔ1 | 15.5 | 14.5 | 30.5 ± 3 | 19.1 | 29.9 (25.9) | 27.4 | 33.0 | 1.112 | 0.922 | 0.184 | 0.247 |
| NTAILΔ1Δ2 | 9.8 | 10 | 22.8 ± 3 | 16.7 | 24.6 (21.2) | 23.6 | 27.2 | 0.965 | 0.837 | 0.421 | 0.485 |
| NTAILΔ4 | 15.3 | 15.7 | 31.4 ± 3 | 19.7 | 31.3 (26.4) | 28.4 | 34.5 | 1.107 | 0.909 | 0.211 | 0.285 |
| NTAILΔ3Δ4 | 11.1 | 10.3 | 21.3 ± 3 | 16.8 | 25.3 (21.0) | 23.9 | 27.7 | 0.891 | 0.770 | 0.590 | 0.683 |
| NiV | |||||||||||
| NTAIL | 14.9a | 14.9 | 28.0b ± 3 | 19.4 | 30.8 (25.7) | 27.7 | 33.6 | 1.009 | 0.834 | 0.391 | 0.475 |
| NTAILΔ1 | 14.0 | 14.3 | 31.2 ± 3 | 19.0 | 29.9 (25.0) | 27.2 | 32.8 | 1.145 | 0.952 | 0.115 | 0.197 |
| NTAILΔ1Δ2 | 10.0 | 9.8 | 25.3 ± 3 | 16.6 | 24.6 (20.2) | 23.5 | 27.0 | 1.078 | 0.936 | 0.166 | 0.230 |
| NTAILΔ4 | 15.5 | 15.5 | 27.0 ± 3 | 19.6 | 31.3 (25.5) | 28.1 | 34.2 | 0.959 | 0.790 | 0.492 | 0.569 |
| NTAILΔ3Δ4 | 10.5 | 10.1 | 21.9 ± 3 | 16.8 | 25.3 (20.7) | 23.8 | 27.5 | 0.921 | 0.797 | 0.521 | 0.625 |
On the other hand, the finding that both HeV and NiV NTAILΔ1 proteins are the least compact proteins is not in agreement with the in silico analyses, although these latter predict Henipavirus NTAILΔ1 proteins to be less compact than the parental, full-length forms (see Fig. 2).
The compaction trend of HeV NTAILΔ1Δ2 and HeV NTAILΔ4 is inversed with respect to that of the corresponding NiV proteins: indeed, while HeV NTAILΔ1Δ2 has a compaction slightly higher than that of the parental full-length form and HeV NTAILΔ4 is less compact, the opposite scenario is observed in the case of the NiV proteins (see Table 1). This indicates that while in the case of HeV NTAIL, removal of Box4 leads to a more extended form, in the case of NiV NTAIL it is the removal of Box1 plus Box2 which leads to a decrease in compaction. This suggests the occurrence of differences in these boxes and/or in the remainder of the polypeptide chain in terms of local structural propensities and/or abilities to form tertiary contacts in the HeV and NiV NTAIL proteins.
Altogether, these data support a major role in both HeV and NiV NTAIL proteins for Box1 as a determinant of protein compaction. It is conceivable that Box1 could exert this effect by establishing long-range tertiary contacts and/or by adopting a transiently populated regular conformation that would result in a PMG-like form. The role of Box2 and Box4 is more dubious, given the poor agreement between the hydrodynamic and the in silico analyses and the inconsistencies between the two viruses. By contrast, experimental and computational data for both viruses consistently indicate that removal of Box3 plus Box4 results in a more compact form. The possibility that the shortening of polypeptide chain length might be a determinant of protein compaction is ruled out by the observation that the NiV NTAILΔ1Δ2 protein, which has an even slightly shorter chain (see Table S2, ESI‡), adopts a more extended conformation with respect to both HeV and NiV NTAILΔ3Δ4 proteins.
Spectroscopic properties and folding propensities of NTAIL deletion proteins
In order to further investigate the local structural propensities of the various boxes, we carried out far-UV circular dichroism (CD) studies. As expected for proteins that are truncated forms of an IDP where putative transiently structured regions have been removed, the far-UV circular dichroism spectra of the NTAIL deletion proteins at neutral pH are all typical of predominantly unstructured proteins, as seen by their large negative ellipticity at 198 nm and very low ellipticity at 190 nm (Fig. 4). These results indicate that the HeV and NiV NTAILΔ3Δ4 proteins, although predicted to have a borderline order and found to be more compact in hydrodynamic studies, yet adopt a mostly disordered conformation, lacking highly populated, stable regular secondary structure elements. | ||
| Fig. 4 Far-UV CD spectra of HeV (A) and NiV (B) NTAIL proteins in 10 mM sodium phosphate pH 7 in the absence or in the presence of either 20% TFE or 3 M TMAO. The protein concentration was 0.1 mg mL−1. Spectra were recorded at 20 °C. Due to high background noise at lower wavelength (beyond 210 nm), the spectra in the presence of TMAO are shown to the point up to which the dyna voltage was in the permissible range. Data are representative of one out of two independent acquisitions. | ||
Trifluoroethanol (TFE) and trimethylamine N-oxide (TMAO) are secondary structure stabilizers that can be used as probes of hidden structural propensities of peptides and proteins. TMAO and other osmolytes may fold unstructured proteins due to the osmophobic effect, a solvophobic thermodynamic force, arising from the unfavorable interaction between the osmolyte and the peptide backbone.63–66 Although both TMAO and TFE act on the peptide backbone, the molecular mechanisms underlying their effects are different. Both solutes act by mimicking the hydrophobic environment experienced by proteins in protein–protein interactions, and can thus be used as probes to unveil regions having a propensity to undergo induced folding.67–70 It has long been known that TFE increases the propensity of amino acids to form an α-helix, presumably by strengthening peptide hydrogen bonds in TFE/H2O mixtures and through favorable interactions of hydrophobic amino acid side chains with TFE.71,72 Peptide hydrogen bonds in helices are believed to be stabilized indirectly by weakening the hydrogen bonding of water molecules to the peptide backbone in the coil form.72 As a result of weakening the hydrophobic interactions within the protein interior, TFE might promote helical structure in most peptides and proteins, even though this helical structure may be non-native.73–75 In contrast, TMAO increases the driving forces for protein folding due to its solvophobic effect on the backbone, forcing thermodynamically unstable proteins to fold without altering the rules for folding to a native-like conformation.64 Furthermore, in opposition to TFE solutions, the propensities of hydrophobic groups to interact with solvent are essentially the same in water as they are in TMAO solutions.76,77 Thus, due to the weakening of hydrophobic interactions, the dominant effect of TFE on proteins is protein denaturation accompanied by the preferential formation of α-helices as a result of the strengthening of peptide hydrogen bonds. Contrarily to that, TMAO promotes folding of unfolded proteins by providing an additional force for folding that has no preference for any particular secondary structure.66 It should be pointed out however, that even if TFE is known to stabilize α-helices more than β-strands, gain of α-helicity in the presence of TFE is not a general rule: for instance, (i) the acidic activator domain of GCN4 forms little or no α-helix in TFE concentrations as high as 30% and folds mostly as β-sheets in 50% TFE,78 (ii) the intrinsically disordered dehydrin Rab18 has an α-helical content as low as 2% in the presence of 90% TFE79 and (iii) the intrinsically disordered rat seminal vesicle protein IV exclusively undergoes β transitions in the presence of TFE (P. Palladino, S. Vilasi, R. Ragone and F. Rossi, personal communication). Furthermore, in the case of measles virus NTAIL, we have recently shown that TFE promotes α-helical folding of the 488–502 region only, with the downstream region only becoming slightly less mobile while retaining an extended conformation in the presence of 20% TFE.80 These observations attest the ability of TFE to show β propensities.
In order to assess the folding potential of the truncated NTAIL proteins, and taking into account the fact that both HeV and NiV NTAIL possess a predicted MoRE of the dubious β state, namely Box2,29 we recorded the CD spectra of NTAIL proteins in the presence of either TFE or TMAO, this latter being used with the specific purpose of unveiling possible β propensities. The TFE concentration in these studies was set to 20%, this choice being dictated by previous CD studies focused on full-length Henipavirus NTAIL proteins showing that most unstructured-to-structured transitions take place at this concentration.29
TMAO experiments were initially carried out with full-length proteins and the concentration of the osmolyte was gradually increased from 0 to 5 M (with 1 M steps). At TMAO concentrations as high as 4 and 5 M, protein precipitation occurred and the spectra were extremely noisy thus precluding meaningful analyses. Because of this observation and also taking into account the fact that previous studies carried out by others showed that 3 M TMAO was found to promote most dramatic structural transitions in various IDPs (for examples see ref. 81–83), we carried out subsequent experiments at 3 M TMAO, a concentration where significant structural transitions were observed and no protein precipitation occurred. Under these conditions, the spectra show a poor signal-to-noise ratio due to the strong absorption of TMAO in the far-UV region. This high background noise prevented meaningful analyses of the spectra beyond 210 nm, as the dyna voltage above this wavelength was found to fall in the non-permissible range.
All the NTAIL proteins, including the full-length forms (see Fig. S1, ESI‡ and ref. 29), show a loss of unordered structure upon addition of 20% TFE, as indicated by the characteristic increase in ellipticity at 190 and 198 nm (Fig. 4 and Fig. S1, ESI‡). Likewise, and in spite of the noisy nature of the spectra in the presence of TMAO, this latter was found to trigger a disorder-to-order transition in all NTAIL proteins, as judged from the drop in the ellipticity in the 210–240 nm region (Fig. 4 and Fig. S1, ESI‡). For both HeV and NiV NTAIL proteins, and irrespective of whether TFE or TMAO was added, the disorder-to-order transition is more pronounced for NTAILΔ1 and NTAILΔ1Δ2 than for NTAILΔ4 and NTAILΔ3Δ4, suggesting a role for Box3 and Box4 in the gain of regular secondary structure. Interestingly, NiV NTAILΔ1Δ2 exhibits a lower ability to undergo a structural transition in the presence of the osmolytes as compared to the HeV NTAILΔ1Δ2 protein (Fig. 4). This observation suggests the existence of subtle differences in the inherent folding propensities of Box1 and Box2 between the HeV and the NiV NTAIL proteins, with these differences having been already unveiled by hydrodynamic studies.
In order to achieve a quantitative assessment of the TFE and TMAO impact, the CD spectra were deconvoluted (see Experimental), thereby allowing the percentage of the various secondary structures to be inferred (Fig. 5). Although deconvolution approaches notoriously lead to estimations that cannot be taken as fully reliable, i.e. they often significantly deviate from the actual content in secondary structure as observed in experimentally determined structures (for examples see ref. 48 and 84), the estimated α-helical content is nevertheless very useful for comparative purposes. Note that the spectra recorded in the presence of TMAO could not be deconvoluted, as the wavelength range in which the dyna voltage was in the permissible range (namely, 260–210 nm) was too small to allow deconvolution by any of the programs implemented in the DICHROWEB server.
 | ||
| Fig. 5 Secondary structure content of HeV (A) and NiV (B) NTAIL proteins in 0 and 20% TFE. The percentages of α-helices, β-strands, turns and unordered regions were calculated using CDSSTR. Values for full-length HeV and NiV NTAIL proteins were derived from spectra shown in Fig. S1 (ESI‡). | ||
As shown in Fig. 5, spectral deconvolution pointed out that for both viruses, NTAILΔ3Δ4 is the NTAIL protein with the lowest α-helical content, followed by the NTAILΔ1Δ2 proteins. In both viruses, NTAILΔ1 and NTAILΔ4 were found to have an α-helical content similar to that of the full-length form, arguing for a poor, if any, ability of Box1 and Box4 to adopt an α-helical conformation in water. On the other hand, data support a role for Box3 and, to a minor extent, for Box2 in sampling, at least transiently, α-helical conformations. Consistent with the hypothesis that Box1 poorly contributes to the overall α-helical content of both HeV and NiV NTAIL proteins, the addition of 20% TFE to both HeV and NiV NTAILΔ1 proteins pointed out a gain in α-helicity similar to that of the full-length form (see Fig. 5). Conversely, and in agreement with a possible involvement of Box3 and Box2 in α-helical folding, addition of TFE to Henipavirus NTAILΔ3Δ4 and NTAILΔ1Δ2 proteins led to a dramatic or significant drop, respectively, in the ability to undergo α-helical folding as compared to the full-length form (see Fig. 5). In particular, for both viruses, NTAILΔ3Δ4 is the NTAIL protein with the lowest ability to undergo α-helical folding, as judged from the finding that the HeV and NiV NTAILΔ3Δ4 proteins have the lowest α-helical content in 20% TFE. Surprisingly however, in both viruses, NTAILΔ4 does not gain α-helicity in the presence of TFE (see Fig. 5). This behavior, which suggests a role for Box4 in α-helical folding, was quite unexpected based on the observation that this latter form has an α-helical content similar to that of the parental, full-length form in the absence of osmolytes. Notably, for all NTAIL proteins, the addition of 20% TFE triggers a decrease in the β content, arguing for the lack of strong β propensities within the HeV and NiV NTAIL proteins (see Fig. 5). This observation, together with the finding that Box2 has an α-helical propensity, suggests that Box2 likely adopts an α-helical conformation, thus shedding light on the hitherto dubious state of this Box.29
Taking into account however the fact that TFE may favor (non-native) α-helical folding, we also sought at performing a quantitative analysis of spectra recorded in the presence of TMAO. To this endeavor, since deconvolution was not feasible for spectra recorded in the presence of TMAO, we derived the α-helical content of the various NTAIL proteins from the ellipticity at 222 nm as described in ref. 85 (see Fig. S2, ESI‡). In order to allow meaningful comparisons with spectra recorded in the absence of any osmolyte, as well as with spectra recorded in TFE, we also used this method to infer the α-helical content of the spectra of the NTAIL proteins alone and in the presence of 20% TFE (see Fig. S2, ESI‡). The absolute values of the α-helical content derived by this analysis turned out to be different from those provided by the deconvolution (cf.Fig. 5 and Fig. S2, ESI‡). Comparison of data obtained in the presence of TFE with data obtained in the presence of TMAO revealed that this latter is less efficient than TFE in promoting α-helical folding, as judged from the lower α-helical content of all NTAIL proteins in the presence of TMAO as compared to TFE. This finding is in agreement with previous reports (for examples see ref. 81 and 86). It should be pointed out however that the lower folding potential of 3 M TMAO with respect to 20% TFE may also arise from the possibility that this TMAO concentration does not correspond to the condition where the midpoint transition takes place.
In spite of the numerical differences between the two calculations, an overall similar trend, with a few exceptions, was observed. In agreement with the results obtained by deconvoluting the spectra, the data shown in Fig. S2 (ESI‡) show that (i) the NTAILΔ3Δ4 proteins were found to possess the lowest α-helical content in the absence of osmolytes and to have the lowest ability to undergo α-helical folding in the presence of TFE or TMAO, (ii) the NTAILΔ1Δ2 proteins have a borderline (HeV) or significantly reduced (NiV) ability to undergo α-helical folding in both TFE and TMAO as compared to the parental full-length forms, and (iii) the NTAILΔ4 proteins were found to possess an α-helical content similar (NiV) or slightly higher (HeV) than that of the parental proteins. In contrast with the data obtained by deconvoluting the spectra, the data shown in Fig. S2 (ESI‡) support the ability of both HeV and NiV NTAILΔ4 proteins to undergo α-helical folding in the presence of osmolytes, consistent with the lack of strong α-helical propensities within Box4. Another notable difference between the data obtained from the deconvolution of the whole CD spectra or from the sole ellipticity at 222 nm concerns NTAILΔ1. Based on the ellipticity at 222 nm, both HeV and NiV NTAILΔ1 proteins were found to possess an even higher ability to undergo α-helical folding than the parental, full-length proteins, although their α-helical content in the absence of osmolytes is comparable to that of the full-length protein (see Fig. S2, ESI‡). It should be pointed out however that the values of α-helical content obtained upon deconvoluting the spectra recorded over the whole wavelength range are likely to be more reliable than the data inferred from a single ellipticity value, with this being especially true for highly noisy spectra such as those recorded in the presence of TMAO.
Having in mind the reported ability of TMAO to unveil possible β propensities, we also attempted at obtaining insights about the β content from the spectra recorded in the presence of TMAO. To this end, we checked those spectra for the presence of a possible minimum at 218 nm, with a pronounced negative peak at this wavelength being a hallmark of β structure.87 None of the NTAIL spectra in the presence of TMAO displays such a minimum, with perhaps the only exception of HeV and NiV NTAILΔ4 proteins that have a spectral shape reminiscent of that of a partly β folded protein (see Fig. 4). Since however this spectral signature was not found in the spectra of the parental, full-length proteins in the presence of TMAO (see Fig. S1, ESI‡), we concluded that the present data do not support any pronounced β propensity within Henipavirus NTAIL.
Altogether, the present data support a role for Box3, and to a minor extent, for Box2 in α-helical folding of NTAIL. By contrast, Box1 was found to be devoid of α-helical propensities and not to be involved in α-helical folding. The observation that Box2 has an α, rather than a β, inherent structural propensity provides insights into this MoRE, the structural state of which has been uncertain so far.29 The finding that Box3 plays a major role in the osmolyte-induced α-helical transition provides support to our previous hypothesis that Box3 could be the region implicated in the PXD-induced α-helical folding of Henipavirus NTAIL.21 Data are also consistent with the lack of pronounced α-helical propensity within Box4, thus providing additional support to our previous computational analyses that predicted the occurrence of an irregular MoRE (I-MoRE) within residues 523–532. We can speculate that the removal of Box4 could reduce the ability of NTAIL to undergo α-helical folding through a mechanism that is independent from the inherent folding ability of this box and that rather reflects a possible stabilization of the α-helices within Box2 and Box3 through long-range tertiary contacts.
Combining computational and spectroscopic analyses
Although the CD spectra of all NTAIL proteins are typical of predominantly unfolded proteins, the observed ellipticity values at 200 and 222 nm (Θ200 and Θ222) of the NTAIL proteins are consistent with the existence of some residual secondary structure, as observed in IDPs adopting a PMG conformation (Fig. 6A). Indeed, Uversky noticed that IDPs can be subdivided in PMG-like and random coil-like (RC-like) forms as a function of their Θ200 and Θ222 values.34 Strikingly, except for NiV NTAILΔ3Δ4 that falls in a position close to the RC-like region of the plot, the other NTAIL proteins are all located in the twilight zone between RC-like and PMG-like proteins (Fig. 6A). This suggests that while these latter are endowed with some transiently populated structure, the NiV NTAILΔ3Δ4 protein would possess less residual regular secondary structure. | ||
| Fig. 6 (A) 222–200 ellipticity plot modified from Uversky.34 The mean residue ellipticity at 222 nm (Θ222) of a set of well-characterized unfolded, random coil-like (RC-like) or premolten globule-like (PMG-like) proteins (from ref. 34) has been plotted against the mean residue ellipticity at 200 nm (Θ200). The position in the plot of Henipavirus NTAIL proteins is highlighted. (B) Plot of the ratio between the ellipticity at 222 nm and the ellipticity at 200 nm (Θ222/Θ200) of the same set of well-characterized RC-like or PMG-like proteins shown in (A). The position in the plot of Henipavirus NTAIL proteins is highlighted. The dashed line corresponds to the boundary between RC-like and PMG-like proteins. (C) CH-ellipticity plot of Henipavirus NTAIL proteins. For each NTAIL protein, the distance from the boundary in the CH plot, referred to as CH distance, has been plotted as a function of the distance from the boundary in the Θ222/Θ200 plot shown in (B). | ||
We then plotted the ratio between the Θ222 and Θ200 of the NTAIL proteins and of a set of well-characterized RC-like and PMG-like proteins34 (Fig. 6B). This ratio has the advantage over the individual Θ222 and Θ200 of being less affected by errors in estimations of protein concentrations, and of providing a unified parameter reflecting the content in both unordered and α-helical structure. We then arbitrarily set a threshold of the Θ222/Θ200 (see the dashed line in Fig. 6B) that allows RC-like IDPs to be discriminated from IDPs adopting a PMG-like conformation.
We then generated a plot in which the distance of each NTAIL protein from this threshold was plotted as a function of its CH-distance (Fig. 6C). This analysis is intended to combine, and hence extend, the two methods previously introduced by Uversky.34,58 Indeed, it combines a computational and a spectroscopic analysis and is in principle meant to allow RC-like forms to be distinguished from PMG-like forms among proteins predicted to be intrinsically disordered by the hydropathy/charge method. In this plot, increasingly negative CH distances designate proteins with increasing disorder, while increasingly positive θ222/θ200 distances designate IDPs becoming progressively more collapsed, as a consequence of an increased content in regular secondary structure. Thus, the left bottom quadrant is expected to correspond to IDPs adopting a RC-like conformation, while the right bottom quadrant is supposed to designate IDPs adopting a PMG conformation.
Surprisingly however, in the case of the NTAIL proteins, this analysis revealed that the HeV and NiV NTAILΔ3Δ4 proteins, which are predicted to be the most ordered by the charge/hydropathy method and which additionally were also shown to be the most compact forms by the hydrodynamic analyses (see Table 1), are the NTAIL proteins exhibiting the lowest distance from the θ222/θ200 boundary (see Fig. 6B) and hence expected to adopt the most extended conformation (see Fig. 6C). Thus, this analysis would wrongly predict the HeV and NiV NTAILΔ3Δ4 proteins to be more extended than the parental, full-length forms by virtue of their increased content in unordered structure and decreased content in α-helical structure. CD studies allowed the lower α-helical content of the HeV and NiV NTAILΔ3Δ4 proteins to be unambiguously accounted for by the lack of Box3, with the MoRE located within Box3 having been shown to be the one with the most pronounced α-helical propensity (Fig. 5 and Fig. S2, ESI‡).
The present data suggest that the amount of regular secondary structure is not seemingly a major determinant of protein compaction, with IDPs depleted in MoREs being nevertheless able to adopt a collapsed state. The bottom line of these observations is that the distance from the θ222/θ200 boundary, while being informative in terms of the relative content in unordered and α-helical structure, is not a reliable indicator of the extent of compaction of the Henipavirus NTAIL proteins. Whether this is a unique property of the Henipavirus NTAIL proteins or rather a general rule of IDPs remains to be established.
Binding affinities between PXD and NTAIL deletion proteins
In order to precisely assess the role of the various boxes in binding to PXD, we investigated the binding abilities of the NTAIL deletion proteins using isothermal titration calorimetry (ITC), an approach that gives access to the stoichiometry, the equilibrium association constant, and the variation in enthalpy and entropy.88 Purified NTAIL proteins were loaded into the calorimeter sample cell, and titrated with the corresponding PXD, achieving a molar ratio of 2 (2.5 for the NiV NTAILΔ1Δ2/PXD pair) at the end of the titration (Fig. 7). The data, following integration and correction for the heats of dilution, were fit with a standard model allowing for a set of independent and equivalent binding sites (Fig. 7). The equilibrium dissociation constant (KD) was derived from the KA, as the reciprocal of this latter. The estimates for the model parameters (see Table 2) confirm a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, as already reported for the parental, full-length NTAIL proteins.21 Results revealed that the binding reactions are both enthalpy and entropy-driven, with the only exception of the NiV NTAILΔ1Δ2–PXD and NiV NTAILΔ4–PXD pairs for which an unfavorable (−4.50 cal mol−1 deg−1) and small favorable (0.615 cal mol−1 deg−1) entropic contributions, respectively, were found. Interestingly, this was also the case of the binding reaction of the full-length NiV NTAIL protein, that was found to have a similarly unfavorable (−4.84 cal mol−1 deg−1) entropic contribution.21 For both viruses, the measured KD for the NTAILΔ1, NTAILΔ1Δ2 and NTAILΔ4 proteins are close to the values found for the parental, full-length proteins (2.1 μM for the NiV pair and 8.7 μM for the HeV pair) (cf.Tables 2 and 3), arguing for the lack of involvement of Box1, Box2 and Box4 in PXD binding. Conversely, under similar experimental conditions, no binding was observed for the NTAILΔ3Δ4 proteins, thereby precluding estimation of the KD (Fig. 7). Note that experiments with 1.5 mM NTAILΔ3Δ4 and 15 mM PXD, which expectedly might have corresponded to an experimental condition suitable for obtaining interpretable binding curves, were not feasible because of limitations in protein solubility. A rough estimation of the KD between NTAILΔ3Δ4 and PXD that would reflect as low as 1–10% of complex formation, which could indeed escape detection, can however be derived from the following equation:
1 stoichiometry, as already reported for the parental, full-length NTAIL proteins.21 Results revealed that the binding reactions are both enthalpy and entropy-driven, with the only exception of the NiV NTAILΔ1Δ2–PXD and NiV NTAILΔ4–PXD pairs for which an unfavorable (−4.50 cal mol−1 deg−1) and small favorable (0.615 cal mol−1 deg−1) entropic contributions, respectively, were found. Interestingly, this was also the case of the binding reaction of the full-length NiV NTAIL protein, that was found to have a similarly unfavorable (−4.84 cal mol−1 deg−1) entropic contribution.21 For both viruses, the measured KD for the NTAILΔ1, NTAILΔ1Δ2 and NTAILΔ4 proteins are close to the values found for the parental, full-length proteins (2.1 μM for the NiV pair and 8.7 μM for the HeV pair) (cf.Tables 2 and 3), arguing for the lack of involvement of Box1, Box2 and Box4 in PXD binding. Conversely, under similar experimental conditions, no binding was observed for the NTAILΔ3Δ4 proteins, thereby precluding estimation of the KD (Fig. 7). Note that experiments with 1.5 mM NTAILΔ3Δ4 and 15 mM PXD, which expectedly might have corresponded to an experimental condition suitable for obtaining interpretable binding curves, were not feasible because of limitations in protein solubility. A rough estimation of the KD between NTAILΔ3Δ4 and PXD that would reflect as low as 1–10% of complex formation, which could indeed escape detection, can however be derived from the following equation: | (1) |
 | ||
| Fig. 7 ITC studies of homologous complex formation between PXD and HeV (A) or NiV (B) NTAIL deletion proteins. Data are representative of at least two independent experiments where the initial concentrations of NTAIL proteins in the microcalorimeter cell and of the homologous PXD in the microsyringe were slightly tuned. Panels (A) and (B) show the data obtained using 150 μM NTAIL and 1.5 mM PXD. Graphs shown in the bottom of each panel correspond to integrated and corrected ITC data fit to a single set of sites model (all sites identical and equivalent). The filled squares represent the experimental data, whereas the solid line corresponds to the model. The derived equilibrium dissociation constant (KD) as well as the stoichiometric number are shown. | ||
| Stoichiometry, n | K D (μM) | Binding enthalpy, ΔH (cal mol−1) | ΔS (cal mol−1 deg−1) | |
|---|---|---|---|---|
| HeV NTAILΔ1 | 0.92 ± 0.015 | 5.18 ± 0.85 | −2771 ± 69.87 | 14.7 |
| HeV NTAILΔ1Δ2 | 0.93 ± 0.010 | 3.98 ± 0.48 | −4082 ± 61.41 | 10.8 |
| HeV NTAILΔ4 | 0.72 ± 0.015 | 7.14 ± 1.10 | −4222 ± 119.9 | 9.14 |
| NiV NTAILΔ1 | 1.31 ± 0.036 | 9.00 ± 2.10 | −2305 ± 95.52 | 15.2 |
| NiV NTAILΔ1Δ2 | 0.75 ± 0.016 | 7.24 ± 1.09 | −8184 ± 241.6 | −4.5 |
| NiV NTAILΔ4 | 0.91 ± 0.02 | 9.00 ± 1.63 | −6586 ± 238.4 | 0.615 |
| Stoichiometry, n | K D (μM) | Binding enthalpy, ΔH (cal mol−1) | Binding entropy, ΔS (cal mol−1 deg−1) | |
|---|---|---|---|---|
| a Data were taken from ref. 21. | ||||
| HeV NTAIL–HeV PXDa | 1.37 ± 0.01 | 8.7 ± 0.55 | −5584 ± 62 | 4.09 |
| NiV NTAIL–HeV PXD | 1.02 ± 0.007 | 7.6 ± 0.46 | −6001 ± 56.38 | 2.92 |
| NiV NTAIL–NiV PXDa | 0.93 ± 0.07 | 2.1 ± 0.24 | −9034 ± 108.9 | −4.84 |
| HeV NTAIL–NiV PXD | 1.12 ± 0.21 | 6.9 ± 1.32 | −2494 ± 81.24 | 15.1 |
The present results support a role for Box3 as a major determinant of PXD binding. These findings validate the previously proposed involvement of Box3 in complex formation, and provide additional support to our structural models of the Henipavirus NTAIL–PXD complexes.21
Having shown that Box3 is the NTAIL region indispensable for binding to PXD, we sought at assessing whether the Box3 regions could be functionally interchanged between the two viruses. We therefore carried out ITC studies in which the parental full-length NTAIL proteins were loaded in the cell and then titrated with the heterologous PXD protein (Fig. S3, ESI‡ and Table 3). As shown in Table 3, replacing HeV NTAIL by NiV NTAIL in the binding reaction with HeV PXD led to binding parameters quite comparable (i.e. within the error bars) to those observed for the homologous pair. Substituting NiV NTAIL with HeV NTAIL in the binding reaction to NiV PXD yielded a slight, though significant, increase in the KD with a concomitant variation in the enthalpic and entropic contributions: while the enthalpic contribution is reduced for the heterologous pair, the entropic penalty is suppressed and the ΔS becomes favorable, possibly reflecting a less pronounced disorder-to-order transition. In support of the significance of the interaction, it should be noted that no interaction was found between Henipavirus NTAIL and measles virus PXD (Habchi et al., unpublished).
These data suggest that the HeV and NiV Box3 regions can be functionally replaced by each other in binding to PXD, consistent with previous studies showing that Henipavirus N–P can form heterologous complexes.23 The HeV PXD scaffold seems to be more tolerant (i.e. equally able to accommodate the homologous and the heterologous Box3 region) than the NiV PXD. We can speculate that this difference could reflect the higher buried surface area (and hence the higher surface complementarity) of the NiV NTAIL–PXD complex as compared to the HeV one.21
PXD-induced folding of NTAIL deletion proteins
In order to be able to draw definitive conclusions about the role of Box3 not only in osmolyte-induced folding but also in PXD-induced folding, we carried out far-UV CD studies where the various NTAIL deletion proteins were mixed with the homologous PXD. The experimental design was the same already used to document PXD-induced α-helical folding of parental, full-length NTAIL proteins.21 Briefly, after recording the CD spectra of individual PXD and NTAIL proteins, for each NTAIL protein we also recorded the CD spectrum of a mixture containing a two-fold molar excess of the homologous PXD. Under these experimental conditions, the NTAIL and PXD concentrations (350 μM and 700 μM, respectively) were well above the estimated KD (and hence expected to lead to 100% complex formation) of the various NTAIL–PXD pairs, with the only exception of the NTAILΔ3Δ4–PXD pair for which the KD could not be determined. In the case of the NTAILΔ1, NTAILΔ1Δ2 and NTAILΔ4 proteins from both viruses, the observed CD spectra of the mixtures differed from the corresponding theoretical average curves calculated from the individual NTAIL and PXD spectra (Fig. 8). Since the theoretical average curves correspond to the spectra that would be expected if no structural variations occur, deviations from these curves indicate structural transitions. The experimentally observed spectra of the mixtures support an α-helical transition, as judged from the more pronounced minima at 208 and 222 nm, and by the higher ellipticity at 190 nm of the experimentally observed spectra compared to the corresponding theoretical average curves (Fig. 8). By contrast, PXD was found to be unable to trigger such an α-helical transition in both HeV and NiV NTAILΔ3Δ4 proteins, as judged from the good superimposition of the spectra of NTAIL + PXD mixtures with the corresponding average spectra (Fig. 8). These results indicate that Box3 is critical for NTAIL to undergo PXD-induced α-helical folding, since a form devoid of this box fails to fold onto PXD. | ||
| Fig. 8 Far-UV CD studies of HeV (A) and NiV (B) NTAIL proteins either alone (black line) or in the presence of a two-fold molar excess of the homologous PXD protein (full circles). The CD spectra of PXD proteins alone (grey line) are also shown, as are the theoretical average curves (empty circles) calculated by assuming that no structural variations occur (see Experimental). Spectra were recorded at 20 °C in 10 mM Tris/HCl pH 7.5, NaCl 200 mM. Protein concentrations were 10 mg mL−1. In NTAIL/PXD mixtures the NTAIL concentration was 350 μM, while that of PXD was 700 μM. The path length was 0.01 mm. Data are representative of at least two independent experiments. | ||
Conclusions
In the present work, we carried out an in-depth biochemical characterization of Henipavirus NTAIL deletion proteins that bear different combinations of four putative MoREs.The first interesting observation that we made concerns the anomalous electrophoretic behavior of the NTAIL proteins, which could not be exclusively accounted for by their content in acidic residues, suggesting that other additional parameters can be responsible for the abnormal electrophoretic migration of IDPs in SDS-PAGE. A possible hint in this regard is provided by the behavior of the NiV NTAILΔ1Δ2 protein. Indeed, this protein was found to display the most aberrant electrophoretic mobility. Besides, its electrophoretic behavior is more anomalous than that of the cognate HeV NTAIL protein. While the two proteins have the same content in acidic residues, the NiV NTAILΔ1Δ2 protein was found to adopt a more extended conformation than the corresponding HeV protein. This observation suggests that the degree of protein extension in solution could be a possible additional parameter affecting the electrophoretic mobility of IDPs. In further support of this hypothesis, analysis of the NTAIL proteins by native polyacrylamide gel electrophoresis (see Fig. S4, ESI‡) clearly shows that the NiV NTAILΔ1Δ2 protein migrates much more slowly than the HeV NTAILΔ1Δ2 protein, consistent with the lower compaction of the NiV NTAILΔ1Δ2 protein as compared to the HeV one. In the same vein, the NiV NTAILΔ4 protein migrates much faster than the HeV NTAILΔ4 protein, reflecting the higher extent of compaction of the NiV protein as compared to the HeV one.
By combining computational, hydrodynamic and spectroscopic analyses, we could unveil regions playing a major role in protein compaction, as well as regions sampling transiently populated α-helices (Fig. 9A). In particular, the spectroscopic studies herein described revealed the structural state of Box2. The present results also support the occurrence within Henipavirus NTAIL of two I-MoREs (e.g. Box1 and Box4) and of two α-MoREs (e.g. Box2 and Box3), thus arguing for the lack of a transiently populated β structure within Box2 (Fig. 9A). Among the two α-MoREs, Box3 was found to be a major determinant of α-helical folding and ITC studies showed that this region is strictly required for binding to PXD (Fig. 9A). These results confirm the previously postulated role of Box3 in binding to PXD and provide direct experimental support for the proposed Henipavirus NTAIL–PXD complexes.21 In addition, ITC studies in which the full-length NTAIL proteins were titrated with the heterologous PXD, showed that the two Box3 regions are functionally interchangeable between the two viruses, consistent with previous immunoprecipitation studies showing that Henipavirus N–P can form heterologous complexes.23 The HeV PXD was found to be seemingly more tolerant than the NiV PXD (i.e. equally able to accommodate the homologous and the heterologous Box3 region), with this property possibly reflecting a higher surface complementarity in the case of the NiV NTAIL–PXD complex as compared to the HeV one.21
 | ||
| Fig. 9 (A) Sequence alignment between the HeV and NiV NTAIL proteins. Hydrophobic residues (L, I, V, F, W and M) are shown in green, acidic residues in red and basic residues in blue. Asterisks and colons below the alignment denote identity and similarity, respectively, while dots indicate weakly similar residues. Lack of symbol below the alignment denotes strongly different residues. The front numbers correspond to the amino acid position in the N sequence. Dots above the alignment indicate intervals of 10 residues. The four boxes are shown, as is the location of the transiently populated α-helices, as unveiled by CD studies. (B) Schematic diagram summarizing the data obtained by computational and hydrodynamic analyses. The red and green arrows highlight the discrepancies found between the two viruses in terms of compaction properties of NTAILΔ1Δ2 and NTAILΔ4 proteins. | ||
Strikingly, and in agreement with previous studies,60 the content in regular secondary structure was found not to be a major determinant of protein compaction, with NTAIL proteins depleted in MoREs being nevertheless able to adopt a collapsed state (Fig. 9B). Indeed, removal of Box3 plus Box4, where Box3 has a strong inherent α-helical propensity as judged from CD studies, led to a more compact form (see Fig. 9B). This finding suggests that the region downstream Box3 (i.e. residues 493–532) is endowed with a large conformational freedom that confers to the full-length protein an overall extended character. Thus, the occurrence of a transiently populated α-helix within Box3 would not impart strong compaction constraints to NTAIL and would be unable to compensate for the high flexibility of the downstream region. Lack of a strict relationship between the content in regular secondary structure and protein compaction is further supported by hydrodynamic and spectroscopic studies focused on NTAILΔ1, which showed that Box1 plays a role as a major determinant of compaction, with this property being quite independent from its ability to adopt a regular secondary structure: indeed CD studies unveiled that removal of Box1 does not lead to any significant reduction in the α-helical content nor does it affect the α-helical folding abilities of NTAILΔ1 as compared to the full-length form (see Fig. 9). Therefore, in the case of the NTAIL proteins, the PMG state likely mainly arises from the occurrence of either long-range or short-range tertiary contacts rather than from local constraints imposed by transiently populated regular secondary structure elements. The occurrence of long-range tertiary contacts within IDPs has been already well documented (for examples see ref. 89–97), suggesting that long-range contacts may serve as major determinants of protein compaction not only in Henipavirus NTAIL proteins but in IDPs in general. The present findings are expected to stimulate future studies in this field and hence to shed light on the conformational behavior of IDPs and on how this latter is encoded by the amino acid sequence.
We also devised a method that combines computational and spectroscopic analyses, which was meant to allow RC-like IDPs to be discriminated from PMG-like IDPs. This method, which is a combination of two methods previously introduced by Uversky,34,58 revealed once again that the content in unordered and α-helical structure is a poor predictive criterion for the prediction of protein compaction. These findings somehow challenge the well-established notion that PMG-like IDPs have more residual secondary structures than RC-like IDPs.34
Given the poor agreement between the hydrodynamic and the in silico analyses and the inconsistencies between the two viruses in the case of the NTAILΔ1Δ2 and NTAILΔ4 proteins (see Fig. 9B), no definitive conclusions could be drawn as to the role of Box2 and Box4 in determining protein compaction. We reasoned that the observed discrepancies might be accounted for by differences in the NTAIL sequence leading to a different pattern in the tertiary contacts (whether long- or short-range) in the two viruses. In order to get insights into this issue, we compared the amino acid sequences of the HeV and NiV NTAIL proteins in view of highlighting possible differences. As shown in Fig. 9A, the NTAIL sequence is highly conserved in the two viruses (76% identity and 90% similarity). As expected for disordered regions not predicted to be involved in partner recognition, the regions connecting Box1 to Box2 and Box3 to Box4 are the most variable ones. The local sequence variability between Box3 and Box4 might account for conformational differences between HeV and NiV NTAILΔ1Δ2. On the other hand, the conformational differences between the HeV and NiV NTAILΔ4 proteins are more difficult to explain. Indeed, the possibility that the differences occurring in the region between Box1 and Box2 could be responsible for the differences between HeV and NiV NTAILΔ4 can be ruled out, based on the conformational similarity between HeV and NiV NTAILΔ3Δ4. Likewise, since the variable region between Box3 and Box4 does not seemingly lead to any significant difference in the conformational behavior of the HeV and NiV NTAILΔ1 proteins, it is difficult to explain how differences in this region could be responsible for the observed conformational differences between HeV and NiV NTAILΔ4. More subtle parameters are certainly at play, which remain however elusive so far.
As for Box3, while residues 474–487 are fully conserved in the two viruses, the last five residues are not (Fig. 9A). Given the functional equivalence of Box3 between the two viruses, this observation may imply that either residues 488–493 are not as much involved in PXD binding as the upstream residues, or that the partner can equally accommodate the IKESTA and AKEAAS stretches.
In conclusion, the present studies provide the first detailed mapping of the PXD binding region within NTAIL and provide the first quantitative estimation of the ability of Henipavirus NTAIL and PXD to form heterologous complexes. These studies designate specific Henipavirus NTAIL regions as targets for future site-directed mutagenesis studies and/or for the design of inhibitors capable to block the NTAIL–PXD interaction.
Experimental
Bioinformatics analyses
Sequences for this study were obtained from the VaZyMolO data base.98 Sequence accession numbers for NTAIL are VaZy82 (HeV) and VaZy1 (NiV). Disorder predictions were carried out using the MeDor metaserver for disorder prediction55 and the PONDR server (http://www.disorder.wsu.edu/) using the default integrated predictor VL-XT.56,57 Access to PONDR was provided by Molecular Kinetics (P.O. Box 2475 CS, Pullman, WA 99165-2475; E-mail: mkinetics@turbonet.com) under license from the WSU Research Foundation. PONDR is copyright ©1999 by the WSU Research Foundation, all rights reserved.The charge–hydropathy (CH) plot was generated as described in ref. 34. The CH plot is divided into two regions by a line, which corresponds to the equation H = (R + 1.151)/2.785, where R is the mean net charge and H is the mean hydrophobicity. In the left part of the diagram (where H < (R + 1.151)/2.785), a protein is predicted as disordered, whereas it is predicted as ordered in the right part.34HBoundary was computed according to ref. 34: HBoundary = (R + 1.15)/2.785. The mean net charge (R) of a protein is defined as the absolute value of the difference between the number of positively and negatively charged residues at pH 7 divided by the total number of amino acid residues. It was calculated using the program ProtParam at the EXPASY server (http://www.expasy.ch/tools). The mean hydrophobicity (H) is the sum of normalized hydrophobicities of individual residues divided by the total number of amino acid residues minus 4 residues (to take into account fringe effects in the calculation of hydrophobicity). Individual hydrophobicities were determined using the Protscale program at the EXPASY server (http://www.expasy.ch/tools), using the options “Hphob/Kyte & Doolittle”, a window size of 5, and normalizing the scale from 0 to 1.
The net charge per residue (NCR) of the various NTAIL truncated proteins was calculated as described in ref. 59 as the difference between the fraction of positively charged residues (f+) and the fraction of negatively charged residues (f−).
The pairwise sequence alignment between the HeV and NiV NTAIL proteins was obtained using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html).99 Sequence similarity and identity were calculated using the Emboss program100 (http://www.ebi.ac.uk/Tools/emboss/align/index.html).
Construction of expression plasmids
The Henipavirus pDest14/NTAILHN constructs, encoding residues 400–532 of either HeV or NiV N with a hexahistidine tag fused to their N-termini, and the pDest14/PXD constructs, encoding residues 660–709 (NiV) or 657–707 (HeV) of P with a hexahistidine tag fused to their C-termini, have already been described.21,29All NTAIL constructs were obtained by PCR using Phusion (Finnzymes) polymerase and synthetic N genes (GenScript) optimized for the expression in E. coli, as templates. Primers (Operon) were designed to introduce an AttB1 and AttB2 sites at the 5′ and 3′ ends, respectively, and to amplify the desired part of the NTAIL ORF. After digestion with DpnI (New England Biolabs) to remove the methylated DNA template, and purification (Nucleospin Extract II, Macherey-Nagel), the PCR products were cloned using the Gateway® recombination system (Invitrogen) into the pDest17OI expression vector, a modified pDest17® (Invitrogen) to which LacO and LacI were inserted to allow a better control of protein expression.101 The resulting constructs encode N-terminally histidine tagged proteins in which the native sequence is preceded by a vector-encoded MSYYHHHHHHLESTSLYKKAGS amino acid stretch. The pDest17OI/NTAILΔ1 and pDest17OI/NTAILΔ1Δ2 gene constructs encode residues 423–532 and 465–532 of Henipavirus N proteins, respectively. The pDest17OI/NTAILΔ4 and pDest17OI/NTAILΔ3Δ4 gene constructs encode residues 400–522 and 400–472 of Henipavirus N proteins, respectively.
Selection and amplification of DNA constructs were carried out using CaCl2-competent E. coli TAM1 cells (Active Motif). The sequence of the coding region of all expression plasmids was verified by sequencing (GATC Biotech) and found to conform to expectations.
Bacterial expression and purification of Henipavirus PXD and NTAIL constructs
The E. coli Rosetta [DE3] pLysS strain (Novagen) was used for the expression of all the Henipavirus PXD and NTAIL constructs. This strain, which is optimized for the expression of recombinant proteins, also carries the lysozyme gene thus allowing a tight regulation of the expression of the recombinant gene, as well as a facilitated lysis. Cultures were grown overnight to saturation in 2YT medium containing 100 μg mL−1 ampicillin and 34 μg mL−1 chloramphenicol. An aliquot of the overnight culture was diluted 1/25 in 2YT medium and grown at 37 °C. When the optical density at 600 nm (OD600) reached 0.6–0.8, isopropyl β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM, and the cells were grown at 37 °C for 3 additional hours. The induced cells were harvested, washed and collected by centrifugation (5000 g, 10 min). The resulting pellets were frozen at −20 °C.The NTAIL truncated proteins were purified using the protocol already described for Henipavirus wt NTAIL proteins29 with minor modifications. Briefly, cellular pellets were resuspended in 5 volumes (v/w) buffer A (10 mM Tris/HCl pH 8, 300 mM NaCl, 10 mM Imidazole, 1 mM phenyl-methyl-sulfonyl-fluoride – PMSF) supplemented with lysozyme 0.1 mg mL−1, DNAse I 10 μg mL−1, 20 mM MgSO4 and protease inhibitor cocktail (Roche, one tablet per 50 mL of bacterial lysate). After a 20 min incubation with gentle agitation, the cells were disrupted by sonication (using a 750 W sonicator and 4 cycles of 30 s each at 45% power output). The lysate was clarified by centrifugation at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 30 min. Starting from a 1 L culture, the clarified supernatant was incubated for 1 h with gentle shaking with 4 mL Chelating Sepharose Fast Flow Resin preloaded with Ni2+ ions (GE, Healthcare), previously equilibrated in buffer A. The resin was washed with buffer A supplemented with 20 mM imidazole, and the recombinant protein was eluted in buffer A supplemented with 250 mM imidazole. Eluates were analyzed by SDS-PAGE for the presence of the desired protein product. The fractions containing the recombinant protein were combined, and then loaded onto a Superdex 200 HR 16/60 column (GE, Healthcare). The elution buffer was 50 mM Tris/HCl pH 7.5, supplemented with either 300 mM (NTAILΔ1 and NTAILΔ4) or 500 mM (NTAILΔ1Δ2 and NTAILΔ3Δ4) NaCl. Purification of Henipavirus PXD and full-length NTAIL samples was carried out as described.21
000 g for 30 min. Starting from a 1 L culture, the clarified supernatant was incubated for 1 h with gentle shaking with 4 mL Chelating Sepharose Fast Flow Resin preloaded with Ni2+ ions (GE, Healthcare), previously equilibrated in buffer A. The resin was washed with buffer A supplemented with 20 mM imidazole, and the recombinant protein was eluted in buffer A supplemented with 250 mM imidazole. Eluates were analyzed by SDS-PAGE for the presence of the desired protein product. The fractions containing the recombinant protein were combined, and then loaded onto a Superdex 200 HR 16/60 column (GE, Healthcare). The elution buffer was 50 mM Tris/HCl pH 7.5, supplemented with either 300 mM (NTAILΔ1 and NTAILΔ4) or 500 mM (NTAILΔ1Δ2 and NTAILΔ3Δ4) NaCl. Purification of Henipavirus PXD and full-length NTAIL samples was carried out as described.21
The proteins were concentrated using Centricon Plus-20 (molecular cutoff of 3000 Da for PXD, and of 5000 Da for NTAIL) (Millipore). All proteins were stored at −20 °C. Dialysis was used to exchange the buffer and adjust it to the ensuing analyses. All purification steps, except for gel filtrations, were carried out at 4 °C.
Protein concentrations of NTAIL truncated forms were derived using the theoretical absorption coefficients at 280 nm, as obtained using the program ProtParam at the EXPASY server (http://www.expasy.ch/tools). In the case of PXD samples, protein concentrations were estimated using the BCA protein assay reagent (Pierce), since estimations based on the theoretical absorption coefficients at 280 nm turned out to be not fully reliable.21
The purified NTAIL proteins were also analyzed by native polyacrylamide gel electrophoresis. In those experiments, migration was carried out in the absence of SDS. In addition, the sample was not boiled before being subjected to electrophoresis and the loading buffer was devoid of both SDS and β-mercaptoethanol. Proteins were thus separated as a function of their inherent charge and compaction.
Mass spectrometry (MALDI-TOF)
The identity of all purified Henipavirus NTAIL proteins was confirmed by mass spectral analysis of tryptic fragments. The latter was obtained by digesting (0.25 μg trypsin) 1 μg of purified recombinant protein obtained after separation onto SDS-PAGE. The tryptic peptides were analyzed using an Autoflex II TOF/TOF. Spectra were acquired in the linear mode. Samples (0.7 μL containing 15 pmol) were mixed with an equal volume of sinapinic acid matrix solution, spotted on the target, then dried at room temperature for 10 min. The tryptic fragments were analyzed in the Autoflex matrix-assisted laser desorption ionization/time of flight (MALDI-TOF)1 (Bruker Daltonics, Bremen, Germany). Peptide fingerprints were obtained and compared with in silico protein digest (Biotools, Bruker Daltonics, Germany). The mass standards were either autolytic tryptic peptides or peptide standards (Bruker Daltonics).Analytical SEC combined with on-line multi-angle laser light scattering (MALLS) and refractometry (RI) and calculations of hydrodynamic radii
Analytical SEC was carried out on a high pressure liquid chromatography (HPLC) system (Alliance 2695, Waters) using silica-based columns (Shodex). A 15 mL KW-802.5 column was used for the characterization of NTAIL truncated proteins. Proteins were eluted with a 50 mM Tris/HCl pH 7.3, 200 mM NaCl buffer at a flow of 0.5 mL min−1. Separation was performed at room temperature. Typically, 30 μL of a protein solution at 12–23 mg mL−1 in 50 mM Tris/HCl pH 7.5, supplemented with either 300 mM (NTAILΔ1 and NTAILΔ4) or 500 mM (NTAILΔ1Δ2 and NTAILΔ3Δ4) NaCl, were injected. Detection was performed using a triple-angle light-scattering detector (MiniDAWN™ TREOS, Wyatt Technology), a quasi-elastic light-scattering instrument (Dynapro™, Wyatt Technology) and a differential refractometer (Optilab® rEX, Wyatt Technology). Molecular mass and RS determination was performed by the ASTRA V software (Wyatt Technology) using a dn/dc value of 0.185 mL g−1. The column was calibrated with proteins of known Stokes radii and molecular masses.The theoretical Stokes radii (RS, in Å) expected for a natively folded (RNFS), fully unfolded random coil state in urea (RUS) and natively unfolded PMG (RPMGS) protein with an expected molecular mass (MMtheo) (in Daltons) were calculated according to ref. 61:
| log(RNFS) = 0.369 × log(MMtheo) − 0.254 | (2) |
| log(RUS) = 0.521 × log(MMtheo) − 0.649 | (3) |
| log(RPMGS) = 0.403 × log(MMtheo) − 0.239 | (4) |
The hydrodynamic radius of a natively unfolded protein with N residues was also calculated according to ref. 60 using either the simple power-law model:
| RIDPS = R0Nν | (5) |
| RIDPSseq = (APPro + B) × (C|Q| + D)Shis × R0Nν | (6) |
The compaction index (CI) is expressed as:
| CI = (RUS − RobsS)/(RUS − RNFS) | (7) |
Circular dichroism (CD)
CD spectra were recorded at 20 °C on a Jasco 810 dichrograph, equipped with a Peltier thermoregulation system. Spectra of NTAIL truncated proteins in the presence of 20% TFE (Sigma-Aldrich) or 3 M TMAO were recorded using 1 mm thick quartz cells. When recording the CD spectra of NTAIL + PXD mixtures, 0.01 mm thick quartz cells were used. In the case of spectra recorded in the presence of TFE or TMAO, the buffer was 10 mM sodium phosphate pH 7. In experiments where the ability of NTAIL proteins to undergo PXD-induced folding was assessed, the buffer was 10 mM Tris/HCl pH 7.5, NaCl 200 mM. CD spectra were measured between 190 and 260 nm, with a scanning speed of 20 nm min−1 and a data pitch of 0.2 nm. Spectra were averaged from three scans. Moreover, for each protein sample, at least two independent acquisitions were carried out so as to estimate the experimental error arising from sample preparation. The contribution of buffer was subtracted from experimental spectra. Spectra were smoothed using the “means-movement” smoothing procedure implemented in the SpectraManager package. Structural variations in NTAIL proteins were measured as a function of changes in the initial CD spectrum upon addition of either 20% TFE or 3 M TMAO or a two-fold molar excess of PXD to NTAIL proteins.Mean ellipticity values per residue ([Θ]) were calculated as [Θ] = 3300 m ΔA/(lcn), where l (path length) in cm, n = number of residues, m = molecular mass in daltons and c = protein concentration expressed in mg mL−1. Number of residues (n) are 132 for NTAILΔ1 proteins, 90 for NTAILΔ1,2 proteins, 145 for NTAILΔ4 proteins, 95 for NTAILΔ3,4 proteins, 58 for HeV PXD and 57 for NiV PXD. Mass (m) values for HeV NTAIL proteins are 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 519 Da (NTAILΔ1), 10
519 Da (NTAILΔ1), 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 Da (NTAILΔ1Δ2), 15
012 Da (NTAILΔ1Δ2), 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 775 Da (NTAILΔ4), and 10
775 Da (NTAILΔ4), and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 319 Da (NTAILΔ3Δ4). Mass values for NiV NTAIL proteins are 14
319 Da (NTAILΔ3Δ4). Mass values for NiV NTAIL proteins are 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 286 Da (NTAILΔ1), 9862 Da (NTAILΔ1Δ2), 15
286 Da (NTAILΔ1), 9862 Da (NTAILΔ1Δ2), 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 482 Da (NTAILΔ4), and 10
482 Da (NTAILΔ4), and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 Da (NTAILΔ3Δ4). Mass values for PXD proteins are 6871 Da for HeV and 6733 Da for NiV. Protein concentrations of 0.1 mg mL−1 were used when recording spectra of NTAIL proteins either in the absence or in the presence of 20% TFE or 3 M TMAO.
176 Da (NTAILΔ3Δ4). Mass values for PXD proteins are 6871 Da for HeV and 6733 Da for NiV. Protein concentrations of 0.1 mg mL−1 were used when recording spectra of NTAIL proteins either in the absence or in the presence of 20% TFE or 3 M TMAO.
When 0.01 mm thick quartz cells were used, protein concentrations as high as 10 mg mL−1 could be used. Use of high protein concentrations ensured achievement of saturation in induced folding experiments in the presence of the PXD homologous partner. In the case of protein mixtures (i.e. NTAIL + PXD), mean ellipticity values per residue ([Θ]) were calculated as [Θ] = 3300 ΔA/{[(C1n1/m1) + (C2n2/m2)]l}, where l (path length) = 0.001 cm, n1 or n2 = number of residues, m1 or m2 = molecular mass in daltons and C1 or C2 = protein concentration expressed in mg mL−1 for each of the two proteins in the mixture. The theoretical average ellipticity values per residue ([Θ]Ave), assuming that neither unstructured-to-structured transitions nor secondary structure rearrangements occur, were calculated as follows: [Θ]Ave = [([Θ]1n1) + ([Θ]2n2R)]/(n1 + n2R), where [Θ]1 and [Θ]2 correspond to the measured mean ellipticity values per residue, n1 and n2 to the number of residues for each of the two proteins, and R to the excess molar ratio of protein 2.
The experimental data in the 190–260 nm range were analyzed using the DICHROWEB website (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml) which was supported by grants to the BBSRC Centre for Protein and Membrane Structure and Dynamics.102,103 The CDSSTR deconvolution method was used to estimate the content in secondary structure using the reference protein set 7. For the spectra recorded from TMAO-containing mixtures, which could be recorded in the 210–260 nm range only, the α-helical content was derived from the ellipticity at 222 nm as described in ref. 85.
Isothermal titration calorimetry (ITC)
ITC experiments were carried out on a ITC200 isothermal titration calorimeter (Microcal, Northampton, USA) at 20 °C. In these studies, the concentration of NTAIL proteins was initially adjusted to 150–180 μM in the microcalorimeter cell (0.2 mL). The corresponding PXD protein (stock solution at 1.5–1.8 mM) was added from a computer-controlled 40 μL microsyringe via a total of 19 injections of 2 μL each at intervals of 180 s. Whatever the binding reaction being studied, the pair of proteins used in each binding assay were dialyzed against a 10 mM Tris/HCl pH 7.5 buffer, supplemented with 300 mM NaCl, to minimize undesirable buffer-related effects. The dialysis buffer was used in all preliminary equilibration and washing steps.Heat dilution of the ligand was taken into account from peaks measured after full saturation of the protein sample contained in the microcalorimeter cell by the ligand. A theoretical titration curve was fitted to the experimental data using the ORIGIN software (Microcal). This software uses the relationship between the heat generated by each injection and ΔH° (enthalpy change in cal mole−1), KA (association binding constant in M−1), n (number of binding sites per monomer), total protein concentration and free and total ligand concentrations. The variation in the entropy (ΔS in cal mol−1 deg−1) of each binding reaction was inferred from the variation in the free energy (ΔG), where this latter was calculated from the following relation: ΔG = −RTLn 1/KA.
Funding
This work was carried out with the financial support of the Agence Nationale de la Recherche, specific program “Physico-Chimie du Vivant”, ANR-08-PCVI-0020-01, and with the partial support of the CNRS. DB is supported by a joint doctoral fellowship from the Direction Générale de l'Armement (DGA) and the CNRS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.Acknowledgements
We wish to thank Nicholas Armstrong and the mass spectrometry platform of the IFR48 of Marseille for mass spectrometry analyses. We also thank Catarina Rodrigues for help with SEC-MALLS measurements. We are very grateful to Vladimir Uversky for kindly providing us with the ellipticity values of a set of well-characterized premolten globule-like and random coil-like proteins.References
- R. Stone, Science, 2011, 331, 1128–1131 Search PubMed.
- B. T. Eaton, J. S. Mackenzie and L. F. Wang, Henipaviruses, in Fields Virology, ed. B. N. Fields, D. M. Knipe and P. M. Howley, 5th edn, Lippincott-Raven, Philadelphia, 2007, pp. 1587–1600 Search PubMed.
- J. F. Drexler, V. M. Corman, F. Gloza-Rausch, A. Seebens, A. Annan, A. Ipsen, T. Kruppa, M. A. Muller, E. K. Kalko, Y. Adu-Sarkodie, S. Oppong and C. Drosten, PLoS One, 2009, 4, e6367 Search PubMed.
- M. Enserink, Science, 2004, 303, 1121 Search PubMed.
- D. Butler, Nature, 2004, 429, 7 Search PubMed.
- V. P. Hsu, M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee and R. F. Breiman, Emerging Infect. Dis., 2004, 10, 2082–2087 Search PubMed.
- N. Homaira, M. Rahman, M. J. Hossain, J. H. Epstein, R. Sultana, M. S. Khan, G. Podder, K. Nahar, B. Ahmed, E. S. Gurley, P. Daszak, W. I. Lipkin, P. E. Rollin, J. A. Comer, T. G. Ksiazek and S. P. Luby, Epidemiol. Infect., 2010, 138, 1630–1636 Search PubMed.
- J. J. Sejvar, J. Hossain, S. K. Saha, E. S. Gurley, S. Banu, J. D. Hamadani, M. A. Faiz, F. M. Siddiqui, Q. D. Mohammad, A. H. Mollah, R. Uddin, R. Alam, R. Rahman, C. T. Tan, W. Bellini, P. Rota, R. F. Breiman and S. P. Luby, Ann. Neurol., 2007, 62, 235–242 Search PubMed.
- L. F. Wang, M. Yu, E. Hansson, L. I. Pritchard, B. Shiell, W. P. Michalski and B. T. Eaton, J. Virol., 2000, 74, 9972–9979 CrossRef CAS.
- B. T. Eaton, C. C. Broder, D. Middleton and L. F. Wang, Nat. Rev. Microbiol., 2006, 4, 23–35 Search PubMed.
- D. Karlin, S. Longhi and B. Canard, Virology, 2002, 302, 420–432 CrossRef CAS.
- S. Longhi, V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet and B. Canard, J. Biol. Chem., 2003, 278, 18638–18648 CrossRef CAS.
- G. Schoehn, M. Mavrakis, A. Albertini, R. Wade, A. Hoenger and R. W. Ruigrok, J. Mol. Biol., 2004, 339, 301–312 CrossRef CAS.
- D. Bhella, Measles virus nucleocapsid structure, conformational flexibility and the rule of six, in Measles virus nucleoprotein, ed. S. Longhi, Nova Publishers Inc., Hauppage, NY, 2007 Search PubMed.
- D. Bhella, A. Ralph, L. B. Murphy and R. P. Yeo, J. Gen. Virol., 2002, 83, 1831–1839 Search PubMed.
- D. Bhella, A. Ralph and R. P. Yeo, J. Mol. Biol., 2004, 340, 319–331 CrossRef CAS.
- M. Ringkjøbing Jensen, G. Communie, E. D. Ribeiro Jr., N. Martinez, A. Desfosses, L. Salmon, L. Mollica, F. Gabel, M. Jamin, S. Longhi, R. W. Ruigrok and M. Blackledge, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9839–9844 CrossRef.
- W. S. Tan, S. T. Ong, M. Eshaghi, S. S. Foo and K. Yusoff, J. Med. Virol., 2004, 73, 105–112 CrossRef CAS.
- M. Eshaghi, W. S. Tan, S. T. Ong and K. Yusoff, J. Clin. Microbiol., 2005, 43, 3172–3177 Search PubMed.
- S. T. Ong, K. Yusoff, C. L. Kho, J. O. Abdullah and W. S. Tan, J. Gen. Virol., 2009, 90, 392–397 Search PubMed.
- J. Habchi, S. Blangy, L. Mamelli, M. Ringkjobing Jensen, M. Blackledge, H. Darbon, M. Oglesbee, Y. Shu and S. Longhi, J. Biol. Chem., 2011, 286, 13583–13602 CrossRef CAS.
- K. Halpin, B. Bankamp, B. H. Harcourt, W. J. Bellini and P. A. Rota, J. Gen. Virol., 2004, 85, 701–707 Search PubMed.
- Y. P. Chan, C. L. Koh, S. K. Lam and L. F. Wang, J. Gen. Virol., 2004, 85, 1675–1684 Search PubMed.
- M. Omi-Furutani, M. Yoneda, K. Fujita, F. Ikeda and C. Kai, J. Virol., 2010, 84, 9793–9799 Search PubMed.
- Z. Lou, Y. Xu, K. Xiang, N. Su, L. Qin, X. Li, G. F. Gao, M. Bartlam and Z. Rao, FEBS J., 2006, 273, 4538–4547 Search PubMed.
- T. A. Bowden, M. Crispin, D. J. Harvey, A. R. Aricescu, J. M. Grimes, E. Y. Jones and D. I. Stuart, J. Virol., 2008, 82, 11628–11636 Search PubMed.
- T. A. Bowden, A. R. Aricescu, R. J. Gilbert, J. M. Grimes, E. Y. Jones and D. I. Stuart, Nat. Struct. Mol. Biol., 2008, 15, 567–572 Search PubMed.
- T. A. Bowden, M. Crispin, D. J. Harvey, E. Y. Jones and D. I. Stuart, J. Virol., 2010, 84, 6208–6217 Search PubMed.
- J. Habchi, L. Mamelli, H. Darbon and S. Longhi, PLoS One, 2010, 5, e11684 CrossRef.
- J. Habchi and S. Longhi, Mol. BioSyst., 2011 10.1039/c1mb05204g.
- J. Habchi, L. Mamelli and S. Longhi, Structural disorder within the nucleoprotein and phosphoprotein from measles, Nipah and Hendra viruses, in Flexible viruses: structural disorder in viral proteins, ed. V. N. Uversky and S. Longhi, John Wiley and Sons, Hoboken, New Yersey, 2011 Search PubMed.
- A. K. Dunker, J. D. Lawson, C. J. Brown, R. M. Williams, P. Romero, J. S. Oh, C. J. Oldfield, A. M. Campen, C. M. Ratliff, K. W. Hipps, J. Ausio, M. S. Nissen, R. Reeves, C. Kang, C. R. Kissinger, R. W. Bailey, M. D. Griswold, W. Chiu, E. C. Garner and Z. Obradovic, J. Mol. Graphics Modell., 2001, 19, 26–59 CrossRef CAS.
- A. K. Dunker and Z. Obradovic, Nat. Biotechnol., 2001, 19, 805–806 CrossRef CAS.
- V. N. Uversky, Protein Sci., 2002, 11, 739–756 CrossRef CAS.
- V. N. Uversky, Cell. Mol. Life Sci., 2003, 60, 1852–1871 CrossRef CAS.
- H. J. Dyson and P. E. Wright, Nat. Rev. Mol. Cell Biol., 2005, 6, 197–208 CrossRef CAS.
- P. Radivojac, L. M. Iakoucheva, C. J. Oldfield, Z. Obradovic, V. N. Uversky and A. K. Dunker, Biophys. J., 2007, 92, 1439–1456 CAS.
- A. K. Dunker, C. J. Oldfield, J. Meng, P. Romero, J. Y. Yang, J. W. Chen, V. Vacic, Z. Obradovic and V. N. Uversky, BMC Genomics, 2008, 9(Suppl 2), S1 CrossRef.
- A. K. Dunker, I. Silman, V. N. Uversky and J. L. Sussman, Curr. Opin. Struct. Biol., 2008, 18, 756–764 CrossRef CAS.
- V. N. Uversky, J. Biomed. Biotechnol., 2010, 2010, 568068 Search PubMed.
- V. N. Uversky and A. K. Dunker, Biochim. Biophys. Acta, 2010, 1804, 1231–1264 CAS.
- K. K. Turoverov, I. M. Kuznetsova and V. N. Uversky, Prog. Biophys. Mol. Biol., 2010, 102, 73–84 CrossRef CAS.
- T. Chouard, Nature, 2011, 471, 151–153 Search PubMed.
- C. J. Oldfield, Y. Cheng, M. S. Cortese, P. Romero, V. N. Uversky and A. K. Dunker, Biochemistry, 2005, 44, 12454–12470 CrossRef CAS.
- A. Mohan, C. J. Oldfield, P. Radivojac, V. Vacic, M. S. Cortese, A. K. Dunker and V. N. Uversky, J. Mol. Biol., 2006, 362, 1043–1059 CrossRef CAS.
- V. Vacic, C. J. Oldfield, A. Mohan, P. Radivojac, M. S. Cortese, V. N. Uversky and A. K. Dunker, J. Proteome Res., 2007, 6, 2351–2366 CrossRef CAS.
- M. Fuxreiter, P. Tompa and I. Simon, Bioinformatics, 2007, 23, 950–956 CrossRef CAS.
- K. Johansson, J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard and S. Longhi, J. Biol. Chem., 2003, 278, 44567–44573 CrossRef CAS.
- R. L. Kingston, D. J. Hamel, L. S. Gay, F. W. Dahlquist and B. W. Matthews, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8301–8306 CrossRef CAS.
- D. Karlin, S. Longhi, V. Receveur and B. Canard, Virology, 2002, 296, 251–262 CrossRef CAS.
- J. M. Bourhis, V. Receveur-Bréchot, M. Oglesbee, X. Zhang, M. Buccellato, H. Darbon, B. Canard, S. Finet and S. Longhi, Protein Sci., 2005, 14, 1975–1992 CrossRef CAS.
- I. Sambi, P. Gatti-Lafranconi, S. Longhi and M. Lotti, FEBS J., 2010, 277, 4438–4451 CrossRef CAS.
- P. Tompa, Trends Biochem. Sci., 2002, 27, 527–533 CrossRef CAS.
- L. M. Iakoucheva, A. L. Kimzey, C. D. Masselon, R. D. Smith, A. K. Dunker and E. J. Ackerman, Protein Sci., 2001, 10, 1353–1362 Search PubMed.
- P. Lieutaud, B. Canard and S. Longhi, BMC Genomics, 2008, 9, S25 CrossRef.
- X. Li, P. Romero, M. Rani, A. K. Dunker and Z. Obradovic, Genome Inform. Ser., 1999, 10, 30–40 Search PubMed.
- P. Romero, Z. Obradovic, X. Li, E. C. Garner, C. J. Brown and A. K. Dunker, Proteins, 2001, 42, 38–48 CrossRef CAS.
- V. N. Uversky, J. R. Gillespie and A. L. Fink, Proteins, 2000, 41, 415–427 CrossRef CAS.
- A. H. Mao, S. L. Crick, A. Vitalis, C. L. Chicoine and R. V. Pappu, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 8183–8188 CrossRef CAS.
- J. A. Marsh and J. D. Forman-Kay, Biophys. J., 2010, 98, 2383–2390 CrossRef CAS.
- V. N. Uversky, Eur. J. Biochem., 2002, 269, 2–12 CrossRef CAS.
- S. Brocca, L. Testa, F. Sobott, M. Samalikova, A. Natalello, E. Papaleo, M. Lotti, L. De Gioia, S. M. Doglia, L. Alberghina and R. Grandori, Biophys. J., 2011, 100, 2243–2252 CrossRef CAS.
- I. Baskakov, A. Wang and D. W. Bolen, Biophys. J., 1998, 74, 2666–2673 Search PubMed.
- I. Baskakov and D. W. Bolen, J. Biol. Chem., 1998, 273, 4831–4834 CrossRef CAS.
- I. V. Baskakov, R. Kumar, G. Srinivasan, Y. S. Ji, D. W. Bolen and E. B. Thompson, J. Biol. Chem., 1999, 274, 10693–10696 Search PubMed.
- D. W. Bolen and I. V. Baskakov, J. Mol. Biol., 2001, 310, 955–963 CrossRef CAS.
- G. Tell, L. Perrone, D. Fabbro, L. Pellizzari, C. Pucillo, M. De Felice, R. Acquaviva, S. Formisano and G. Damante, Biochem. J., 1998, 329, 395–403 Search PubMed.
- Q. X. Hua, W. H. Jia, B. P. Bullock, J. F. Habener and M. A. Weiss, Biochemistry, 1998, 37, 5858–5866 CrossRef CAS.
- K. Dahlman-Wright and I. J. McEwan, Biochemistry, 1996, 35, 1323–1327 CrossRef CAS.
- K. Watt, T. J. Jess, S. M. Kelly, N. C. Price and I. J. McEwan, Biochemistry, 2005, 44, 734–743 Search PubMed.
- P. Luo and R. L. Baldwin, Biochemistry, 1997, 36, 8413–8421 CrossRef CAS.
- A. Cammers-Goodwin, T. J. Allen, S. L. Oslick, K. F. McClure, J. H. Lee and D. S. Kemp, J. Am. Chem. Soc., 1996, 118, 3082–3090 CrossRef CAS.
- M. Buck, H. Schwalbe and C. M. Dobson, Biochemistry, 1995, 34, 13219–13232 CrossRef CAS.
- P. Fan, C. Bracken and J. Baum, Biochemistry, 1993, 32, 1573–1582 CrossRef CAS.
- Y. Luo and R. L. Baldwin, J. Mol. Biol., 1998, 279, 49–57 CrossRef CAS.
- V. N. Uversky, V. P. Kutyshenko, N. Protasova, V. V. Rogov, K. S. Vassilenko and A. T. Gudkov, Protein Sci., 1996, 5, 1844–1851 CrossRef CAS.
- A. Wang and D. W. Bolen, Biochemistry, 1997, 36, 9101–9108 CrossRef CAS.
- M. Van Hoy, K. K. Leuther, T. Kodadek and S. A. Johnston, Cell, 1993, 72, 587–594 Search PubMed.
- J. M. Mouillon, P. Gustafsson and P. Harryson, Plant Physiol., 2006, 141, 638–650 Search PubMed.
- V. Belle, S. Rouger, S. Costanzo, E. Liquiere, J. Strancar, B. Guigliarelli, A. Fournel and S. Longhi, Proteins: Struct., Funct., Bioinf., 2008, 73, 973–988 Search PubMed.
- I. V. Baskakov, R. Kumar, G. Srinivasan, Y. S. Ji, D. W. Bolen and E. B. Thompson, J. Biol. Chem., 1999, 274, 10693–10696 Search PubMed.
- R. Kumar, J. M. Serrette, S. H. Khan, A. L. Miller and E. B. Thompson, Arch. Biochem. Biophys., 2007, 465, 452–460 Search PubMed.
- L. B. Dyksterhuis, E. A. Carter, S. M. Mithieux and A. S. Weiss, Arch. Biochem. Biophys., 2009, 487, 79–84 Search PubMed.
- M. Couturier, M. Buccellato, S. Costanzo, J. M. Bourhis, Y. Shu, M. Nicaise, M. Desmadril, C. Flaudrops, S. Longhi and M. Oglesbee, J. Mol. Recognit., 2010, 23, 301–315 CAS.
- J. K. Myers, C. N. Pace and J. M. Scholtz, Biochemistry, 1997, 36, 10923–10929 Search PubMed.
- M. Foucault, K. Mayol, V. Receveur-Brechot, M. C. Bussat, C. Klinguer-Hamour, B. Verrier, A. Beck, R. Haser, P. Gouet and C. Guillon, Proteins, 2009, 78, 1441–1456 Search PubMed.
- S. M. Kelly and N. C. Price, Curr. Protein Pept. Sci., 2000, 1, 349–384 CrossRef CAS.
- M. W. Freyer and E. A. Lewis, Methods Cell Biol., 2008, 84, 79–113 Search PubMed.
- P. Bernado, C. W. Bertoncini, C. Griesinger, M. Zweckstetter and M. Blackledge, J. Am. Chem. Soc., 2005, 127, 17968–17969 CrossRef CAS.
- M. M. Dedmon, K. Lindorff-Larsen, J. Christodoulou, M. Vendruscolo and C. M. Dobson, J. Am. Chem. Soc., 2005, 127, 476–477 CrossRef CAS.
- C. W. Bertoncini, Y. S. Jung, C. O. Fernandez, W. Hoyer, C. Griesinger, T. M. Jovin and M. Zweckstetter, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 1430–1435 CrossRef CAS.
- C. W. Bertoncini, C. O. Fernandez, C. Griesinger, T. M. Jovin and M. Zweckstetter, J. Biol. Chem., 2005, 280, 30649–30652 CrossRef CAS.
- P. Vise, B. Baral, A. Stancik, D. F. Lowry and G. W. Daughdrill, Proteins, 2007, 67, 526–530 Search PubMed.
- D. F. Lowry, A. C. Hausrath and G. W. Daughdrill, Proteins, 2008, 73, 918–928 Search PubMed.
- D. F. Lowry, A. Stancik, R. M. Shrestha and G. W. Daughdrill, Proteins, 2008, 71, 587–598 Search PubMed.
- M. K. Cho, G. Nodet, H. Y. Kim, M. R. Jensen, P. Bernado, C. O. Fernandez, S. Becker, M. Blackledge and M. Zweckstetter, Protein Sci., 2009, 18, 1840–1846 Search PubMed.
- L. Salmon, G. Nodet, V. Ozenne, G. Yin, M. R. Jensen, M. Zweckstetter and M. Blackledge, J. Am. Chem. Soc., 2010, 132, 8407–8418 CrossRef CAS.
- F. Ferron, C. Rancurel, S. Longhi, C. Cambillau, B. Henrissat and B. Canard, J. Gen. Virol., 2005, 86, 743–749 Search PubMed.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS.
- P. Rice, I. Longden and A. Bleasby, Trends Genet., 2000, 16, 276–277 CrossRef CAS.
- R. Vincentelli, S. Canaan, V. Campanacci, C. Valencia, D. Maurin, F. Frassinetti, L. Scappucini-Calvo, Y. Bourne, C. Cambillau and C. Bignon, Protein Sci., 2004, 13, 2782–2792 Search PubMed.
- L. Whitmore and B. A. Wallace, Nucleic Acids Res., 2004, 32, W668–W673 CrossRef CAS.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89, 392–400 CrossRef CAS.
Footnotes |
| † Published as part of a Molecular BioSystems themed issue on Intrinsically Disordered Proteins; Guest Editor: M. Madan Babu. |
| ‡ Electronic supplementary information (ESI) available: Fig. S1–S4 and Tables S1 and S2. See DOI: 10.1039/c1mb05401e |
| This journal is © The Royal Society of Chemistry 2012 |
