Open Access Article
Open Access ArticleA series of water-soluble photosensitizers based on 3-cinnamoylcoumarin for in vitro antimicrobial photodynamic inactivation†
Zhiyuan Sun‡
ac,
Shaona Zhou‡b,
Haixia Qiub,
Ying Gub and
Yuxia Zhao *a
*a
aTechnical Institute of Physics and Chemistry, Chinese Academy of Sciences, Beijing 100190, P. R. China. E-mail: yuxia.zhao@mail.ipc.ac.cn; Tel: +86 10 82543569
bDepartment of Laser Medicine, Chinese PLA General Hospital, Beijing, 100853, P. R. China
cUniversity of Chinese Academy of Sciences, Beijing 100049, P. R. China
First published on 9th May 2018
Abstract
A series of novel water-soluble photosensitizers (PSs; M1–M5) based on 3-cinnamoylcoumarin derivatives, incorporating carboxylic acid salt (M1, M2), pyridine salt (M3, M4) and quaternary ammonium salt (M5) groups, were designed and synthesized. Their photophysical and photochemical properties and in vitro antimicrobial photodynamic inactivation (PDI) were investigated. M2, modified with two carboxylic acid salts, was unstable in phosphate-buffered saline (PBS). The four other PSs all showed higher binding/uptake to methicillin-resistant Staphylococcus aureus (MRSA), A. baumannii and C. albicans compared with the clinical drug methylene blue (MB), except for M1 to A. baumannii. Furthermore, the three cationic PSs (M3–M5) exhibited equivalent antibacterial PDI efficacies against MRSA and A. baumannii compared with MB. The antifungal efficacies of M4 and M5 to C. albicans were both significantly higher than that of MB, especially for M5, indicating that the quaternary ammonium-salt-modified coumarin derivative has substantial potential for antifungal PDI.
Introduction
Since their discovery in the early 20th century, antibiotics have been widely used to treat a variety of infectious diseases in clinical practice all over the world.1–3 However, with the extensive use of antibiotics, many drug-resistant or multidrug-resistant bacteria, even superbacteria, have emerged.4,5 Consensus has now been reached on the importance of reducing the abuse of antibiotics and finding new methods of treating infection diseases.6–9 Antimicrobial photodynamic inactivation (PDI) uses light to excite a disease-localized photosensitizer (PS) to generate reactive oxygen species (ROS), such as singlet oxygen (1O2), which can inactivate bacteria, viruses and fungi in situ.10–12 Unlike antibiotics, ROS has strong oxidizing properties with respect to all organic substrates, thus avoiding the problem of drug resistance.13–15Because of the wide variety of microorganisms, there is no clear guide for the selection of PSs for antimicrobial PDI at present. Generally, the outer walls of Gram-positive bacteria are relatively porous, so can readily be penetrated by cationic, anionic, neutral or nonionic PSs.16 Gram-negative bacteria have an additional and more negatively charged outer layer, which serves as a barrier to prevent the entry of outer species,17 meaning that only cationic PSs can easily be taken up. However, fungi are different from bacteria. Although fungi have an even thicker negatively charged outer wall than Gram-negative bacteria, anionic PSs still can inactivate them effectively.18 The available evidence shows that antifungal photodynamic therapy (PDT) involves more complex mechanisms, such as the presence of drug efflux pumps, the induction of enzymes and the formation of biofilms.19,20
In our previous work, a series of PSs modified by poly(ethylene glycol) (PEG), pyridinium or carboxylic acid salt based on benzylcyclopentanone were synthesized and investigated.21 The results showed that pyridinium- and carboxylic-acid-salt-modified PSs both had better antimicrobial PDI efficacies.22 Additionally, quaternary ammonium-salt-modified PSs have been used as antibacterial agents for a long time, and most could effectively inactivate bacteria.23–25 Coumarin is a natural substance found in many plants.26 Many clinical drugs contain coumarin or its derivatives.27,28 In cancer chemotherapy, coumarin and 7-hydroxycoumarin act as cytostatic agents against renal cell carcinoma.29,30 Furthermore, coumarins exhibit potent antitumor activities at different stages of cancer formation through various mechanisms, such as inducing cell apoptosis or inhibiting DNA-associated enzymes.31 In antitumor PDT, it was also proved that coumarin derivatives have good curative effects.32,33
In this article, we designed and synthesized five PSs based on 3-cinnamoylcoumarin derivatives through introduction of carboxylic acid salt (M1 and M2), cationic pyridinium salt (M3 and M4) and quaternary ammonium salt (M5) groups. Their photophysical and photochemical properties and in vitro antimicrobial PDI toward Gram-positive bacteria methicillin-resistant Staphylococcus aureus (MRSA), negative bacteria A. baumannii and fungus C. albicans were investigated.
Experimental
Synthesis and characterization
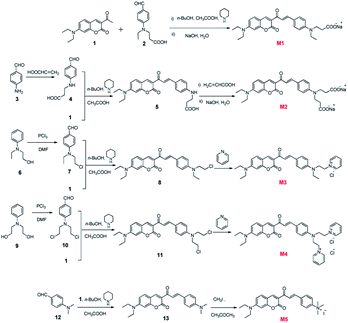
The synthetic routes towards M1–M5 are shown in Fig. 1. 3-Acetyl-7-(dimethylamino)-2H-chromen-2-one (1) and 3-(ethyl(4-formylphenyl)amino) propanoic acid (2) were synthesized according to our previous work.33 4-Aminobenzaldehyde (3), 2-(ethyl(phenyl)amino)ethanol (6), 2,2′-(phenylazanediyl) diethanol (9) and 4-(dimethylamino)benzaldehyde (12) were purchased from Energy Chemical. All targeted compounds were obtained by an initial high-yield aldol reaction between 3-acetyl-7-(dimethylamino)-2H-chromen-2-one and the corresponding p-aminobenzaldehyde, followed by the modification of different anions or cations. Synthetic details and characterization data are provided in ESI.†Instruments
1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker AV400 (400 MHz) spectrometer with deuterated reagents, CDCl3 or D2O, using tetramethylsilane as an internal standard. Mass spectra were measured using Bruker APEX 7.0E. Ultraviolet-visible (UV-Vis) spectra were measured using a Hitachi U-3900 spectrophotometer. Steady-state fluorescence was carried out on a Hitachi F-4500 spectrometer. Phosphate-buffered saline (PBS) buffer solution (0.01 M sodium phosphate, pH = 7.2–7.4) was purchased from Solarbio. Other chemical reagents were from Energy Chemical or J&K chemical Ltd. MRSA, A. baumannii and C. albicans were provided by Chinese PLA General Hospital.Singlet oxygen quantum yields (ΦΔ)
The singlet oxygen quantum yield (ΦΔ) was determined using Rose Bengal (RB) as the reference with a yield of 0.47 in dimethylformamide (DMF).34 1,3-Diphenylisobenzofuran (DPBF) was used as the scavenger.35 Mixed solutions of PSs (M1–M5 or RB) and DPBF were prepared. The concentrations of all solutions were adjusted to the same absorbance (0.1) at 473 nm, and the initial concentration of DPBF was consistent. To ensure the saturation of air, all solutions were stirred vigorously during the experimental procedure. The bleaching of the absorption band of DPBF in each solution at 415 nm was monitored under the irradiation of a 473 nm diode laser. The DPBF solution alone was also irradiated to diminish the errors origin from the photo-activation. The ΦΔ of each PS was calculated from the slope of the decay curve.The uptakes of PSs by microorganism cells
The binding of M1, M3, M4 and M5 to the three strains was measured as previously described.36,37 Briefly, the concentrations of bacteria or fungus were maintained at McF = 0.5, 108 cells per mL for a bacteria suspension and 106 cells per mL for a fungus suspension with 20 μM PSs. After 1 h of incubation in the dark at room temperature, the suspension was centrifuged at 5000 rpm for 10 min to harvest the bacteria. The obtained bacteria were washed with PBS three times and then lysed with lysozyme (100 μg mL−1) for 1 h, followed by sonication for 2 h at 37 °C. The lysed bacterial solution was centrifuged again to obtain the supernatant. The fluorescence intensities of the samples were measured for the different strains. Then the uptakes of various PSs were calculated according to the standard curve.In vitro antimicrobial PDI
In the PDI group (PS + laser), MRSA and A. baumannii (∼108 colony-forming unit [CFU] mL−1) and C. albicans cells (∼106 CFU mL−1) were incubated with PSs at different concentrations (2.5, 5, 7.5, 10 and 25 μM) for 1 h in the dark and then irradiated under a laser with an exposure dose of 24 J cm−2 (irradiated for 10 min by a laser with 40 mW cm−2 power density, 457 nm for M1, M3–M5 and 630 nm for methylene blue (MB) as the reference). A blank group and a laser group were also included. Incubation and illumination were carried out in 96-well plates (Costar, USA) with 40 μL of mixtures in each well. The viability of the microbial cells was determined by the dilution plating method, as described previously.38 In brief, 10 μL aliquots of each sample were serially diluted and spread over the surfaces of Sabouraud's dextrose agar plates. All plates were aerobically incubated in an incubator for 24 h. log10 reductions in microbial cells were compared with the untreated control. All experiments were performed three times.Results and discussion
Solubility and octanol–water partition coefficient
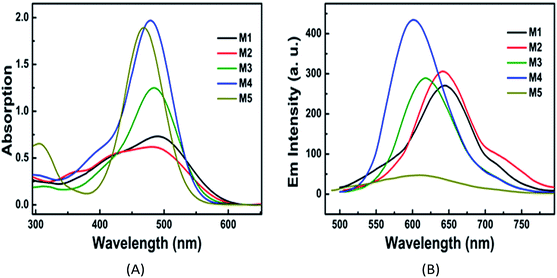
The UV-Vis absorption spectra and fluorescence emission spectra of PSs M1–M5 in PBS are shown in Fig. 2, and their corresponding data are listed in Table 1. All of these PSs have a strong charge transfer (CT) absorption peak at around 470–490 nm. For each PS, both the absorbance maximum λabsmax and the fluorescence emission maximum (λflmax) in PBS present red-shifts compared with the data in DMF, which can be ascribed to the solvent effects of the reduction of excited state energy in polar proton solvents. The octanol–water partition coefficient (log P) are 1.49, −1.20, 0.05, −0.48 and 0.11 for M1–M5, respectively. These results indicate that the water solubility of these PSs is enhanced with the increase in the number of pyridinium or carboxylic acid salts. Additionally, the ΦΔ of PSs M1–M5 were determined as 0.22, 0.10, 0.06, 0.15, and 0.16, respectively, in DMF, which are acceptable for PDT. However, the data in PBS could not be obtained due to the fast decay of singlet oxygen in water or PBS.35| Com | Solvent | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
λabsmax (nm) | εmax (104 M−1 cm−1) | λflmax (nm) | Δνss (10−4 nm−1) | Φf | ΦΔ |
|---|---|---|---|---|---|---|---|---|
a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P is the octanol–water partition coefficient; λabsmax is the absorption band maximum; εmax is molar absorption coefficient at λabsmax; λflmax is fluorescence emission maximum; Δνss is the Stokes shift; Φf is fluorescence quantum yield; ΦΔ is singlet oxygen quantum yield measured by photochemical trap method. P is the octanol–water partition coefficient; λabsmax is the absorption band maximum; εmax is molar absorption coefficient at λabsmax; λflmax is fluorescence emission maximum; Δνss is the Stokes shift; Φf is fluorescence quantum yield; ΦΔ is singlet oxygen quantum yield measured by photochemical trap method. |
||||||||
| M1 | DMF | 1.49 | 469 | 1.63 | 551 | 3.17 | 0.018 | 0.22 |
| PBS | 490 | 1.21 | 641 | 4.81 | 0.0023 | — | ||
| M2 | DMF | −1.20 | 437 | 1.98 | 566 | 5.22 | 0.0087 | 0.10 |
| PBS | 486 | 1.48 | 643 | 5.02 | 0.0031 | — | ||
| M3 | DMF | 0.05 | 473 | 3.46 | 602 | 4.53 | 0.0039 | 0.06 |
| PBS | 485 | 2.60 | 618 | 4.44 | 0.0036 | — | ||
| M4 | DMF | −0.48 | 470 | 5.15 | 529 | 2.37 | 0.011 | 0.15 |
| PBS | 481 | 4.50 | 609 | 4.37 | 0.0039 | — | ||
| M5 | DMF | 0.11 | 464 | 3.94 | 559 | 3.66 | 0.0098 | 0.16 |
| PBS | 470 | 3.86 | 615 | 5.01 | 0.0022 | — | ||
Photostability
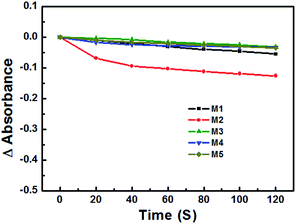
The photostability of these PSs was studied under irradiation of a 473 nm laser. As shown in Fig. 3, compared with the four other PSs, M2 was bleached 15% after 120 s. Additionally, even when kept in the dark, the PBS solution of M2 was still unstable after 8 h, and was therefore excluded in the follow-up in vitro experiments.The binding/uptake of PSs by bacteria and fungus cells
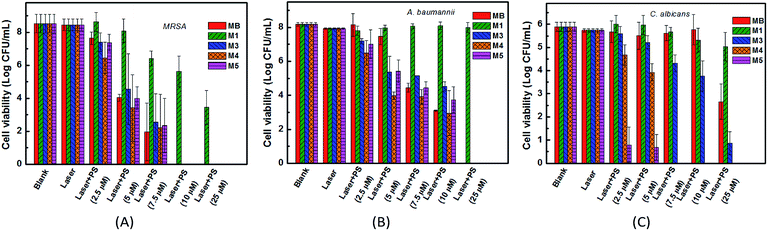
The binding/uptake amounts of these PSs by bacteria (MRSA and A. baumannii) and fungus (C. albicans) were tested. As shown in Fig. 4, M1 and M3–M5 can be uptaken more efficiently by the three strains compared with MB, which has almost no clear uptake of fluorescence signal. The reason may be ascribed to the good water solubility of MB (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.96 (ref. 39 and 40)), which affects its penetration in lipid cell membranes. Moreover, the intakes of M1, M3–M5 by C. albicans (665–3320 pmol/106 cells) are somewhat larger than the corresponding amounts taken by bacteria (2–118 pmol/106 cells), which is probably because of the larger size of the Candida species, about 25–50 times of the bacteria size.41 For the negatively charged cell walls, the cellular intakes of cationic PSs M3 and M5 by both bacteria and fungus are much higher than the corresponding amounts of anionic PS M1, such as 118 pmol/106 cells of M5 versus 38 pmol/106 cells of M1 in the MRSA group and 2260 pmol/106 cells of M3 versus 665 pmol/106 cells of M1 in the C. albicans group. The cellular uptake of M1 and M4 by MRSA and C. albicans are similar, while the cellular uptake of M1 by A. baumannii is obviously lower than that of M4. These results all indicate that the cationic PSs M3–M5 have good potential in antimicrobial PDI. Compared with M3 and M5, the binding/uptake of M4 is lower, which indicates that the charge is not the only determinant. According to our previous report,22 the cellular uptake also correlated with the log
P = −0.96 (ref. 39 and 40)), which affects its penetration in lipid cell membranes. Moreover, the intakes of M1, M3–M5 by C. albicans (665–3320 pmol/106 cells) are somewhat larger than the corresponding amounts taken by bacteria (2–118 pmol/106 cells), which is probably because of the larger size of the Candida species, about 25–50 times of the bacteria size.41 For the negatively charged cell walls, the cellular intakes of cationic PSs M3 and M5 by both bacteria and fungus are much higher than the corresponding amounts of anionic PS M1, such as 118 pmol/106 cells of M5 versus 38 pmol/106 cells of M1 in the MRSA group and 2260 pmol/106 cells of M3 versus 665 pmol/106 cells of M1 in the C. albicans group. The cellular uptake of M1 and M4 by MRSA and C. albicans are similar, while the cellular uptake of M1 by A. baumannii is obviously lower than that of M4. These results all indicate that the cationic PSs M3–M5 have good potential in antimicrobial PDI. Compared with M3 and M5, the binding/uptake of M4 is lower, which indicates that the charge is not the only determinant. According to our previous report,22 the cellular uptake also correlated with the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of PSs. Based on the lower log
P value of PSs. Based on the lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P data of −0.48 for M4 versus 1.49 for M3 and 0.11 for M5, we speculate that the lower binding/uptake of M4 is due to its enhanced water solubility by introducing two cationic groups.
P data of −0.48 for M4 versus 1.49 for M3 and 0.11 for M5, we speculate that the lower binding/uptake of M4 is due to its enhanced water solubility by introducing two cationic groups.
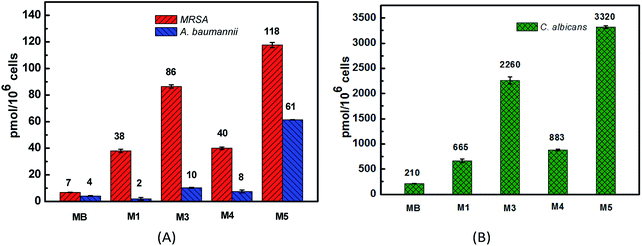 | ||
| Fig. 4 The cellular uptakes of PSs M1, M3–M5 and reference MB by MRSA, A. baumannii (A) and C. albicans (B). | ||
In vitro antimicrobial PDI
The toxicities of PSs toward the three strains with or without laser irradiation were tested. For the dark toxicity, after incubating the microorganisms with 100 μM PSs M1, M3–M5 or MB in the dark for 24 h, the bacteria or the fungus viabilities had no significant decrease in all groups. As shown in Fig. 5, except for the anionic PS M1, the cationic PSs M3–M5 all have equivalent antibacterial PDI efficacies against MRSA and A. baumannii compared with the clinical drug MB, with an over 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log CFU mL−1 decrease of the bacterial viability when the concentration of M3–M5 is up to 5 μM. The results correlate well with the cellular uptake and the ΦΔ of the PSs. For C. albicans, cationic PSs M4 and M5 both exhibited much better antifungal PDI efficacies compared with MB, especially for M5. There was an over 3
log CFU mL−1 decrease of the bacterial viability when the concentration of M3–M5 is up to 5 μM. The results correlate well with the cellular uptake and the ΦΔ of the PSs. For C. albicans, cationic PSs M4 and M5 both exhibited much better antifungal PDI efficacies compared with MB, especially for M5. There was an over 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log CFU mL−1 decrease of the fungal viability when the concentration of M5 reached 2.5 μM. However, the antimicrobial PDI efficacy of the anionic PS M1 was poor, effective only for MRSA, with an over 3
log CFU mL−1 decrease of the fungal viability when the concentration of M5 reached 2.5 μM. However, the antimicrobial PDI efficacy of the anionic PS M1 was poor, effective only for MRSA, with an over 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log CFU mL−1 decrease of the bacterial viability when its concentration reached 25 μM. For C. albicans, the difference may be caused by different mechanisms for various PSs. As mentioned in the introduction, the antifungal PDT process involves more complex mechanisms compared with antibacterial PDT.42 Unfortunately, until now the specific reasons are still not clear. Additionally, based on the comparable intake of M1 and M4 (Fig. 4), the antimicrobial PDI effect of M1 is much lower than that of M4. Except for the difference between cations and anions, we speculate that the antimicrobial PDI effect of a PS may also correlate with its binding site or subcellular localization, which will be more remarkable when the PS is bound onto an important organelle. Obviously, more studies are required in the future.
log CFU mL−1 decrease of the bacterial viability when its concentration reached 25 μM. For C. albicans, the difference may be caused by different mechanisms for various PSs. As mentioned in the introduction, the antifungal PDT process involves more complex mechanisms compared with antibacterial PDT.42 Unfortunately, until now the specific reasons are still not clear. Additionally, based on the comparable intake of M1 and M4 (Fig. 4), the antimicrobial PDI effect of M1 is much lower than that of M4. Except for the difference between cations and anions, we speculate that the antimicrobial PDI effect of a PS may also correlate with its binding site or subcellular localization, which will be more remarkable when the PS is bound onto an important organelle. Obviously, more studies are required in the future.
Conclusions
In conclusion, a series of novel water-soluble PSs (M1–M5) based on 3-cinnamoylcoumarin derivatives were synthesized, and their properties were investigated. The results show that with the introduction of two carboxylic acid salts, the photostability of the PSs declined. The cationic PSs M3–M5 exhibited higher binding/uptake to MRSA, A. baumannii and C. albicans compared with the clinical drug MB. Additionally, the in vitro experiments showed that three cationic PSs (M3–M5) had equivalent antibacterial PDI efficacies against MRSA and A. baumannii compared with MB. The antifungal PDI efficacies of M4 and M5 to C. albicans were both much higher than that of MB, especially for M5. However, anionic PS M1 was only effective for MRSA. Although the cationic PSs M4 and M5 showed an excellent antifungal PDI performance, the specific reasons are still not clear. Further study needs to be carried out to elucidate the mechanism. This work indicates the potential of coumarin derivatives in antimicrobial PDI.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The work was supported by the National Natural Science Foundation of China (No. 61575208).Notes and references
- A. Rodriguez-Rojas, J. Rodriguez-Beltran, A. Couce and J. Blazquez, Int. J. Med. Microbiol., 2013, 303, 293–297 CrossRef CAS PubMed.
- A. J. Alanis, Arch. Med. Res., 2005, 36, 697–705 CrossRef PubMed.
- R. Laxminarayan, A. Duse, C. Wattal, A. K. M. Zaidi, H. F. L. Wertheim, N. Sumpradit, E. Vlieghe, G. L. Hara, I. M. Gould, H. Goossens, C. Greko, A. D. So, M. Bigdeli, G. Tomson, W. Woodhouse, E. Ombaka, A. Q. Peralta, F. N. Qamar, F. Mir, S. Kariuki, Z. A. Bhutta, A. Coates, R. Bergstrom, G. D. Wright, E. D. Brown and O. Cars, Lancet Infect. Dis., 2013, 13, 1057–1098 CrossRef PubMed.
- R. Ferraz, V. Teixeira, D. Rodrigues, R. Fernandes, C. Prudêncio, J. P. Noronha, Ž. Petrovski and L. C. Branco, RSC Adv., 2014, 4, 4301–4307 RSC.
- H. Landecker, Body Soc., 2016, 22, 19–52 Search PubMed.
- X. Ragàs, D. Sánchez-García, R. Ruiz-González, T. Dai, M. Agut, M. R. Hamblin and S. Nonell, J. Med. Chem., 2010, 53, 7796–7803 CrossRef PubMed.
- T. Dai, Y. Y. Huang and M. R. Hamblin, Photodiagn. Photodyn. Ther., 2009, 6, 170–188 CrossRef CAS PubMed.
- H. Nikaido, Science, 1994, 264, 382–388 CAS.
- F. S. Felipe, Y. Y. Huang and M. R. Hamblin, Recent Pat. Anti-Infect. Drug Discovery, 2013, 8, 108–120 CrossRef.
- W. Fan, P. Huang and X. Chen, Chem. Soc. Rev., 2016, 45, 6488–6519 RSC.
- Y. Shen, A. J. Shuhendler, D. Ye, J. J. Xu and H. Y. Chen, Chem. Soc. Rev., 2016, 45, 6725–6741 RSC.
- M. R. Hamblin, Curr. Opin. Microbiol., 2016, 33, 67–73 CrossRef CAS PubMed.
- M. Khurana, H. A. Collins, A. Karotki, H. L. Anderson, D. T. Cramb and B. C. Wilson, Photochem. Photobiol., 2007, 83, 1441–1448 CrossRef CAS PubMed.
- B. W. Henderson, D. A. Bellnier, W. R. Greco, A. Sharma, R. K. Pandey, L. A. Vaughan and T. J. Dougherty, Cancer Res. Treat., 1997, 57, 4000–4007 CAS.
- A. V. Kustov, D. V. Belykh, N. L. Smirnova, E. A. Venediktov, T. V. Kudayarova, S. O. Kruchin, I. S. Khudyaeva and D. B. Berezin, Dyes Pigm., 2018, 149, 553–559 CrossRef CAS.
- M. A. Pereira, M. A. F. Faustino, J. P. C. Tome, M. G. P. M. S. Neves, A. C. Tome, J. A. S. Cavaleiro, A. Cunha and A. Almeida, Photochem. Photobiol. Sci., 2014, 13, 680–690 CAS.
- Q. Chen, J. Li, Y. Wu, F. Shen and M. Yao, RSC Adv., 2013, 3, 13835–13842 RSC.
- G. Jori, C. Fabris, M. Soncin, S. Ferro, O. Coppellotti, D. Dei, L. Fantetti, G. Chiti and G. Roncucci, Lasers Surg. Med., 2006, 38, 468–481 CrossRef PubMed.
- D. Sanglard, Curr. Opin. Microbiol., 2002, 5, 379–385 CrossRef CAS PubMed.
- P. Gonzalez-Parraga, J. A. Hernandez and J. C. Arguelles, Yeast, 2003, 20, 1161–1169 CrossRef CAS PubMed.
- Y. Fang, T. Liu, Q. Zou, Y. Zhao and F. Wu, Sci. Rep., 2016, 6, 28357 CrossRef CAS PubMed.
- Y. Fang, T. Liu, Q. Zou, Y. Zhao and F. Wu, RSC Adv., 2015, 5, 56067–56074 RSC.
- A. C. Engler, N. Wiradharma, Z. Y. Ong, D. J. Coady, J. L. Hedrick and Y.-Y. Yang, Nano Today, 2012, 7, 201–222 CrossRef CAS.
- L. Huang, Y. Y. Huang, P. Mroz, G. P. Tegos, T. Zhiyentayev, S. K. Sharma, Z. Lu, T. Balasubramanian, M. Krayer, C. Ruzie, E. Yang, H. L. Kee, C. Kirmaier, J. R. Diers, D. F. Bocian, D. Holten, J. S. Lindsey and M. R. Hamblin, Antimicrob. Agents Chemother., 2010, 54, 3834–3841 CrossRef CAS PubMed.
- E. R. Kenawy, S. D. Worley and R. Broughton, Biomacromolecules, 2007, 8, 1359–1384 CrossRef CAS PubMed.
- E. Jameel, T. Umar, J. Kumar and N. Hoda, Chem. Biol. Drug Des., 2016, 87, 21–38 CAS.
- S. Emami and S. Dadashpour, Eur. J. Med. Chem., 2015, 102, 611–630 CrossRef CAS PubMed.
- F. G. Medina, J. G. Marrero, M. Macias-Alonso, M. C. Gonzalez, I. Cordova-Guerrero, A. G. Teissier Garcia and S. Osegueda-Robles, Nat. Prod. Rep., 2015, 32, 1472–1507 RSC.
- T. Nasr, S. Bondock and M. Youns, Eur. J. Med. Chem., 2014, 76, 539–548 CrossRef CAS PubMed.
- G. J. Finn, B. S. Creaven and D. A. Egan, Eur. J. Pharm. Sci., 2005, 26, 16–25 CrossRef CAS PubMed.
- T. Zhu, Y. Wang, W. Ding, J. Xu, R. Chen, J. Xie, W. Zhu, L. Jia and T. Ma, Chem. Biol. Drug Des., 2015, 85, 385–393 CAS.
- M. Gangopadhyay, T. Singh, K. K. Behara, S. Karwa, S. K. Ghosh and N. P. Singh, Photochem. Photobiol. Sci., 2015, 14, 1329–1336 CAS.
- Q. Zou, Y. Fang, Y. Zhao, H. Zhao, Y. Wang, Y. Gu and F. Wu, J. Med. Chem., 2013, 56, 5288–5294 CrossRef CAS PubMed.
- X.-L. Wang, Y. Zeng, Y.-Z. Zheng, J.-F. Chen, X. Tao, L.-X. Wang and Y. Teng, Chem.–Eur. J., 2011, 17, 11223–11229 CrossRef CAS PubMed.
- I. E. Kochevar and R. W. Redmond, Methods Enzymol., 2000, 319, 20–28 CAS.
- K. Mizuno, T. Zhiyentayev, L. Huang, S. Khalil, F. Nasim, G. P. Tegos, H. Gali, A. Jahnke, T. Wharton and M. R. Hamblin, J. Nanomed. Nanotechnol., 2011, 2, 1–9 Search PubMed.
- A. Rezusta, P. Lopez-Chicon, M. P. Paz-Cristobal, M. Alemany-Ribes, D. Royo-Diez, M. Agut, C. Semino, S. Nonell, M. J. Revillo, C. Aspiroz and Y. Gilaberte, Photochem. Photobiol., 2012, 88, 613–619 CrossRef CAS PubMed.
- B. D. Jett, K. L. Hatter, M. M. Huycke and M. S. Gilmore, BioTechniques, 1997, 23, 648–650 CAS.
- M. Oz, D. E. Lorke and G. A. Petroianu, Biochem. Pharmacol., 2009, 78, 927–932 CrossRef CAS PubMed.
- S. J. Wagner, A. Skripchenko, D. Robinette, J. W. Foley and L. Cincotta, Photochem. Photobiol., 1998, 67, 343–349 CrossRef CAS PubMed.
- P. Calzavara-Pinton, M. T. Rossi, R. Sala and M. Venturini, Photochem. Photobiol., 2012, 88, 512–522 CrossRef CAS PubMed.
- M. Paardekopper, A. E. V. Gompel, V. J. Steveninck and P. J. Van den Broek, J. Photochem. Photobiol. B, 1995, 62, 561–567 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02557f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |