Themed collection Cross-talk and integrated post-translational modifications

Cross-talk and integrative post-translational modifications
Si Wu and Lindsay Pino introduce this Molecular Omics themed issue on cross-talk and integrated post-translational modifications.

Mol. Omics, 2023,19, 93-94
https://doi.org/10.1039/D2MO90036J
MS-based proteomics for comprehensive investigation of protein O-GlcNAcylation
Protein O-GlcNAcylation plays critical roles in mammalian cells, and here we review MS-based proteomics methods for comprehensive and site-specific analysis of protein O-GlcNAcylation, ranging from enrichment, fragmentation, to quantification.
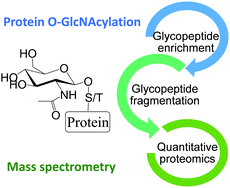
Mol. Omics, 2021,17, 186-196
https://doi.org/10.1039/D1MO00025J
Lipopolysaccharide and statin-mediated immune-responsive protein networks revealed in macrophages through affinity purification spacer-arm controlled cross-linking (AP-SPACC) proteomics
Affinity purification spacer-arm controlled cross-linking mass spectrometry (AP-SPACC-MS) to study LPS and statin mediated inflammatory signaling in macrophages.

Mol. Omics, 2023,19, 48-59
https://doi.org/10.1039/D2MO00224H
Laser capture microdissection-capillary zone electrophoresis-tandem mass spectrometry (LCM-CZE-MS/MS) for spatially resolved top-down proteomics: a pilot study of zebrafish brain
Laser capture microdissection-capillary zone electrophoresis-tandem mass spectrometry (LCM-CZE-MS/MS) for spatially resolved top-down proteomics revealed drastically different proteoform profiles between two LCM sections of zebrafish brain.
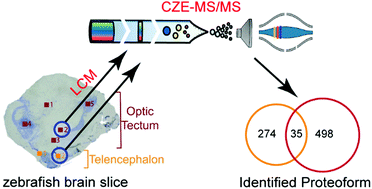
Mol. Omics, 2022,18, 112-122
https://doi.org/10.1039/D1MO00335F
An experimental design to extract more information from MS-based histone studies
Enzymatic treatment of a commercial histone extract as a quantify-first strategy allows isolating ion populations of interest that currently remain in the dark.
Mol. Omics, 2021,17, 929-938
https://doi.org/10.1039/D1MO00201E
Crosslinking mass spectrometry unveils novel interactions and structural distinctions in the model green alga Chlamydomonas reinhardtii
Interactomics is an emerging field that seeks to identify both transient and complex-bound protein interactions that are essential for metabolic functions.
Mol. Omics, 2021,17, 917-928
https://doi.org/10.1039/D1MO00197C
Analysis of pancreatic extracellular matrix protein post-translational modifications via electrostatic repulsion-hydrophilic interaction chromatography coupled with mass spectrometry
Glycosylation and phosphorylation in extracellular matrix proteins from human pancreas tissues were analyzed using a chromatographic simultaneous enrichment strategy followed by mass spectrometry.
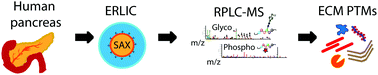
Mol. Omics, 2021,17, 652-664
https://doi.org/10.1039/D1MO00104C
The phosphoproteome of rice leaves responds to water and nitrogen supply
Quantitative phosphoproteomic analysis of rice plants grown with different levels of water supply and nitrogen supplementation revealed changes in phosphorylation of proteins involved in membrane transport, RNA processing and carbohydrate metabolism.

Mol. Omics, 2021,17, 706-718
https://doi.org/10.1039/D1MO00137J
Deciphering the architecture and interactome of hnRNP proteins and enigmRBPs
RNA-binding proteins (RBPs) have conserved domains and consensus sequences that interact with RNAs and other regulatory proteins forming ribonucleoprotein (RNP) complexes. Chemical crosslinking of proteins provides insights into RNP complexes interactome and protein conformations.
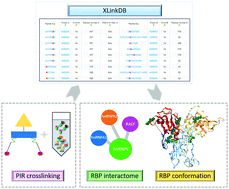
Mol. Omics, 2021,17, 503-516
https://doi.org/10.1039/D1MO00024A
About this collection
- Protein PTM detection methods and functional characterization of protein PTMs, including but not limited to mass spectrometry (MS) based bottom-up/top-down strategies,
- Spaciotemporal measurements of proteins and protein interactions within cells and tissues, including size-exclusion chromatography and proximity labelling,
- Bioinformatic approaches supporting PTM and protein interaction data analysis and integration, including integrated omics approaches for disease study,
- Systematic characterization, prediction, and data integration for protein cross-talk between PTMs, such as in epigenetics and disease.