Systematic analysis of the interactions driving small molecule–RNA recognition†
Abstract
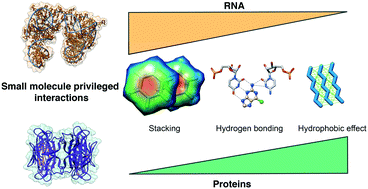
RNA molecules are becoming an important target class in drug discovery. However, the principles for designing RNA-binding small molecules are yet to be fully uncovered. In this study, we examined the Protein Data Bank (PDB) to highlight privileged interactions underlying small molecule–RNA recognition. By comparing this analysis with previously determined small molecule–protein interactions, we find that RNA recognition is driven mostly by stacking and hydrogen bonding interactions, while protein recognition is instead driven by hydrophobic effects. Furthermore, we analyze patterns of interactions to highlight potential strategies to tune RNA recognition, such as stacking and cation–π interactions that favor purine and guanine recognition, and note an unexpected paucity of backbone interactions, even for cationic ligands. Collectively, this work provides further understanding of RNA–small molecule interactions that may inform the design of small molecules targeting RNA.
- This article is part of the themed collection: Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...