Analysis of DNA interactions and GC content with energy decomposition in large-scale quantum mechanical calculations†
Abstract
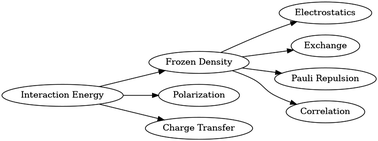
GC content is a contributing factor to the stability of nucleic acids due to hydrogen bonding. More hydrogen bonding generally results in greater stability. Empirical evidence, however, has suggested that the GC content of a nucleic acid is a poor predictor of its stability, implying that there are sequence-dependent interactions besides what its GC content indicates. To examine how much such sequence-dependent interactions affect the interaction energies of double-stranded DNA (dsDNA) molecules, dsDNA molecules of different sequences are generated and examined in silico for variabilities in the interaction energies within each group of dsDNA molecules of the same GC content. Since the amount of hydrogen bonding depends on the GC content, holding the GC content fixed when examining the differences in interaction energies allows sequence-dependent interactions to be isolated. The nature of sequence-dependent interactions is then dissected using energy decomposition analysis (EDA). By using EDA, the components of the interactions that depend on the neighboring base pairs help explain some of the variability in the interaction energies of the dsDNA molecules despite having the same GC content. This work provides a new paradigm and tool for the study and analysis of the distributions of interaction components in dsDNA with the same GC content using EDA within large-scale quantum chemistry calculations.


 Please wait while we load your content...
Please wait while we load your content...