Development of a microfluidic dispensing device for multivariate data acquisition and application in molecularly imprinting hydrogel preparation†
Abstract
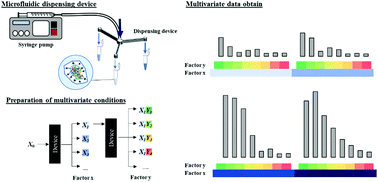
Molecularly imprinted polymers (MIPs) are superior materials with a molecular recognition ability that apply to various applications. In order to get high specificity recognition for target molecules, selecting polymerization conditions, which provide high interaction with the target template, is crucial. However, it requires time and labor to find the optimal polymerization composition, especially for large biomolecules. The advance in the microfluidic field enables researchers to control the flow rate and divide solutions based on the design of microfluidic devices for acquiring multivariate data by simultaneously preparing samples with different conditions. In this work, we fabricated microfluidic dispensing devices with different flow path widths that can give the solution of different flow rates. The accuracy of the flow rate was compared with the simulation value. As a result, the flow rate data showed almost the same data as the simulation value, and the dispensing volume ratio showed high reproducibility. Besides, the multivariate data from mixing the fluorescent molecule and protein solutions prepared by the dispensing device and a micropipette showed no significant difference with existing laboratory equipment. Finally, the dispensing device was used for preparing MIP hydrogels for lysozyme as a template protein. We successfully acquired multivariate data on the adsorption capacity of proteins, as a result, the hydrogels provided a high imprinting factor and adsorption specificity toward lysozymes.
- This article is part of the themed collection: New era in advanced functional materials emerging from molecular imprinting and related techniques


 Please wait while we load your content...
Please wait while we load your content...