An artificial antibody for exosome capture by dull template imprinting technology†
Lukuan
Liu‡
ab,
Jianhui
Liu‡
a,
Wen
Zhou
ac,
Zhigang
Sui
a,
Jing
Liu
b,
Kaiguang
Yang
 *a,
Lihua
Zhang
*a,
Zhen
Liang
a and
Yukui
Zhang
a
*a,
Lihua
Zhang
*a,
Zhen
Liang
a and
Yukui
Zhang
a
aCAS Key Laboratory of Separation Sciences for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China. E-mail: yangkaiguang@dicp.ac.cn; lihuazhang@dicp.ac.cn
bStem Cell Clinical Research Center, The First Affiliated Hospital of Dalian Medical University, Dalian 116011, China
cUniversity of Chinese Academy of Sciences, Beijing, 100049, China
First published on 21st April 2022
Abstract
Exosomes are extracellular vesicles with unique size distribution derived from the parent cells. They are involved in intercellular communication and transport, and are also biomarkers for early diagnosis and prognosis of disease. However, the isolation and characterization of exosomes face the challenges of large sample requirements and low enrichment efficiency of traditional methods. Herein, a simple method is proposed for the preparation of an artificial antibody that has a synergistic effect by featuring nanocavities obtained by dull template imprinting and molecular recognition conferred by electrostatic interaction. With this artificial antibody, highly efficient capture and proteome analysis of exosomes from urine and cell culture media are achieved: for urine, the abundance of Top100 exosomal proteins enriched by this artificial antibody increased from 1.85% to 9.66%. For the cell culture medium, the abundance of the Top100 proteins enriched by this artificial antibody was 28.4%. Moreover, this artificial antibody has a comparable effect to the commercial precipitation-based method in capturing exosomes and has advantages in removing contaminants such as prothymosin alpha (PTMA), demonstrating the superior selectivity of the artificial antibody. Overall, this artificial antibody holds promise to capture exosomes from biofluids and is compatible with subsequent proteome analysis.
Introduction
Exosomes are small extracellular vesicles with a diameter between 40 and 160 nm secreted by most cells. They are widely found in cell culture media, blood, urine and other biofluids.1 Exosomes carry important functional components such as nucleic acids, lipids, metabolites and proteins from parent cells.2 Among them, proteins in exosomes mediate material transport and information exchange between cells, and participate in many physiological and pathological processes such as regulation of cell proliferation and migration, antigen presentation, immune response, and tumor development.3–6 However, biofluids in which exosomes are presented are complex and have many interfering components such as non-exosomal vesicles, apoptotic bodies, protein aggregates, chylomicrons and lipoprotein particles.7 Moreover, the abundance of exosomes is extremely low. For example, the number of extracellular vesicles in platelet-free plasma only accounts for 0.0035% of the total number of particles in plasma. The content of exosomal vesicles is lower and contains only 1 μg protein per 3 × 1010 vesicles.8,9 Therefore, enriching exosomes with high efficiency and selectivity is an important prerequisite for their proteomic research.To date, many techniques for capturing exosomes from biofluids have been developed using the physicochemical and biochemical properties of exosomes.10–14 Among them, ultracentrifugation technology is currently the most widely used method.15 However, this technique requires large amounts of samples, and the recovery rate of exosomes is low, so it cannot be used for exosome isolation and analysis of trace and precious samples. Therefore, researchers developed an exosome isolation method based on the principle of polyethylene glycol (PEG) precipitation.16 Compared to ultracentrifugation, the PEG precipitation technique requires lower sample volumes and achieves higher recovery rates. However, this technique suffers from co-precipitation of other non-exosomal contaminants, such as lipoproteins, leading to a low purity of exosomes. Other exosome isolation techniques, such as size exclusion, dialysis, immunomagnetic beads, microfluidics, and aptamer-based and lipid-based methods, often include complex processes, low throughput, and large sample requirements, and the purity has not been significantly improved.10,17–19 Therefore, it is still necessary to develop a novel and efficient exosome capture method for proteomic analysis.
As a novel artificial antibody preparation technique that combines shape matching and chemical recognition, molecular imprinting can be used to recognize target molecules independent of antibodies and is more versatile and easier to tailor.20 Our previous work has shown that molecular imprinting has been successfully used to capture tumor cells by using fixed cells as templates.20–22 Conceivably, by using exosomes as templates, molecular imprinting could provide nanocavities that precisely match the shape and chemical properties of the exosomes. These nanocavities could act as antibodies to capture exosomes from biofluids. However, it is difficult to obtain adequate and pure exosomes to be used as templates due to the limitation of current exosome capture methods. Therefore, we speculate that it may be a good choice to prepare artificial antibodies for exosome capture by synthesizing nanoparticles with similar size distribution and surface properties to exosomes as dull templates.
It has been proven that the exosome membrane is negatively charged and can interact with positively charged molecules. The anion exchange resin was successfully used to capture exosomes.23 However, this method requires collection of different effluent fractions, and contaminants and exosomes may co-elute, thus reducing the recovery rate and purity. Predictably, the use of electrostatic interaction is expected to provide further enhancement of exosome-specific capture sensitivity when the cationic molecules are integrated into the molecular imprinting process.
In this paper, an artificial antibody (molecule imprinting polymer, MIP) for exosome capture was constructed based on dull template imprinting and electrostatic interaction. The MIP possesses nanocavities that precisely match the shape and molecular binding features of the target exosomes. By the synergistic effect of conformational recognition and electrostatic interaction, the MIP has been used to capture exosomes from urine and cell conditioned media. The enrichment effect of the MIP on exosomes was further evaluated at the protein level by using a quantitative proteomic method and Gene Ontology analysis.
Experimental
Materials
Methanol was purchased from Thermo Fisher (PA, USA). Trypsin, ammonia solution (28%), tetraethyl orthosilicate (TEOS, 98%), methylenebisacrylamide (MBA), acrylamide (AAm), 2-(diethylamino)ethyl acrylate, sodium dodecyl sulfonate (SDS), azodiisobutylamidine hydrochloride (AIBA), dithiothreitol (DTT), urea, iodoacetamide (IAA) and ammonium bicarbonate (ABC) were purchased from Sigma-Aldrich (MO, USA). Hydrofluoric acid was purchased from Tianjin Kemiou Chemical Reagent Co., Ltd (Tianjin, China). The TEI kit was purchased from Thermo Fisher (CA, USA). Ultrafiltration centrifuge tubes (10 kDa) were purchased from Sartorius (Goettingen, Germany). The deionized water used in the experiments was purified using a Milli-Q system (MA, USA).Cell culture and pre-treatment of the cell culture medium
HeLa cells were cultured for at least six passages using stable isotope labeling with amino acids in cell culture (SILAC) (Lys6, Arg10). When the cells grew to 80–90% confluence, the SILAC-labeled medium containing fetal bovine serum (exosomes removed) was collected. The obtained culture medium was centrifuged successively to remove cells (500g) and cell debris (3000g). The supernatant was collected and filtered with a 0.22 μm sterile filter to obtain the HeLa-CCM for exosome capture.Preparation of silica nanoparticles
50 mL of methanol, 7.74 mL of water, and 5 mL of 28% ammonia water were mixed, and then 1.90 mL of TEOS was added rapidly. The solution was stirred at 40 °C for 30 min and then centrifuged at 6000g to obtain silica nanoparticles. Finally, the silica nanoparticles were washed three times with water and dispersed in 1 mL of water to use them as dull templates. The particle size distribution and ζ-potential in the aqueous solution were measured by dynamic light scattering using a Nano-ZS90 particle sizer (Malvern, UK).Preparation of the artificial antibody for exosome capture by the dull template method
0.50 g of MBA, 0.01 g of AAm, 0.01 g of 2-(diethylamino)ethyl acrylate and 100 μL of silica nanoparticle solution were dissolved and dispersed into 6 mL of water. Then, 0.03 g of AIBA was added to initiate polymerization at 60 °C for 24 h. After that, the polymer was dried and the silica nanoparticles were etched by using hydrofluoric acid. Finally, the polymer was washed with water three times and dried to obtain the artificial antibody (MIP). The morphology of the MIP was characterized using a JSM-7800F scanning electron microscope.Capture of exosomes from urine and HeLa-CCM using the MIP
The MIP dispersed in methanol was loaded into pipette tips (200 μL) and then centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) for 10 min until the height of the MIP in the pipette tips reached about 1 cm. Then the pipette tips were washed three times with water for capturing exosomes from urine and HeLa-CCM. 1 mL of urine or 3 mL of HeLa-CCM was loaded into the above pipette tips and centrifuged (16
000g) for 10 min until the height of the MIP in the pipette tips reached about 1 cm. Then the pipette tips were washed three times with water for capturing exosomes from urine and HeLa-CCM. 1 mL of urine or 3 mL of HeLa-CCM was loaded into the above pipette tips and centrifuged (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) for 20 min. The MIP was aspirated by using 4% SDS solution and then centrifuged at 16
000g) for 20 min. The MIP was aspirated by using 4% SDS solution and then centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. The supernatant was obtained for later usage. A PEG-based commercial TEI kit (KIT) was used as the control and processed according to the instructions.
000g for 10 min. The supernatant was obtained for later usage. A PEG-based commercial TEI kit (KIT) was used as the control and processed according to the instructions.
Proteomic analysis of MIP-isolated exosomes from urine and HeLa-CCM by mass spectrometry
In this paper, the filter-assisted sample pretreatment method (FASP) was used to prepare peptides for mass spectrometry (MS) analysis. The above supernatants were reduced with 100 mM DTT at 95 °C for 10 min and diluted with 8 M urea. Then the diluted samples were loaded in the filter unit and centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min. Then the proteins were alkylated with 50 mM IAA at room temperature in the dark and then rinsed with urea and ammonium bicarbonate successively. Then the proteins were digested with trypsin at 37 °C for 18 h. The tryptic peptides were collected through centrifugation.
000g for 15 min. Then the proteins were alkylated with 50 mM IAA at room temperature in the dark and then rinsed with urea and ammonium bicarbonate successively. Then the proteins were digested with trypsin at 37 °C for 18 h. The tryptic peptides were collected through centrifugation.
The tryptic peptides were vacuum-dried and analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, USA) coupled with an EASY-nLC 1200 (Thermo Scientific, USA). The injected digest was separated on a reverse phase (RP) C18 analytical column (14 cm × 150 μm i.d.) at a flow rate of 600 nL min−1. The separation gradient was employed as follows: from 4% to 25% buffer B (80% ACN/0.1% FA) for 61 min, from 25% to 50% buffer B for 10 min, from 50% to 95% buffer B for 1 min, and from 95% to 95% buffer B for 3 min. The mass spectrometry data were acquired in the data dependent mode. The scan range for full scan was set from 350 m/z to 1500 m/z in the Orbitrap with a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (at 200 m/z). The AGC target was set as 4 × 105, and the maximum injection time was set as 50 ms. Within the cycle time of 3 s, ions were sequentially isolated with a 1.6 m/z isolation window and fragmented by HCD with a collision energy of 30%. The tandem mass spectra were obtained using Orbitrap (resolution: 15
000 (at 200 m/z). The AGC target was set as 4 × 105, and the maximum injection time was set as 50 ms. Within the cycle time of 3 s, ions were sequentially isolated with a 1.6 m/z isolation window and fragmented by HCD with a collision energy of 30%. The tandem mass spectra were obtained using Orbitrap (resolution: 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at 200 m/z) with the AGC target set at 5 × 104 and the maximum injection time at 30 ms.
000 at 200 m/z) with the AGC target set at 5 × 104 and the maximum injection time at 30 ms.
The obtained raw data were searched using MaxQuant (v1.6.17.0) against the Swiss-Prot human protein database (UniProt Proteome, release 2022_02, 42 358 entries). Fully tryptic cleavage constraints were set with up to two cleavage sites. Cysteine carbamidomethylation (+57.0215 Da) was set as fixed modification. Methionine oxidation (+15.9949 Da) and protein N-terminal acetylation (+42.0106 Da) were set as variable modifications. For HeLa cell culture medium samples, SILAC-labeled lysine and arginine (Lys6, Arg10) were added as variable modifications. The mass tolerances were 10 ppm for the full scan and 20 ppm for the tandem mass spectra. The intensity-based absolute quantification (iBAQ) approach was used to calculate protein abundances. The intensity-based fraction of total (iFOT) was used to represent the normalized abundance of a protein across experiments.24 The option ‘match between runs’ was enabled with a 1 min time window. Gene Ontology (GO) term enrichment analysis and protein–protein network analysis were based on DAVID 2021 (Dec. 2021) and STRING (version 11.5) databases.
Results and discussion
Preparation and characterization of the MIP
To achieve highly selective capture of exosomes, we developed an artificial antibody (MIP) with electrostatic and steric synergistic effects for exosome capture by dull template imprinting (Fig. 1).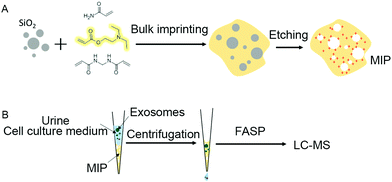 | ||
| Fig. 1 Preparation of the artificial antibody (MIP) by using silica templates (A) and the MIP tips fabricated for capturing exosomes in urine and the cell culture medium (B). | ||
Molecularly imprinted materials mainly use the imprinted sites left by templates to recognize target molecules, but the premise of conventional preparation methods is to use target molecules with high purity as templates. Due to the limitation of exosome capture methods, all the currently obtained exosomes contain non-vesicular contaminants such as lipoproteins and protein aggregates to a certain extent. If the currently available exosomes are directly used as templates, the imprinted sites from contaminants will inevitably be introduced, resulting in the inability to improve the capture specificity of exosomes substantially.
Therefore, in order to capture exosomes specifically, we synthesized silica nanoparticles as dull templates with similar size distribution properties to exosomes, and the distribution range is between 40 nm and 160 nm (Fig. 2A). The silica nanoparticles in the aqueous solution were negatively charged, which was similar to exosomes, and the ζ-potential was about −39.6 mV (Fig. 2B).25 Then, the cationic functional monomer 2-(diethylamino)ethyl acrylate was introduced during the polymerization process to form an electrostatic adsorption effect for the negatively charged exosome membrane. Moreover, the hydrophilic monomer acrylamide and the hydrophilic cross-linker N,N′-methylene diacrylamide were used as the imprinting system to reduce nonspecific adsorption and improve biocompatibility. After polymerization and template removal, imprinted sites similar in size to exosomes were left inside the MIP (Fig. 2C). As shown in Fig. 1B, the synthesized MIP was packed into pipette tips to capture exosomes in complex samples and the captured particles were further used for proteome and GO analysis to determine whether these particles contain proteins, which are representative of exosomes.
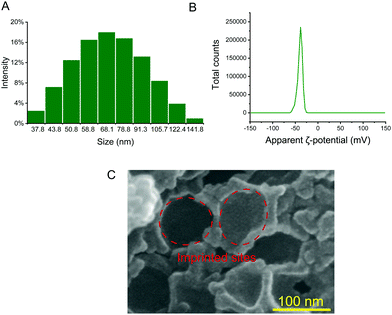 | ||
| Fig. 2 Size distribution of silica nanoparticles (A); ζ-potential of silica nanoparticles in aqueous solution (B); and scanning electron micrograph of the MIP (C). | ||
Nanoparticle tracking analysis (NTA) of MIP-isolated exosomes from urine and HeLa-CCM
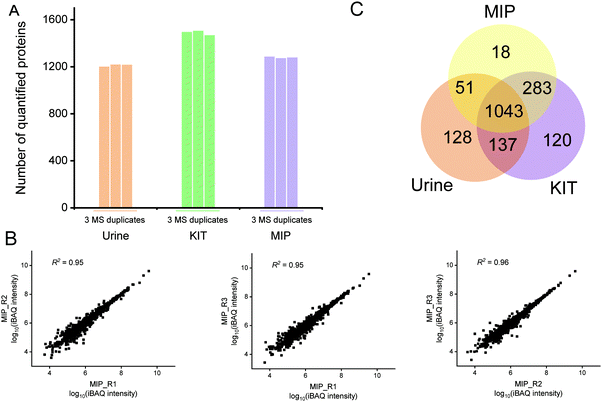
We analyzed the size distribution of particles isolated by the MIP from urine and HeLa-CCM by performing NTA. As shown in Fig. S1 (ESI†), for experiments that used urine as a source of exosomes, particles isolated by the MIP have a concentration of 1.30 × 1010 particles per mL. For experiments that used HeLa-CCM as a source of exosomes, particles isolated by the MIP have a concentration of 2.70 × 109 particles per mL. These results also demonstrate that the MIP is able to isolate particles within the size range of exosomes from human urine and HeLa-CCM.Proteomic analyses of MIP-isolated exosomes from urine
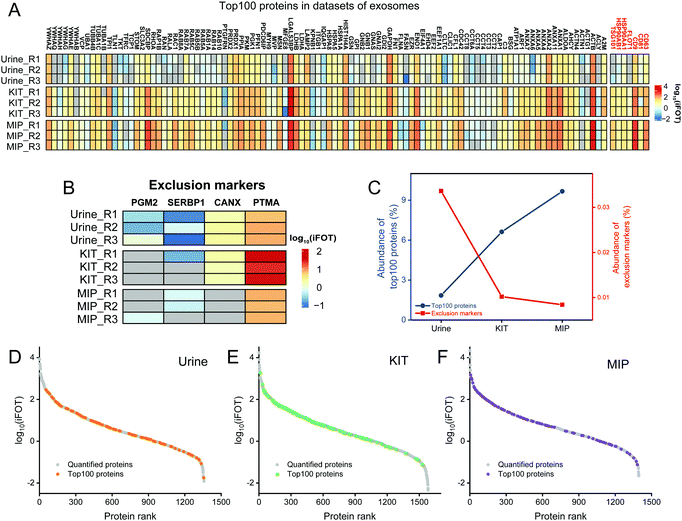
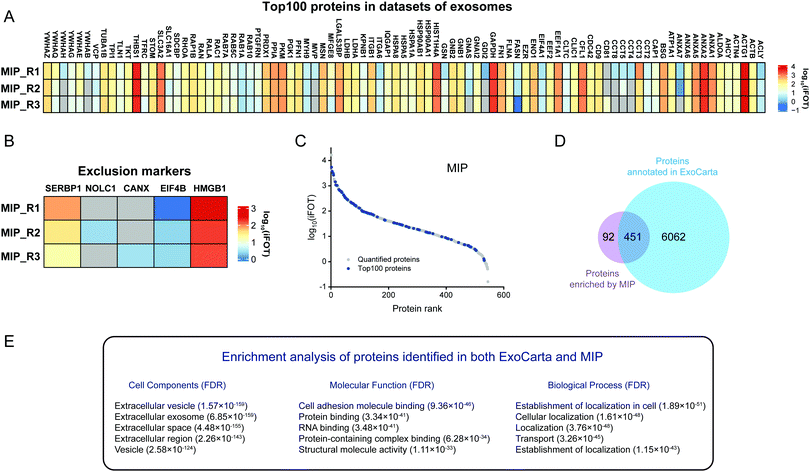
In addition to the enrichment of Top100 proteins, the removal of contaminants is also an important indicator for evaluating the enrichment effect of exosomes. The frequently used exclusion markers such as HMGB1, CANX, EIF4B, COX5B, PDAP1, HMGB2, SKP1, EIF4H, MAPRE1, PGM2, PTMA, HMGB3, SERBP1 and SLIRP were also studied.26 As shown in Fig. 4B, both SERBP1 and PTMA were present in untreated urine, KIT-isolated and MIP-isolated exosomes, which needed to be removed by developing new methods. The abundances of PGM2, SERBP1 and CANX were decreased and not even quantified after enrichment by the MIP and KIT. Notably, the abundance of PTMA in KIT-isolated exosomes was significantly higher than that in MIP-isolated exosomes. Qualitatively, these results indicate that the MIP has a comparable effect to the PEG-based method in capturing exosomes and has advantages in removing contaminants, such as PTMA.
Furthermore, in order to evaluate the purity of exosomes with the information of protein abundance, the quantitative information of exosome marker proteins and exclusion markers was also analyzed. As shown in Fig. 4C, iBAQ quantitative analysis of the identified proteins showed that the abundance of the Top100 proteins identified in untreated urine was only 1.85%, which was mainly due to the fact that the majority of the high abundance proteins in urine were not exosome-derived proteins. After the enrichment of the commercialized KIT, the abundance of the Top100 proteins increased to 6.62%. In contrast, after the enrichment of the MIP, the abundance of the Top100 proteins increased to 9.66%, and the abundance of exclusion markers decreased to 0.0084%, indicating that electrostatic interactions and steric effects can significantly increase the abundance of the exosome-derived proteins.
In order to investigate the protein composition of exosomes and the abundance distribution of Top100 proteins, we ranked the identified proteins using the intensity information obtained by the iBAQ quantitative method. The exosome proteins were averagely divided into three categories by their abundance and we define the most abundant range as high-abundance proteins. For the Top100 proteins from urine, KIT-isolated and MIP-isolated exosomes, the numbers in the high-abundance protein range were 29, 66 and 65, respectively (Fig. 4D–F). The significantly increased abundance of Top100 proteins indicated that the MIP has a good effect in increasing the abundance of exosome-derived proteins.
We further comparatively analyzed the proteomes of KIT-isolated and MIP-isolated exosomes. 95.2% of the proteins in the MIP-isolated exosomes were also found in the KIT-isolated exosomes (Fig. 5C). Prominent GO categories of the 67 proteins identified in MIP-isolated exosomes but not in KIT-isolated exosomes were also related to the extracellular region, extracellular space and extracellular exosomes (Fig. 5D). These results further support the conclusion that the MIP can isolate exosomes effectively from urine. The above results show that, in urine samples, from the protein level, the MIP can not only increase the enrichment abundance of exosome marker proteins, but also increase the proportion of proteins localized in exosomes.
Proteomic analyses of MIP-isolated exosomes from HeLa-CCM
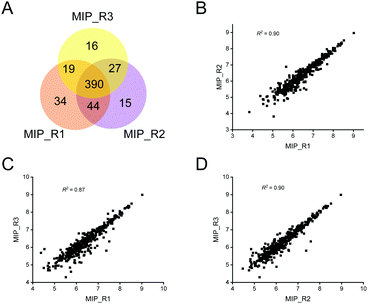
In view of the excellent enrichment ability of the MIP for exosomes in urine, we further introduced the MIP to the enrichment of exosomes in HeLa-CCM. The enrichment ability of the MIP to selectively capture exosomes in HeLa-CCM was evaluated by qualitative and quantitative proteome analysis of exosomes derived from the SILAC-labeled HeLa cells. Protein samples from MIP-isolated exosomes in HeLa-CCM were analyzed in triplicate. As shown in Fig. 6A, over 450 proteins were quantified in three MS replicates from MIP-isolated exosomes with good reproducibility (R2 ≥ 0.87, Fig. 6B–D). Exosomes are derived from labeled cells, so the ratio of the labeled proteins determines the enrichment ability of the MIP. In this study, the labeled proteins accounted for more than 89.5% of the total identified proteins, indicating that the MIP has a significant effect in removing non-cellular contaminants.To further analyze the enrichment effect of MIP-isolated exosomes from HeLa-CCM, the expression profiles of the Top100 proteins and exclusion markers in datasets of extracellular vesicles are depicted in Fig. 7A and B. The results showed that 72% (90/125) of the Top100 proteins including the commonly used exosome markers such as CD9, CD81, HSP90AA1 and HSP90AB1 were found in the MIP-isolated exosomes from HeLa-CCM, and the abundance of the Top100 proteins was 28.4%. Among them, 52.2% (47/90) of the Top100 proteins were highly abundant (Fig. 7C). The exclusion markers such as HMGB1, EIF4B, SERBP1, CANX and NOLC1 were identified in the MIP-isolated exosomes from HeLa-CCM, but their abundance was only 0.24%. The exclusion markers such as COX5B, PDAP1, HMGB2, SKP1, EIF4H, MAPRE1, PGM2, PTMA, HMGB3 and SLIRP were not identified.
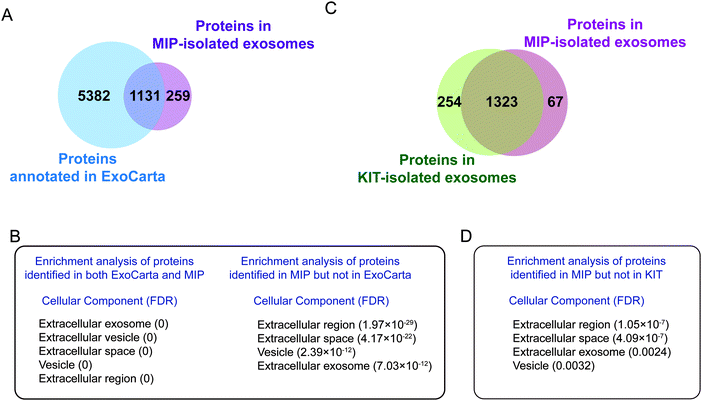
To confirm that the proteins identified in the MIP-isolated exosomes from HeLa-CCM were bona fide exosome-associated proteins, we compared the proteomic data of our MIP-isolated exosomes with the human protein data annotated in the ExoCarta database, followed by GO analysis. As shown in Fig. 7D, 83.1% of the proteins in the MIP-isolated exosomes matched the human proteins annotated in the ExoCarta database. Protein–protein interaction (PPI) analysis of the subset of 451 proteins quantified in both MIP-isolated exosomes and the ExoCarta database revealed a highly interconnected network (Fig. S3B, ESI†). Prominent GO categories of the 451 proteins were related to extracellular exosomes, exhibited molecular functions such as cell adhesion molecule binding, protein binding and RNA binding, and participated in biological processes such as establishment of localization and transport (Fig. 7E). The above results indicate that the MIP can efficiently enrich exosomes from CCM. Prominent GO categories of the 92 proteins identified in MIP-isolated exosomes but not in the ExoCarta database were located in the extracellular region, Golgi apparatus, and extracellular space. These proteins participated in biological processes such as extracellular matrix organization and cellular responses to the vascular endothelial growth factor stimulus (Table S1, ESI†). It is more likely that these 92 proteins are derived from the Golgi apparatus.
To investigate the differences in exosome proteins between the urine and HeLa-CCM samples, we compared the proteins identified in both ExoCarta and the MIP from the two samples. As shown in Fig. S4A (ESI†), 252 proteins were identified in both the urine and HeLa-CCM. These proteins perform binding functions such as cell adhesion molecule binding, protein binding and cadherin binding, and participate in biological processes such as regulated exocytosis, export from cells and secretion by cells (Fig. S4B, ESI†). These proteins reflected the basic characteristics of exosomes as cellular secretion, which may be ubiquitous in different samples. In contrast, the 879 exosome proteins uniquely identified in urine mainly perform functions such hydrolase activity and catalytic activity (Fig. S4C, ESI†). The 199 exosome proteins uniquely identified in the HeLa-CCM mainly perform functions such as RNA binding and structural molecule activity (Fig. S4C, ESI†). These differential proteins reflect the specificity of exosomes in different samples.
Conclusions
In summary, we put forward a simple method for the preparation of an artificial antibody for exosome capture by dull template imprinting technology. Due to the synergistic effect of topographical matching featured by molecular imprinting and the electrostatic interaction between the MIP and exosome membranes, the MIP had high capture efficiency and selectivity for exosomes in urine and cell culture media without the assistance of antibodies. Additionally, exosomes isolated from the MIP can be used for proteome analysis. Thus, the MIP holds promise in the exosome isolation and characterization fields.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful for the financial support from the National Key R&D Programme of China (2017YFA0505003), the National Natural Science Foundation (21725506, 21775149, 22004012 and 21874131), the Doctoral Research Start-up Fund Project of Liaoning Province (2020-BS-190), the Talent innovation support program of Dalian (2019CT07), and the Innovation Fund Project of Dalian Institute of Chemical Physics (DICP I202030). K. Y. is an excellent member of the Youth Innovation Promotion Association, CAS (Y2021058), and China Postdoctoral Science Foundation (2020M670804).References
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, eaau6977 CrossRef CAS.
- G. van Niel, G. D'Angelo and G. Raposo, Nat. Rev. Mol. Cell Biol., 2018, 19, 213–228 CrossRef CAS.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Théry, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS.
- G. Rodrigues, A. Hoshino, C. M. Kenific, I. R. Matei, L. Steiner, D. Freitas, H. S. Kim, P. R. Oxley, I. Scandariato, I. Casanova-Salas, J. Dai, C. R. Badwe, B. Gril, M. Tešić Mark, B. D. Dill, H. Molina, H. Zhang, A. Benito-Martin, L. Bojmar, Y. Ararso, K. Offer, Q. LaPlant, W. Buehring, H. Wang, X. Jiang, T. M. Lu, Y. Liu, J. K. Sabari, S. J. Shin, N. Narula, P. S. Ginter, V. K. Rajasekhar, J. H. Healey, E. Meylan, B. Costa-Silva, S. E. Wang, S. Rafii, N. K. Altorki, C. M. Rudin, D. R. Jones, P. S. Steeg, H. Peinado, C. M. Ghajar, J. Bromberg, M. de Sousa, D. Pisapia and D. Lyden, Nat. Cell Biol., 2019, 21, 1403–1412 CrossRef CAS.
- C. Liu, J. Zhao, F. Tian, L. Cai, W. Zhang, Q. Feng, J. Chang, F. Wan, Y. Yang, B. Dai, Y. Cong, B. Ding, J. Sun and W. Tan, Nat. Biomed. Eng., 2019, 3, 183–193 CrossRef CAS PubMed.
- J. Jiang, J. Mei, Y. Ma, S. Jiang, J. Zhang, S. Yi, C. Feng, Y. Liu and Y. Liu, Exploration, 2022, 2, 20210144 CrossRef.
- N. Karimi, A. Cvjetkovic, S. C. Jang, R. Crescitelli, M. A. Hosseinpour Feizi, R. Nieuwland, J. Lötvall and C. Lässer, Cell. Mol. Life Sci., 2018, 75, 2873–2886 CrossRef CAS PubMed.
- Y. Tian, M. Gong, Y. Hu, H. Liu, W. Zhang, M. Zhang, X. Hu, D. Aubert, S. Zhu, L. Wu and X. Yan, J. Extracell. Vesicles, 2019, 9, 1697028 CrossRef PubMed.
- J. Webber and A. Clayton, J. Extracell. Vesicles, 2013, 2, 19861 CrossRef PubMed.
- F. A.-W. Coumans, A. R. Brisson, E. I. Buzas, F. Dignat-George, E. E.-E. Drees, S. El-Andaloussi, C. Emanueli, A. Gasecka, A. Hendrix, A. F. Hill, R. Lacroix, Y. Lee, T. G. van Leeuwen, N. Mackman, I. Mäger, J. P. Nolan, E. van der Pol, D. M. Pegtel, S. Sahoo, P. R.-M. Siljander, G. Sturk, O. de Wever and R. Nieuwland, Circ. Res., 2017, 120, 1632–1648 CrossRef CAS PubMed.
- A. Kumar, S. R. Dhadi, N. N. Mai, C. Taylor, J. W. Roy, D. A. Barnett, S. M. Lewis, A. Ghosh and R. J. Ouellette, J. Extracell. Vesicles, 2021, 10, e12138 CAS.
- T. Takeuchi, K. Mori, H. Sunayama, E. Takano, Y. Kitayama, T. Shimizu, Y. Hirose, S. Inubushi, R. Sasaki and H. Tanino, J. Am. Chem. Soc., 2020, 142, 6617–6624 CrossRef CAS PubMed.
- G. Li, N. Zhu, J. Zhou, K. Kang, X. Zhou, B. Ying, Q. Yi and Y. Wu, J. Mater. Chem. B, 2021, 9, 2709–2716 RSC.
- N. Zhu, G. Li, J. Zhou, Y. Zhang, K. Kang, B. Ying, Q. Yi and Y. Wu, J. Mater. Chem. B, 2021, 9, 2483–2493 RSC.
- C. Gardiner, D. D. Vizio, S. Sahoo, C. Théry, K. W. Witwer, M. Wauben and A. F. Hill, J. Extracell. Vesicles, 2016, 5, 32945 CrossRef PubMed.
- M. Ding, C. Wang, X. Lu, C. Zhang, Z. Zhou, X. Chen, C.-Y. Zhang, K. Zen and C. Zhang, Anal. Bioanal. Chem., 2018, 410, 3805–3814 CrossRef CAS PubMed.
- N. Zhang, N. Sun and C. Deng, Talanta, 2021, 221, 121571 CrossRef CAS PubMed.
- Y. Wan, G. Cheng, X. Liu, S.-J. Hao, M. Nisic, C.-D. Zhu, Y.-Q. Xia, W.-Q. Li, Z.-G. Wang, W.-L. Zhang, S. J. Rice, A. Sebastian, I. Albert, C. P. Belani and S.-Y. Zheng, Nat. Biomed. Eng., 2017, 1, 0058 CrossRef CAS PubMed.
- A. Kumar, S. R. Dhadi, N. N. Mai, C. Taylor, J. W. Roy, D. A. Barnett, S. M. Lewis, A. Ghosh and R. J. Ouellette, J. Extracell. Vesicles, 2021, 10, e12138 CAS.
- L. Liu, C. Dong, X. Li, S. Li, B. Ma, B. Zhao, X. Li, Z. Liang, K. Yang, L. Zhang and Y. Zhang, Small, 2020, 16, 1904199 CrossRef CAS PubMed.
- L. Liu, K. Yang, H. Gao, X. Li, Y. Chen, L. Zhang, X. Peng and Y. Zhang, Anal. Chem., 2019, 91, 2591–2594 CrossRef CAS PubMed.
- L. Liu, K. Yang, Z. Dai, Z. Liang, L. Zhang, X. Peng and Y. Zhang, Chin. Chem. Lett., 2019, 30, 672–675 CrossRef CAS.
- N. Heath, L. Grant, T. M. De Oliveira, R. Rowlinson, X. Osteikoetxea, N. Dekker and R. Overman, Sci. Rep., 2018, 8, 5730 CrossRef PubMed.
- X. Li, C. Zhang, T. Gong, X. Ni, J. E. Li, D. Zhan, M. Liu, L. Song, C. Ding, J. Xu, B. Zhen, Y. Wang and J. Qin, Nat. Commun., 2018, 9, 4910 CrossRef PubMed.
- H. Malhotra, N. Sheokand, S. Kumar, A. S. Chauhan, M. Kumar, P. Jakhar, V. M. Boradia, C. I. Raje and M. Raje, J. Biomed. Nanotechnol., 2016, 12, 1101–1114 CrossRef CAS PubMed.
- F. G. Kugeratski, K. Hodge, S. Lilla, K. M. McAndrews, X. Zhou, R. F. Hwang, S. Zanivan and R. Kalluri, Nat. Cell Biol., 2021, 23, 631–641 CrossRef CAS PubMed.
- S. Mathivanan, C. J. Fahner, G. E. Reid and R. J. Simpson, Nucleic Acids Res., 2011, 40, D1241–D1244 CrossRef PubMed.
- M. Pathan, P. Fonseka, S. V. Chitti, T. Kang, R. Sanwlani, J. Van Deun, A. Hendrix and S. Mathivanan, Nucleic Acids Res., 2019, 47, D516–D519 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb00494a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |