Open Access Article
Open Access ArticleBioscaffold arrays decorated with Ag nanoparticles as a SERS substrate for direct detection of melamine in infant formula
Nan Zhao ,
Hefu Li
,
Hefu Li *,
Cunwei Tian,
Yanru Xie,
Zhenbao Feng,
Zongliang Wang,
Xunling Yan,
Wenjun Wang and
Huishan Yu*
*,
Cunwei Tian,
Yanru Xie,
Zhenbao Feng,
Zongliang Wang,
Xunling Yan,
Wenjun Wang and
Huishan Yu*
School of Physics Science and Information Technology, Shandong Key Laboratory of Optical Communication Science and Technology, Liaocheng University, Liaocheng 252059, China. E-mail: lihefu.2009@163.com; 13906350692@163.com
First published on 15th July 2019
Abstract
Three-dimensional (3D) plasmonic structures have been intensively investigated as high performance surface enhanced Raman scattering (SERS) substrates. Here, we demonstrate a 3D biomimetic SERS substrate prepared by deposition of silver nanoparticles (Ag NPs) on the bioscaffold arrays of cicada wings using laser molecular beam epitaxy. This deposition method can offer a large number of nanoparticles with average diameter of ∼10 nm and nanogaps of sub-10 nm on the surface of chitin nanopillars to generate a high density of hotspots. The prepared 3D Ag/cicada SERS substrate shows a limit of detection (LOD) for Rhodamine 6G as low as 10−7 M, high enhancement factor of 1.09 × 105, and excellent signal uniformity of 6.8%. Moreover, the molecular fingerprints of melamine in infant formula can be directly extracted with an LOD as low as 10 mg L−1, without the need for functional modification. The prepared SERS-active substrate, due to its low cost, high-throughput, and good detection performance, can be widely used in applications such as food safety and environmental monitoring.
Introduction
Melamine as an important raw material has been widely used for the production of melamine resins. Because of its high nitrogen content (66% by mass), it has been illegally added into milk products to falsely increase apparent protein content based on conventional Kjeldahl test.1 Melamine ingested into the human body automatically generates insoluble melamine cyanurate crystals in kidneys, causing serious damage to health.2 In 2008, the reason for the outbreak of acute kidney injury of infants in China was found to be due to the infant formula being contaminated by melamine.3 In short, it is of great importance to develop a reliable detection method that can realize direct and convenient detection of melamine in infant formula.Surface-enhanced Raman scattering (SERS),4,5 a powerful label-free spectroscopic technique, has been widely used in the fields of food safety, chemical analysis and environmental monitoring.6–9 Due to the localized surface plasmon resonance, the plasmonic nanostructures can provide strong near-field enhancement of electromagnetic field, and therefore significantly magnify the Raman signal of molecules adsorbed on the substrate surface. It should be noted that quantitative and ultrasensitive detection of melamine has been carried out in recent years.10–15 The fabrication of SERS-active substrates usually uses the assembly method, and the prepared substrates can achieve high sensitivity in SERS measurements, even down to the 1 ppb level.10 However, the assembly method is time-consuming and it is difficult to precisely control the gaps between adjacent nanoparticles. SERS substrates based on nanoparticles created by deposition method have been widely used because of their high-throughput and low-cost. Especially, compared with one- and two-dimensional plasmonic nanostructures,16,17 three-dimensional (3D) plasmonic platforms exhibit more obvious SERS effects because they can not only generate more effective hotspots, but also increase plenty of binding sites for probing molecules.18–25 Recently, 3D SERS substrates based on natural creatures (such as butterfly wing, rose petal, taro leaf) with noble metal NPs deposition have emerged as promising flexible platforms.26–28 Especially, the cicada wing, originating from a very common arthropod, possesses all the advantages of the 3D SERS substrate.29–31 In addition, the cicada wing as a biological material is flexible, cheap, easily-obtained and environment-friendly. Therefore, economical preparation of 3D biomimetic SERS substrates is desirable and has broad application prospects.
Here, we demonstrate a 3D biomimetic SERS substrate based on naturally cone-shaped nanopillars on the cicada wing, and highlight its capability for detecting and differentiating the fingerprints of melamine in infant formula. Compared with other deposition methods, the Ag NPs decorated on the bioscaffold arrays are realized at lower temperature by laser molecular beam epitaxy, thus protecting bioscaffold arrays from damage during sputtering. The number, size and gap of Ag NPs on the side surface of nanopillars can be effectively controlled by adjusting the deposition time. With the optimized deposition time, the 3D biomimetic SERS substrate with plentiful nanogaps (sub-10 nm) shows strong SERS enhancement (1.09 × 105) and small relative standard deviation of SERS intensity (6.8%). By comparing the Raman signal of the standard melamine solution and the mixed solution of melamine and infant formula, we demonstrate the substrate's capability for direct detection of melamine in infant formula. This SERS substrate without additional background molecules is particularly attractive because of its highly selective enhancement of molecular fingerprint information of melamine.
Experiments
2.1 Materials and instruments
Cryptotympana atrata fabricius were purchased from a coherent orchardist in Zhejiang Province of China. Rhodamine 6G (R6G) was purchased from Sigma-Aldrich. Deionized water was purchased from the School of Chemistry and Chemical Engineering of Liaocheng University. Melamine was purchased from Tongda Chemical Co., Ltd. Jinan Century. Infant formula was purchased from local supermarket in Liaocheng city. Silver target (99.99%) was purchased from Zhongnuo Advanced Material (Beijing) Technology Co., Ltd.The surface morphologies of cicada wing were obtained by the SPM system (Solver P47-PRO, NT-MDT, Russia) in tapping mode. The surface information of the Ag/cicada wing SERS substrate was achieved by the field emission scanning electron microscopy (FESEM) (JEOL, model JSM-6380). The UV-vis absorption spectra of the original cicada wing and Ag/cicada wing were measured by the HITACHI U-3310 spectrophotometer. Ag NPs-decorated chitin nanopillars arrays of the wing were prepared by using the laser molecular beam epitaxy (LMBE) growth system (model LMBE 450, SKY Company, China). The contact angle was measured with the OCA20 machine (Data Physics, Germany). The Raman spectra of samples were recorded with the Renishaw (Gloucestershire, UK) RM-2000 Laser Raman Spectrometer.
2.2 Sample preparation
All cicada wings were firstly cleaned in acetone, ethanol and deionized water in turn, each for 10 min, to remove residues on the surface, followed by drying in air and segmenting into 1 × 1 cm2 sections using scissors. Next, the pretreated sections were fixed as bioscaffolds on glass slides with double-sided adhesive tape using tweezers. Finally, the as-prepared bioscaffolds were decorated with Ag NPs with different deposition times in the LMBE growth system. During the deposition process, the pulse energy of the laser beam of wavelength 248 nm was up to 110 mJ, the laser pulse repetition rate was 5 Hz, and the vacuum was 2.0 × 10−4 Pa. In our experiments, high-purity Ag targets were sputtered under the same conditions except with different deposition times. Unless otherwise stated, 3D biomimetic SERS substrates refer to chitin nanopillar arrays decorated with 30 min Ag sputtering. In the SERS measurement, 10 mg melamine and 500 mg infant formula were added into 100 mL deionized water to prepare the 100 mg L−1 standard melamine solution and the 5 g L−1 infant formula solution, respectively. The melamine solution in infant formula (10 mg L−1 and 50 mg L−1) was prepared by mixing both in appropriate proportions.2.3 Raman measurements
The Raman signals of R6G, melamine, and melamine-contaminated infant formula were obtained after the droplet (5 μL) evaporated naturally, and all signals were obtained at room temperature. The Raman spectrometer used a 785 nm laser as the excitation source and a CCD detector with 1 cm−1 resolution. The SERS spectra were obtained by focusing a laser spot of about 2 μm on the sample using a 20× objective lens. The spectra were recorded with an accumulation time of 10 s. The incident power was kept low (∼0.03 mW) to get the strongest Raman spectra without destruction of SERS substrates. The accumulation time, the laser power and the sample volume were the same for all Raman spectra.Results and discussions
3.1 Morphology of the as-prepared 3D Ag/cicada wing
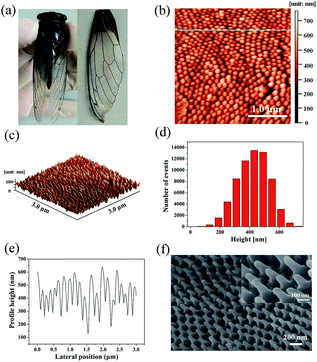
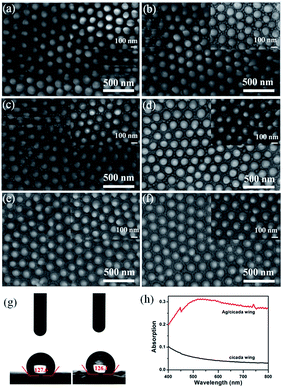
The Cryptotympana atrata, a widespread insect, can be found on a number of deciduous trees in the summer. Evolutionary pressure has pushed the wings of cicada species to develop unique surface nanostructures (chitin nanopillar arrays) that enable functionalities such as superhydrophobicity and antibacterial.32,33 Fig. 1a shows the top-view photograph of the cicada species and its detached forewings. In order to effectively observe the wing surface topography, scanning probe microscope and scanning electron microscope were used. Because the wing was thin and very soft, to obtain the best SPM results, it was necessary to fix the wing to a hard substrate using double-sided glue. As shown in Fig. 1b and c, the 2D and 3D AFM images of the wing surface show the existence of high-density cone-shaped nanopillar arrays. The shape profile of the nanopillars was approximately uniform in height, diameter, and center-to-center spacing. In addition, the histogram in Fig. 1d for height measurement demonstrates a narrow height range for the ordered nanostructures. Moreover, cross-section profiling of the nanostructures shown in Fig. 1e shows the uniformity as well as the relative sharpness of the nanopillars. The average values of the basal diameter, the top diameter, and the top spacing of the protrusions were estimated to be about 150, 40, and 180 nm respectively. Due to the finite size of the AFM tip, the exact value for the pillar height was difficult to obtain. Hence, high angle (45°) FE-SEM imaging was also utilized to obtain the nanopillar height. The sloping-view FE-SEM images of the cicada wing surfaces deposited with Ag NPs were given in Fig. 1f, from which, the pillar height was estimated to be about 230 nm. More importantly, the results also demonstrated that the cicada wing substrate was successfully decorated with Ag NPs without damaging the structure of the cicada wing, and both the tops and the sides of the protrusions were covered with Ag NPs.As is well known, compared with one-dimensional (1D) and two-dimensional (2D) SERS substrates, the 3D SERS-active substrates can not only provide abundant hotspots which play an important role in producing strong local field enhancement, but also provide large surface area for the deposition of metallic nanoparticles and the absorption of analyte molecules. The natural cicada wing with large-area arrays of vertically aligned nanopillars directly acts as the 3D substrate template, which greatly simplifies the complex experimental operations during the process of template fabrication,34–37 and can be achieved directly from naturally existing raw materials, resulting in considerable reduction in production costs of SERS substrates. In addition, the distinctive morphology of nanopillars—cone-shape with sharp top, wide separation at the neighboring top, and narrow separation at the neighboring bottom—can facilitate the sputtering of a large number of Ag NPs onto the side surfaces of nanopillars to form 3D plasmonic nanostructures. Considering the SERS signal can be obviously improved by optimizing the feature of plasmonic nanostructures, we investigated the effect of the deposition time on the morphology of the Ag/cicada wing by extending the deposition time from 5 to 30 min with an incremental step of 5 min. As shown in Fig. 2a, with a very short sputtering duration of 5 min, only a few small Ag NPs of about 12 nm in diameter were sputtered in the gap between the neighboring bottoms of the nanopillars; by increasing the sputtering duration to 10 min (Fig. 2b), Ag NPs with a slightly larger size of about 25 nm in diameter were formed on the gap area. For the sputtering duration of 15 min, the change in the number and size of the Ag NPs grown on the gap area was not obvious, but the Ag NPs were began to be formed on the side surface of the nanopillars (Fig. 2c). For a further prolongation of the sputtering deposition time (20 min and 25 min), the number and size of the Ag NPs grown on the gap area increased, and more Ag NPs reached the side surfaces of the cone-shaped nanopillars, as shown in Fig. 2d and e, respectively. When the sputtering duration increases to 30 min, as shown in Fig. 2f, the Ag NPs on the gap area between the bottoms of neighboring nanopillars were aggregated to form a film. Small Ag NPs were distributed randomly but uniformly on the whole side surfaces of the cone-shaped nanopillars. The size of these small Ag NPs was about 10 nm in diameter, with an average separation of less than 10 nm between adjacent Ag NPs, which is critical to create high SERS activity excited by the surface plasmon coupling between the narrow gaps of the neighboring Ag NPs. The large-scale and orderly distribution of protrusions coated with Ag NPs on the surfaces of the cicada wings also indicate that the substrates might have good reproducibility during the SERS detection.
Meanwhile, as shown in Fig. 2g, the high values of contact angle (127.6° and 126.1°) indicate that the deposition of Ag NPs does not reduce the hydrophobic characteristic of the cicada wing. This hydrophobic surface will induce an accumulation of analyte molecules, further enhancing the SERS effect.38 Compared with the naked cicada wing, the Ag/cicada wing of 30 min deposition time exhibited obvious enhanced capability of light absorption in the range of 400–800 nm, as shown in Fig. 2h. A broad plasmon band centered at ∼750 nm appeared, confirming the deposition of Ag NPs onto the cicada wing. Although there was a mismatch between the surface plasmon wavelength (750 nm) and the source wavelength (785 nm), the prepared SERS substrate showed strongly enhanced Raman signals. This may be because UV-vis absorption spectra offer only a rough indication on the matching between surface plasmon resonance and incident laser. Therefore, by optimizing the deposition time, this deposition technology can conveniently achieve a large number of hotspots. In addition, one-step decoration of abundant cicada wings as the SERS substrates in less than 0.5 h can improve the productivity significantly, thus dramatically increasing the capability of the substrates.
3.2 SERS performance and EF calculation
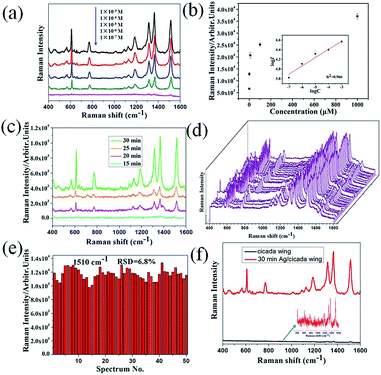
To investigate the SERS performance of the 3D biomimetic substrate, R6G, due to its well-established vibrational features, was used as the probe molecule. Fig. 3a shows the SERS spectra of R6G droplets (5 μL) with different concentrations in the range of 10−3 to 10−7 M, which was dropped onto the Ag/cicada wing substrate (of 30 min deposition time) and detected after being evaporated naturally. The Raman characteristic peaks of the R6G molecule at 611, 780 and 1187 cm−1 were related to C–C–C ring in-plane bending, out-of-plane bending and C–H in-plane bending vibrations, respectively, and the other characteristic peaks at 1311, 1361, and 1510 cm−1 all stemmed from the aromatic C–C stretching vibrations. These Raman peaks of R6G can be quite clearly observed, even at a low concentration of 10−7 M. Apart from the 3D plasmonic nanostructures, the lower concentration limit of detection may be also due to the surface hydrophobicity as shown in Fig. 2g. When a drop of liquid was dropped onto the hydrophobic SERS substrate, the droplet was easy to form a nearly spherical shape. As it evaporated naturally, the analyte molecules would be condensed into a certain region, thus improving the lower concentration limit of detection of the SERS substrate. Based on the experimental data in Fig. 3a, the relationship between the SERS intensity of the peak centered at 1510 cm−1 and the concentration was demonstrated in Fig. 3b. Clearly, a higher R6G concentration leaded to a more intense Raman signal. When logarithm-scale axes are used, the relation between the logarithmic integrated signal intensity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I) and the logarithmic concentration (log
I) and the logarithmic concentration (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C) is approximately linear (average and standard deviation from five samples), as shown in the inset of Fig. 3b. The linear relationship between log
C) is approximately linear (average and standard deviation from five samples), as shown in the inset of Fig. 3b. The linear relationship between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I and log
I and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C was previously confirmed in ref. 39, where the adsorption of analyte molecules follows Langmuir isotherm. Obviously, the linear relationship can be used to determine unknown concentrations of R6G solution on this substrate.
C was previously confirmed in ref. 39, where the adsorption of analyte molecules follows Langmuir isotherm. Obviously, the linear relationship can be used to determine unknown concentrations of R6G solution on this substrate.
Fig. 3c shows the variation of the SERS intensity of R6G (1 μM) at 1510 cm−1, with different deposition times, which create distinctive morphological characteristics of SERS substrates. When the deposition time is less than 15 min, only a few Ag NPs could be observed in the substrates so that, the SERS spectra of R6G cannot be detected (and thus no result is presented in the figure). With extension of the deposition time up to 30 min, both the number and the size of the Ag NPs on the side surfaces of the nanopillars increased, leading to the emergence of ∼10 nm nanogaps, and thus enhancing the maximum SERS intensity. These results demonstrated that enhancement of the SERS intensity can be influenced by both the number of nanoparticles and the density of nanogaps. For practical applications, the signal reproducibility and uniformity are crucial to the detection performance of SERS substrates. In order to evaluate spot-to-spot reproducibility, the SERS spectra of 50 random points on the 3D biomimetic SERS substrates functionalized with 10−6 M R6G were collected, as shown in Fig. 3d. To obtain a statistically meaningful result, the relative standard deviation (RSD) of the intensity at the 1510 cm−1 peak of R6G was calculated, which turns out to be 6.8%, as shown in Fig. 3e. Fig. 3d and e fully illustrated that the prepared substrate had excellent uniformity and reproducibility over the entire surface, which is comparable to or even better than many recent studies.30,31
To calculate the SERS enhancement factor (EF) of the 3D biomimetic substrate, the original cicada wing was chosen as a conventional Raman substrate, and the EF was evaluated via the widely accepted formula: EF = (ISERS/IRaman) × (NRaman/NSERS),40 where ISERS and IRaman are peak intensities of the same band in the SERS and conventional Raman spectra, NRaman refers to the amount of R6G molecules probed in a bulk sample, and NSERS is the number of monolayer R6G molecules adsorbed on the substrate surface under the laser spot area. Here, the band centered at 1510 cm−1 and obtained from SERS spectra of 10−3 M R6G on the corresponding substrates was chosen to calculate I values, as shown in Fig. 3f. In the SERS measurement, for the original substrate, the integrated peak intensity IRaman was measured to be ∼490 after the droplet evaporated naturally; for the SERS substrate, the value of ISERS was measured to be ∼1.48 × 104. NRaman refers to the amount of R6G molecules probed in a bulk sample, and NSERS is the number of monolayer R6G molecules adsorbed on the substrate surface under the laser spot area. NRaman was determined based on the concentration (10−3 M) of the R6G solution and the illuminated volume (Villu) of the Raman system. For our Raman setup, the illumination focus had a diameter of ∼2 μm and the penetration depth of laser beam was ∼3 mm. Therefore, the calculated Villu was ∼9.42 × 103 μm3, and the estimated value for NRaman was ∼5.67 × 109. To determine NSERS, we assumed that monolayer R6G molecules were absorbed on the surface of the SERS substrate. The surface area of one R6G molecule (1.37 nm in length, and 1.43 nm in width) is ∼2.0 nm2.41 Dividing the illuminated area by the surface area of one R6G molecule gives NSERS as 1.57 × 106. Accordingly, the EF of the 3D biomimetic substrate is estimated to be as large as ∼1.09 × 105.
3.3 Direct detection of melamine in infant formula
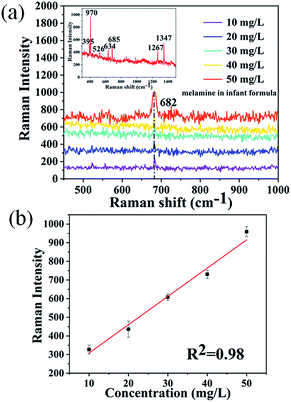
In order to evaluate the potential of the 3D biomimetic SERS substrate, melamine-contaminated infant formula was chosen as the analyte. It is well known that precise analysis of melamine in milk sample is difficult because milk products contain protein and carbohydrate contents, and melamine can form hydrogen-bond with these milk constituents. Therefore, a rational and convenient approach to directly detect melamine in milk powder with high sensitivity is essential. Based on the above results, the promising 3D biomimetic nanosensor can be applied for direct detection of melamine in infant formula. In order to confirm the specificity of this direct detection technology, we prepared the mixed solution of melamine and infant formula with different concentrations. Fig. 4a shows the SERS spectra of infant formula and the melamine mixed solutions with concentrations ranging from 10 mg L−1 to 50 mg L−1. The fingerprint peaks of melamine located at 682 cm−1 (the ring breathing mode), were obviously observed. By comparing the Raman spectra measured from melamine, the Raman spectrum of the infant formula powder are also measured as shown in the inset of Fig. 4a. The most intense peak in the powder spectrum was observed at 395, 970, 526, 634, 685, 1267 and 1347 cm−1. Therefore, the 3D biomimetic SERS substrate showed excellent ability to directly recognize melamine molecules in infant formula without any sample pretreatment. By monitoring the melamine Raman peak at 682 cm−1, it was found that the intensity of this peak was enhanced with increasing concentration of melamine. A linear relationship was found between the peak intensity at 682 cm−1 and melamine concentration with a good determination coefficient (R2) of 0.98, as shown in Fig. 4b. These results proved that this 3D biomimetic SERS substrate has good selectivity and sensitivity for analyzing melamine in infant formula. The reason for directly extracting the characteristic bands of melamine molecules in infant formula may be attributed to the electromagnetic enhancement and chemical enhancement. Especially, due to the latter contribution, bands of melamine molecules become observable in SERS spectra, while other existing bands of the sample may be attenuated beyond the detection limit.42Conclusions
In conclusion, we take advantage of 3D Ag/cicada wing substrate prepared by laser molecular beam epitaxy to develop a low-cost, uniformed SERS substrate for direct detection of melamine in infant formula. The detection performance of the 3D biomimetic SERS substrate can be easily and effectively optimized by the deposition time, which can control the morphology of Ag/cicada wing substrates. The optimal 3D SERS substrate showed an excellent signal reproducibility (6.8%), high EF (1.09 × 105) and low LOD for R6G (10−7 M). Finally, the optimal 3D SERS substrate demonstrates that the dominating characteristic bands of melamine molecules in infant formula can be clearly distinguished even with the concentration as low 10 mg L−1, making it a promising platform for detection of melamine in infant formula and other milk products.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the funding from Natural Science Foundation of Shandong Province under grant numbers ZR2016FQ05, ZR2018MA044, and Scientific Research Starting Foundation of Liaocheng University under grant number 318051542, and Natural Science Foundation of China under grant numbers 11504386, 61775089, and 61574071, and Alliance Fund of Shandong Provincial Key Laboratory under grant number SDKL2016038.Notes and references
- E. Y. Chan, S. M. Griffiths and C. W. Chan, Lancet, 2008, 372, 1444–1445 CrossRef CAS.
- A. K.-C. Hau, T. H. Kwan and P. K.-T. Li, J. Am. Soc. Nephrol., 2009, 20, 245–250 CrossRef CAS.
- Y. Wu and Y. Zhang, Food Chem. Toxicol., 2013, 56, 325–335 CrossRef CAS.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CrossRef CAS.
- A. B. Zrimsek, N. Chiang, M. Mattei, S. Zaleski, M. O. McAnally, C. T. Chapman, A. Henry, G. C. Schatz and R. P. Van Duyne, Chem. Rev., 2017, 117, 7583–7613 CrossRef CAS.
- A. P. Craig, A. S. Franca and J. Irudayaraj, Annu. Rev. Food Sci. Technol., 2013, 4, 369–380 CrossRef CAS.
- T. Yaseen, H. Pu and D. Sun, Trends Food Sci. Technol., 2018, 72, 162–174 CrossRef CAS.
- K. Kneipp, H. Kneipp, I. Itzkan, R. R. Dasari and M. S. Feld, Chem. Rev., 1999, 99, 2957–2975 CrossRef CAS.
- S. Xu, W. Tang, D. B. Chase, D. L. Sparks and J. F. Rabolt, ACS Appl. Nano Mater., 2018, 1, 1257–1264 CrossRef CAS.
- X. Liang, X. J. Zhang, T. T. You, N. Yang, G. S. Wang and P. G. Yin, J. Raman Spectrosc., 2018, 49, 245–255 CrossRef CAS.
- X. Guo, Y. Wu, Y. Ying, Y. Wen, F. Wang, Q. Ke and H. Yang, J. Raman Spectrosc., 2018, 49, 215–221 CrossRef CAS.
- R. Li, G. Yang, J. Yang, J. Han, J. Liu and M. Huang, Food Control, 2016, 68, 14–19 CrossRef CAS.
- N. Dong, Y. Hu, K. Yang and J. Liu, Sens. Actuators, B, 2016, 228, 85–93 CrossRef CAS.
- A. Kaleem, M. Azmat, A. Sharma, G. Shen and X. Ding, Food Chem., 2019, 277, 624–631 CrossRef CAS.
- C. Li, J. Yu, S. Xu, S. Jiang, X. Xiu, C. Chen, A. Liu, T. Wu, B. Man and C. Zhang, Adv. Mater. Technol., 2018, 1800174, 1–11 Search PubMed.
- A. Chen, A. Eugene DePrince III, A. Demortière, A. Joshi-Imre, E. V. Shevchenko, S. K. Gray, U. Welp and V. K. Vlasko-Vlasov, Small, 2011, 7, 2365–2371 CrossRef CAS PubMed.
- A. Shalabney, C. Khare, J. Bauer, B. Rauschenbach and I. Abdulhalim, J. Nanophotonics, 2012, 6, 061605 CrossRef.
- J. Chen, Y. Li, K. Huang, P. Wang, L. He, K. R. Carter and S. R. Nugen, ACS Appl. Mater. Interfaces, 2015, 7, 22106–22113 CrossRef CAS.
- C. Zhang, C. Li, J. Yu, S. Jiang, S. Xu, C. Yang, Y. J. Liu, X. Gao, A. Liu and B. Man, Sens. Actuators, B, 2018, 258, 163–171 CrossRef CAS.
- X. Huang, Y. Liu, J. Barr, J. Song, Z. He, Y. Wang, Z. Nie, Y. Xiong and X. Chen, Nanoscale, 2018, 10, 13202–13211 RSC.
- G. C. Phan-Quang, H. K. Lee, H. W. Teng, C. S. L. Koh, B. Q. Yim, E. K. M. Tan, W. L. Tok, I. Y. Phang and X. Y. Ling, Angew. Chem., Int. Ed., 2018, 57, 5792–5796 CrossRef CAS.
- Z. Li, Z. Du, K. Sun, X. He and B. Chen, RSC Adv., 2017, 7, 53157–53163 RSC.
- K. Min, K. S. Choi, W. J. Jeon, D. K. Lee, S. Oh, J. Lee, J.-Y. Choi and H. K. Yu, RSC Adv., 2018, 8, 12927–12932 RSC.
- A. Purwidyantri, C.-H. Hsu, C.-M. Yang, B. A. Prabowo, Y.-C. Tiang and C.-S. Lai, RSC Adv., 2019, 9, 4982–4992 RSC.
- Y. Zhang, R. J. Liu, X. Ma, X. Y. Liu, Y. X. Zhang and J. Zhang, RSC Adv., 2018, 8, 37750–37756 RSC.
- Y. Tan, J. Gu, L. Xu, X. Zang, D. Liu, W. Zhang, Q. Liu, S. Zhu, H. Su, C. Feng, G. Fan and D. Zhang, Adv. Funct. Mater., 2012, 22, 1578–1585 CrossRef CAS.
- B. Xu, Y. Zhang, W. Zhang, X. Liu, J. Wang, X. Zhang, D. Zhang, H. Jiang, R. Zhang and H. Sun, Adv. Optical Mater., 2013, 1, 56–60 CrossRef.
- P. Kumar, R. Khosla, M. Soni, D. Deva and S. K. Sharma, Sen. Actuators, B, 2017, 246, 477–486 CrossRef CAS.
- F. Shao, Z. Lu, C. Liu, H. Han, K. Chen, W. Li, Q. He, H. Peng and J. Chen, ACS Appl. Mater. Interfaces, 2014, 6, 6281–6289 CrossRef CAS.
- L. Guo, C. X. Zhang, L. Deng, G. X. Zhang, H. J. Xu and X. M. Sun, J. Appl. Phys., 2014, 115, 213101 CrossRef.
- M. Y. Lv, H. Y. Teng, Z. Y. Chen, Y. M. Zhao, X. Zhang, L. Liu, Z. Wu, L. M. Liu and H. J. Xu, Sens. Actuators, B, 2015, 209, 820–827 CrossRef CAS.
- J. Oh, C. E. Dana, S. Hong, J. K. Roman, K. D. Jo, J. W. Hong, J. Nguyen and D. M. Cropek, ACS Appl. Mater. Interfaces, 2017, 9, 27173–27184 CrossRef CAS PubMed.
- S. M. Kelleher, O. Habimana, J. Lawler, B. O. Reilly, S. Daniels, E. Casey and A. Cowley, ACS Appl. Mater. Interfaces, 2016, 8, 14966–14974 CrossRef CAS.
- Z. Huang, G. Meng, Q. Huang, Y. Yang, C. Zhu and C. Tang, Adv. Mater., 2010, 22, 4136–4139 CrossRef CAS.
- M. S. Schmidt, J. Hübner and A. Boisen, Adv. Mater., 2012, 24, 11–18 Search PubMed.
- P. Karadan, S. Aggarwal, A. A. Anappara, C. Narayana and H. C. Barshilia, Sens. Actuators, B, 2018, 254, 264–271 CrossRef CAS.
- X. Wang, S. Park, J. Ko, X. Xiao, V. Giannini, S. A. Maier, D. Kim and J. Choo, Small, 2018, 14, 1801623 CrossRef PubMed.
- H. Li, Q. Yang, J. Hou, Y. Li, M. Li and Y. Song, Adv. Funct. Mater., 2018, 28, 1800448 CrossRef.
- W. Wu, M. Hu, F. S. Ou, R. S. Williams and Z. Li, Proc. SPIE, 2010, 7673, 767301 CrossRef.
- W. Y. Li, P. H. C. Camargo, X. M. Lu and Y. N. Xia, Nano Lett., 2009, 9, 485–490 CrossRef CAS.
- L. Y. Chen, K. H. Yang, H. C. Chen, Y. C. Liu, C. H. Chen and Q. Y. Chen, Analyst, 2014, 139, 1929–1937 RSC.
- S. K. Saikin, Y. Chu, D. Rappoport, K. B. Crozier and A. Aspuru-Guzik, J. Phys. Chem. Lett., 2010, 1, 2740–2746 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |