Themed collection 2014 Emerging Investigators

Contributors to Emerging Investigators 2014
Guest Editors Ulrike Eggert and Michael Washburn introduce this year's Emerging Investigators issue and present the profiles of the contributing authors.

Mol. BioSyst., 2014,10, 1628-1631
https://doi.org/10.1039/C4MB90016B
Discovery of protein–RNA networks
We review the latest advances and future challenges in experimental and computational investigation of protein–RNA networks.
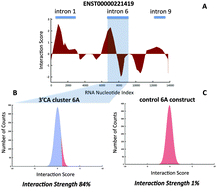
Mol. BioSyst., 2014,10, 1632-1642
https://doi.org/10.1039/C4MB00099D
Engineering reduced evolutionary potential for synthetic biology
Biological devices can be redesigned to slow evolutionary degradation of their functions by altering how they are encoded in DNA sequences and by engineering host organisms with improved genetic stability.
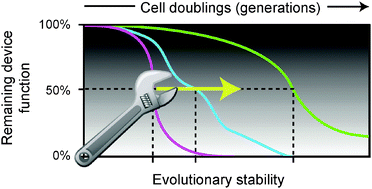
Mol. BioSyst., 2014,10, 1668-1678
https://doi.org/10.1039/C3MB70606K
From cradle to grave: high-throughput studies of aging in model organisms
This review summarizes the development of high-throughput replicative lifespan and longevity aging assays in unicellular and nematode model organisms.
Mol. BioSyst., 2014,10, 1658-1667
https://doi.org/10.1039/C3MB70604D
Crosstalk between kinases and Nedd4 family ubiquitin ligases
Understanding the interplay between kinase and E3 ligase signaling pathways will allow better understanding of therapeutically relevant pathways and the design of small molecule therapeutics targeting these pathways.
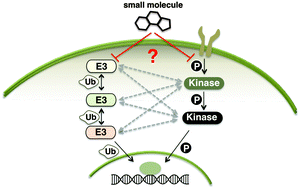
Mol. BioSyst., 2014,10, 1643-1657
https://doi.org/10.1039/C3MB70572B
Improved cyclopropene reporters for probing protein glycosylation
Cyclopropenes have emerged as a new class of bioorthogonal chemical reporters.
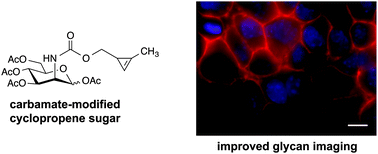
Mol. BioSyst., 2014,10, 1693-1697
https://doi.org/10.1039/C4MB00092G
Patterning of cells through patterning of biology
For the first time, patterns of cells have been constructed by spatially manipulating native gene expression. This control of expression was effected using light activated RNA interference (LARI), a technique in which knockdown of gene expression is modulated through siRNA modified with light cleavable groups.

Mol. BioSyst., 2014,10, 1689-1692
https://doi.org/10.1039/C3MB70587K
One third of dynamic protein expression profiles can be predicted by a simple rate equation
A simple rate equation predicts time-resolved expression for 1/3 of the tested genes and suggest two mutually exclusive modes of regulation.

Mol. BioSyst., 2014,10, 2850-2862
https://doi.org/10.1039/C4MB00358F
Peroxisomes are juxtaposed to strategic sites on mitochondria
Peroxisomes are situated in the cell adjacent to specific subdomains of mitochondria such as the ER/Mitochondria contact site or sites enriched for the pyruvate dehydrogenase complex.

Mol. BioSyst., 2014,10, 1742-1748
https://doi.org/10.1039/C4MB00001C
The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae
The interactome of the CTD phosphatase Rtr1 is regulated by the CTDK-I subunit Ctk1.

Mol. BioSyst., 2014,10, 1730-1741
https://doi.org/10.1039/C4MB00109E
Quantitative phosphoproteomic profiling of PINK1-deficient cells identifies phosphorylation changes in nuclear proteins
A quantitative phosphoproteomic method coupled with siRNA mediated silencing of a kinase associated with Parkinson's disease was applied to measure phosphorylation changes. The results indicate alterations in protein phosphorylation downstream of this kinase, potentially expanding our understanding of its function.
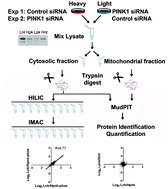
Mol. BioSyst., 2014,10, 1719-1729
https://doi.org/10.1039/C3MB70565J
A comprehensive analysis of the Streptococcus pyogenes and human plasma protein interaction network
A schematic figure for the analysis of bacterial interaction protein, 2 different MS methods are used.
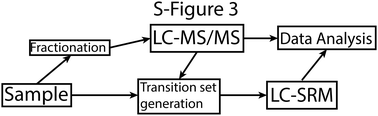
Mol. BioSyst., 2014,10, 1698-1708
https://doi.org/10.1039/C3MB70555B
Creating functional engineered variants of the single-module non-ribosomal peptide synthetase IndC by T domain exchange
Production of indigoidine can be enhanced by swapping a synthetic T domain into the NRPS IndC.
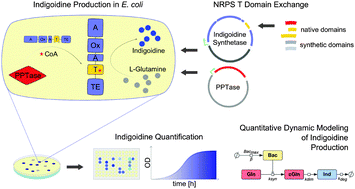
Mol. BioSyst., 2014,10, 1709-1718
https://doi.org/10.1039/C3MB70594C
A red light-controlled synthetic gene expression switch for plant systems
The gene switch can be induced to high expression levels in red light and is inactive in far-red-supplemented white light.
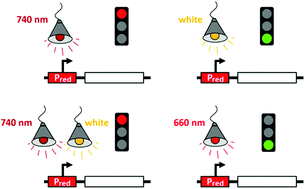
Mol. BioSyst., 2014,10, 1679-1688
https://doi.org/10.1039/C3MB70579J
About this collection
A Molecular BioSystems themed issue guest edited by Ulrike Eggert and Michael Washburn showcasing some of the top research from emerging investigators in chemical biology, systems biology and the -omics sciences, the core areas of Molecular BioSystems .