Themed collection Editor’s Choice: Single-Cell Analysis

Single-cell impedance spectroscopy of nucleated cells
A single-cell impedance spectroscopy with an extended frequency range (550 MHz) for nucleated cells was developed to discriminate changes in cell dielectric parameters.

Lab Chip, 2025,25, 2939-2948
https://doi.org/10.1039/D5LC00111K
Label-free differentiation of living versus dead single yeast cells using broadband electrical impedance spectroscopy
A bio impedance chip focused and trapped a single non-adherent cell between coplanar waveguide electrodes. The cell's electrical properties as measured by broadband electrical impedance spectroscopy differentiated between live and dead cells.

Lab Chip, 2025,25, 1744-1754
https://doi.org/10.1039/D5LC00043B
Precision single cell analysis to characterize host dependent antimicrobial response heterogeneity in physiological medium
We highlight the interindividual heterogeneity in pathogen killing kinetics by physiological concentrations of antibiotics in urine and develop a microfluidic device capable of tracking single-cell trajectories while allowing for reagent exchange.

Lab Chip, 2025,25, 714-728
https://doi.org/10.1039/D4LC00765D
Single and few cell analysis for correlative light microscopy, metabolomics, and targeted proteomics
We combined a single-cell lysis and handover system with mass spectrometry and reverse-phase protein arrays, allowing correlative single- and few-cell analysis combining microscopy with metabolomics and targeted proteomics.

Lab Chip, 2024,24, 4321-4332
https://doi.org/10.1039/D4LC00269E
Opto-combinatorial indexing enables high-content transcriptomics by linking cell images and transcriptomes
We introduce a simple integrated analysis method that links cellular phenotypic behaviour with single-cell RNA sequencing by utilizing a combination of optical indices from cells and hydrogel beads.
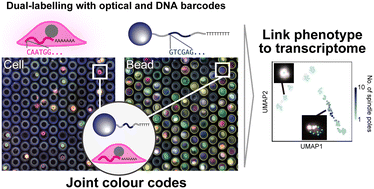
Lab Chip, 2024,24, 2287-2297
https://doi.org/10.1039/D3LC00866E
Label-free cell classification in holographic flow cytometry through an unbiased learning strategy
Unbiased learning pipeline for label-free single-cell classification.

Lab Chip, 2024,24, 924-932
https://doi.org/10.1039/D3LC00385J
About this collection
Handpicked by our Associate Editor, Amy Herr (University of California, Berkeley), we are pleased to highlight select works on single-cell analysis published in recent years. Read what she had to say below:
Microfluidic precision and integration have powered great strides in single-cell analysis tools. So much so, that life sciences researchers do not even expressly recognize the microfluidic core of the powerful but now commonplace analysis tools that they rely on every day (i.e., single-cell sequencing). Two decades ago, an industry leader convivially verbalized a daring (at the time) forecast that incorporation of microfluidic technologies in life sciences tools would mirror the impact of integrated microprocessors in personal computing. For single-cell analysis tools, her forecast was prescient.
If you accept that premise (at least for a moment), then I am delighted to share with you, dear reader, my choice of Lab on a Chip articles that look beyond this hidden but essential core single-cell analysis functionality. Here, I highlight a set of single-cell analysis articles that exemplify the spirit of human creativity and, potentially, high future impact. I loosely explore guided by Uzzi et al. and their criterion that “The highest-impact science is primarily grounded in exceptionally conventional combinations of prior work yet simultaneously features an intrusion of unusual combinations”. Through a curiosity-driven – albeit cursory – analysis of recent Lab on a Chip single-cell analysis articles, I curate for you a few with the hallmark of “being within the mainstream of a research trajectory, where scientists are currently focused, while being distinctive in one’s creativity” of future impact.
Drawn from Lab on a Chip’s single-cell analysis portfolio of publications from the prior year or so, here are three articles that build on a solid foundation of deep field-specific research infused with a surprising degree of out-of-field influence. First, the work of Zou et al. primarily extends the range of frequencies across which impedance cytometry performs well, especially for nucleated cells. With an eye towards forecasting potential for future impact, this study includes a solid corpus of references in the lab-on-a-chip domain, but also draws in substantial work from immunology and cell biology building strong cross-field linkages.
As a second example, Tsuchida et al. links cellular phenotypic behavior with single-cell RNA-seq by creating a system of ‘joint color codes’ that utilize epi-fluorescence imaging and next-generation sequencing. Inherently, this paper bridges microfluidic imaging with genomic analysis meaning that key citation clusters include optical engineering and high-throughput sequencing studies, and the interplay of the two fields.
Thirdly, consider Abe, Lee, Chin et al. and their work which describes a microfluidic platform capable of “[measuring] both bacterial growth and killing within the gel and enables medium exchange to assess the ability of surviving cells to resume growth after antimicrobial removal”, where the referenced gel is a microfluidic gel encapsulation platform. Yang and co-authors cite a broad range of fields, including clusters in microbiology and infectious-disease references and clusters in microfluidics and biosensing. The work builds on many cross-field references, suggesting that this study is deeply interdisciplinary and, thus, has a strong potential to ‘move the needle’ with regards to our understanding or capabilities.
During my cursory Uzzi-inspired analysis, I identified even more single-cell analysis articles that sparkle with this potential for future high impact. The work of Rima, Berchtold, Arnold and co-authors describes an intensive system that allows single-cell imaging coupled with microcapillary sampling of lysate for subsequent mass spectrometry or reverse-phase protein analyses, with cited works balancing core-field microfluidic familiarity with novelty.
Focusing on phenotypic imaging measurements that leverage microfluidic precision, the work of Ciaparrone et al. describes integration of digital holography microscopy with microfluidic tools and AI/ML image analysis with single-cell classification capabilities. To make progress, the authors unite optics/photonics, microfluidic cytometry, and machine learning to generate plentiful cross-field pairs of references (e.g., optics ↔ AI, microfluidics ↔ computational imaging), with a case-study application in drug-resistant endometrial cancer cells.
As a final example in recent the single-cell analysis theme, Favakeh et al. reports on single-cell yeast impedance spectroscopy that builds on a solid conventional core (microfluidics and bioengineering) with notable novel pairings (microbiology) to generate potential cross-field impact.
Now, twenty years after my colleague’s ‘microfluidic prophesy’, we are riding the crest of the artificial intelligence and machine learning (AI/ML) revolution in the life sciences. Looking ahead just as she did that time ago, I forecast that single-cell (and sub-cellular) analysis tools will only grow in importance, especially those that are built upon highly integrated microfluidic platforms. Why? Because analytical chemists, microfluidic design engineers, and measurement scientists will drive the next stages of the AI/ML revolution. Data is now king (or queen). Consequently, precision tools with quantitative outputs that operate at scale are increasingly essential to the quality and size of required data sets. Mind you, not just data sets as training data sets (please look beyond that horizon!). Our role is bigger than just these front-end data sets: when the time comes that AI/ML foundation models are commodities – and that day will come in relatively short order – precision analytical tools operating at scale will be central on the back end, as well as the front end. These Lab on a Chip single-cell analysis tools will be indispensable to model validation and, ultimately, hypothesis testing and refinement. These tools will be essential to the future of biology and biomedicine.