 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mechanistic insights into the base-catalyzed hydrolysis of PCDDs via QSAR and DFT approaches
Kun
Xie
 and
Haiqin
Zhang
and
Haiqin
Zhang
 *
*
College of Environmental Science and Engineering, Liaoning Technical University, Fuxin 123000, China. E-mail: zhanghaiqin@lntu.edu.cn; Fax: +86-418-5110399; Tel: +86-418-5110399
First published on 11th December 2025
Abstract
Polychlorinated dibenzo-p-dioxins (PCDDs) are persistent organic pollutants that pose considerable threats to ecological and human health owing to their high toxicity potential. Understanding the mechanisms for underlying the base-catalyzed hydrolysis of PCDDs in aquatic environments is essential for assessing their environmental behaviour and ecological risks. Herein, we combined quantitative structure–activity relationship (QSAR) models with density functional theory calculations to analyse the base-catalyzed hydrolysis mechanisms of PCDDs. Among the four developed QSAR models, the single-parameter QSAR model based on the lowest unoccupied molecular orbital energy (ELUMO) demonstrated the best performance, achieving a coefficient of determination of 0.89 and a root mean square error of 0.49, indicating superior overall performance. Results indicate that the second-order rate constants for base-catalyzed hydrolysis (kOH) of PCDDs are primarily influenced by ELUMO, molecular polarizability (α), molecular volume (Vm), degree of chlorination (NCl), and chlorine position. Specifically, increases in the α and Vm values of PCDDs lead to higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH values, while an increase in the ELUMO value results in a lower log
kOH values, while an increase in the ELUMO value results in a lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH value. This study investigates the relationship between the molecular structure and the rate of base-catalyzed hydrolysis of PCDDs, providing valuable insight into their environmental fate. Furthermore, this research offers a novel theoretical perspective on the base-catalyzed hydrolysis of PCDDs, which will aid in regulatory assessments and risk management.
kOH value. This study investigates the relationship between the molecular structure and the rate of base-catalyzed hydrolysis of PCDDs, providing valuable insight into their environmental fate. Furthermore, this research offers a novel theoretical perspective on the base-catalyzed hydrolysis of PCDDs, which will aid in regulatory assessments and risk management.
Environmental significancePolychlorinated dibenzo-p-dioxins (PCDDs) are highly toxic and persistent pollutants widely detected in aquatic environments, yet their degradation behaviour under alkaline conditions remains poorly understood. Understanding their base-catalyzed hydrolysis is crucial for predicting environmental fate and guiding remediation strategies. This study reveals how key molecular descriptors—particularly ELUMO, polarizability, and molecular volume—influence hydrolysis rates. Integration of QSAR modelling and quantum chemical analysis provides a predictive framework for assessing PCDDs degradability. These findings enhance mechanistic understanding of PCDDs reactivity and support environmental risk assessments and regulatory management of persistent organic pollutants. |
1 Introduction
Polychlorinated dibenzo-p-dioxins (PCDDs) are a class of highly toxic organic compounds, known as persistent organic pollutants.1 Their structures are bridged by two benzene rings connected by two oxygen atoms, with each benzene ring potentially substituted by 1 to 4 chlorine atoms (Fig. S1). Toxic PCDD congeners are distinguished by the presence of chlorine atoms at the 2, 3, 7, and 8 positions, with 2,3,7,8-T4CDD being the most toxic.2 These compounds primarily exert their toxicity by binding to the aryl hydrocarbon receptor (AhR) in cells, influencing gene expression and disrupting cellular functions, which can lead to various toxic responses.3,4 PCDDs are typically byproducts of industrial processes, including incineration processes,5 chemical manufacturing,6 and metal production.7 Owing to their high bioaccumulation potential, lipophilicity, low water solubility, and semi-volatility, PCDDs do not readily degrade in the environment.8–10 As a result, these compounds can persist in soil, water, and organisms for extended periods, and have also been detected in dairy products, meat, and other foods consumed by humans,11 posing a potential threat to ecosystems and human health. Therefore, understanding the environmental behaviour of PCDDs is essential for assessing their persistence and potential ecological risks.Organic compound hydrolysis is a crucial chemical process in the environment.12 Our recent research has identified base-catalyzed hydrolysis as the primary degradation pathway of PCDDs in aquatic environments. Initial findings suggest that the reactivity of PCDDs hydrolysis is influenced by position and quantity of chlorine atoms on PCDD congeners.13 Given that PCDDs have been detected in aquatic environments at concentrations as low as picograms per litre,14,15 even trace amounts raise concerns regarding their persistence and potential ecological impacts. However, the relationship between the rate of base-catalyzed hydrolysis and the specific molecular structures of PCDDs remains unclear, necessitating further investigation. Due to the time-consuming and labour-intensive nature of experimentally determining this relationship, it is impractical to examine each PCDD congener individually. Therefore, the development of a high-throughput model to evaluate the hydrolysis mechanisms of various PCDDs is essential.
Quantitative structure–activity relationship (QSAR) models have primarily been utilised to investigate the toxicity,16 AhR binding affinity,17 bioconcentration and biodegradability of PCDDs.18,19 The robust scientific foundation of QSAR technology is predicated on the principle that similar chemical structures are likely to exhibit analogous chemical behaviours.20,21 Consequently, QSAR technology can beapplied to examine the relationship between the molecular structure and the base-catalyzed hydrolysis rate of PCDDs, as well as to analyse the mechanisms underlying their base-catalyzed hydrolysis. However, to date, no QSAR model for the base-catalyzed hydrolysis of PCDDs has been reported to date.
Herein, based on the second-order rate constants for base-catalyzed hydrolysis (kOH) of 75 PCDDs, calculated using quantum chemical methods, we utilised multiple linear regression (MLR) to develop four QSAR models. These models aim to explore the mechanisms underlying the base-catalyzed hydrolysis of PCDDs. Developed in accordance with the guidelines of the Organization for Economic Co-operation and Development (OECD),20 and these models serve as a valuable tool for assessing the environmental persistence of organic chemicals.
2 Computational details
2.1 Data collection
The kOH values for 75 PCDDs were derived from our previous DFT/TST calculations under aqueous standard conditions (298.15 K and 1 M), and their reliability has been demonstrated.13 In accordance with OECD guidelines (OECD, 2007), the dataset was divided into a training set consisting of 60 compounds and a validation set comprising 15 compounds, maintaining a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. For analytical purposes, the data were logarithmically transformed (log
1 ratio. For analytical purposes, the data were logarithmically transformed (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH) and standardised to uniform units of mol−1 L h−1. The log
kOH) and standardised to uniform units of mol−1 L h−1. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH values vary from a minimum of −4.28 mol−1 L h−1 (2-M1CDD) to a maximum of 3.39 mol−1 L h−1 (1,2,3,4,6,7,8,9-O8CDD).
kOH values vary from a minimum of −4.28 mol−1 L h−1 (2-M1CDD) to a maximum of 3.39 mol−1 L h−1 (1,2,3,4,6,7,8,9-O8CDD).
2.2 Calculation of molecular descriptors
Three molecular descriptors were considered for model development, including constitutional, geometric, and quantum chemical descriptors, which have been widely applied in QSAR studies to describe molecular size, electronic structure, and reactivity.8,22,23 Among the constitutional descriptors,24 the total number of chlorine atoms (NCl), the number of chlorine atoms at the Cβ position (Nβ), and the number of pairs of meta-position chlorine atoms (Nm) were examined. The selected geometric and quantum chemical descriptors, including molecular volume (Vm), energy of the highest occupied molecular orbital (EHOMO), energy of the lowest unoccupied molecular orbital (ELUMO), molecular polarizability (α) and the most positive partial charge on a hydrogen atom (qH+) etc., were calculated using the Gaussian 16 software package,25 at the M062X/6−31 + G(d,p)/SMD level. This method was selected based on its proven efficacy in investigating hydrolysis mechanisms and kinetics.13,26 For descriptors not available in the output file of Gaussian 16, the wave function file input feature of Multiwfn software was employed for computation and output.27 The meanings of the molecular descriptors involved in models development are provided in Table S1.2.3 Development and evaluation of models
MLR is a conventional statistical method used in the development of QSAR models.28 In this study, stepwise MLR analysis was employed for variable selection and model development using SPSS software (version 26.0). The goodness-of-fit of the models was evaluated using statistical parameters such as the adjusted determination coefficient (Radj2) and the root mean square error (RMSE). Internal validation techniques, including leave-one-out cross-validation (QLOO2), and external explained variance (Qext2) were employed to evaluate the predictive ability of the models. Additional details regarding model validation are provided in the Supplementary Material. The QLOO2 value was calculated as follows: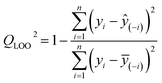 | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH value for the i-th data point, y(−i)is the predicted log
kOH value for the i-th data point, y(−i)is the predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH value for the i-th data point when the model is trained without this point, and y(−i)is the mean calculated log
kOH value for the i-th data point when the model is trained without this point, and y(−i)is the mean calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH value of the remaining n – 1 data points, excluding the i-th data point.
kOH value of the remaining n – 1 data points, excluding the i-th data point.
2.4 Applicability domain of the models
To clarify the scope and limitations of the models, the applicability domain (AD) was characterised using a Williams plot derived from standardised residuals (δ) and leverage values (h).29 Plotting was performed using OriginPro software (version 26.0). PCDDs were considered outliers if |δ| > 3. Additionally, predictions for PCDDs with high leverage values (h > h*, where h* is the warning leverage) were deemed unreliable. The δ, h, and h* values were calculated using the following equations: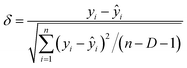 | (2) |
| hi = xiT(XTX)−1xi | (3) |
| h* = 3(D + 1)/n | (4) |
3 Results and discussion
3.1 Development and validation of the QSAR models
First, forward stepwise regression was performed to screen the 15 molecular descriptors of three different types. Then MLR analysis was performed using log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH as the dependent variable and the selected molecular structural parameters as predictor variables, resulting in four QSAR models, as shown in Table 1.
kOH as the dependent variable and the selected molecular structural parameters as predictor variables, resulting in four QSAR models, as shown in Table 1.
| No. | Equation | R adj 2 | RMSEtrab | Q LOO 2 | R ext 2 | RMSEexte | Q ext 2 |
|---|---|---|---|---|---|---|---|
| a Adjusted determination coefficient. b The root mean square error on the training set. c Leave-one-out. d External determination coefficient. e The root mean square error on the validation set. f External explained variance. | |||||||
| 1 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH = −10.15ELUMO − 4.01 kOH = −10.15ELUMO − 4.01 |
0.89 | 0.63 | 0.88 | 0.92 | 0.49 | 0.89 |
| 2 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH = 0.077α − 20.75 kOH = 0.077α − 20.75 |
0.83 | 0.78 | 0.82 | 0.89 | 0.55 | 0.88 |
| 3 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH = 0.114Vm − 17.61 kOH = 0.114Vm − 17.61 |
0.82 | 0.79 | 0.81 | 0.92 | 0.48 | 0.91 |
| 4 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH = 1.32 NCl − 0.48 Nβ + 0.37 Nm − 4.92 kOH = 1.32 NCl − 0.48 Nβ + 0.37 Nm − 4.92 |
0.87 | 0.69 | 0.86 | 0.89 | 0.56 | 0.77 |
A single-parameter QSAR model (1) with ELUMO was developed to investigate the relationship between kOH and molecular structure of PCDDs. The values of the descriptors used in the QSAR model (1), along with the calculated and predicted kOH values, were listed in Table S2. High Radj2 and QLOO2 values of the training set indicated the goodness-of-fit and robustness of the model (1). The linear regression results of predicted versus calculated values for model (1) as well as the Williams plot representing the AD, are shown in Fig. 1. The data points from both the training and validation sets are evenly distributed on both sides of the reference line y = x, indicating that model (1) fit both datasets well. The AD analysis, shown in Fig. 1B, demonstrated the lack of outliers in the validation set, h < h*, and |δ| < 3.
As shown in Table 1, QSAR model (2) was developed to explore the relationship between kOH and α. The relevant data are presented in Table S3. The Radj2 and QLOO2 values of this QSAR model were 0.83 and 0.82, respectively, indicating high goodness-of-fit and good robustness of model (2). The differences (0.01) between the R2 and QLOO2 values was <0.3, indicating no over-fitting of model (2).30 Moreover, this model demonstrated acceptable predictability, with Qext2 = 0.88 and RMSEext = 0.55. The results of the linear fit of the model predictions compared to the calculated values and the Williams plot characterizing AD are presented in Fig. S2. The results of the AD characterization revealed |δ| <3 and h < h*(0.1), indicating that all PCDDs were within the AD.
Among geometric descriptors, Vm is generally used to develop QSAR models for the physicochemical properties of PCDDs,8,31 suggesting that Vm could provide a convenient first estimator of kOH values for PCDDs. According to Table S4, QSAR model (3) demonstrated strong predictive capabilities within the training and external validation sets. The close Radj2 = 0.82 and QLOO2 = 0.81 values for the training set, along with high Qext2 = 0.91 values and lower RMSEext = 0.48 values for the validation set, suggested that model (3) was well-fitted, robust, and had good external predictive ability. Fig. S3A demonstrates the correlation between predicted and calculated values for model (3), indicating the model's good predictive performance. The AD characterization (Fig. S3B) revealed that all PCDDs were within the AD, with no outliers. In the validation sets, there is a ‘good high leverage’ point (1,2,3,4,6,7,8,9-O8CDD) with a higher than the warning h value of 0.1 and |δ| < 3, implying that the model (3) possesses some degree of extrapolating ability.29,32
PCDD congeners with the same number of chlorine atoms can display significantly different chemical and physical properties depending on the positions of the chlorine atoms attached to the parent structure.8 Therefore, different degrees of chlorination and chlorine atoms position is essential in understanding these differences. Three molecular descriptors (NCl, Nβ, and Nm) were included in the QSAR model (4), which demonstrated good fitting, robustness, and predictive capability. NCl was the most significant descriptor that negatively contributed to log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH (t = 10.722, P < 0.001). The variance inflation factor (VIF) values for each descriptor were less than 5, indicating the absence of multicollinearity between the descriptors. The p-values for all descriptors less than 0.05, demonstrating statistically significant contributions to the predictive power of model (4). The fitting results between the predicted and calculated values of the model (4) were good, as shown in Table S5 and Fig. S4A. Additionally, Fig. S4B, illustrates the AD characterisation of model (4), revealing |δ| < 3, which indicates no outliers (Table 2).
kOH (t = 10.722, P < 0.001). The variance inflation factor (VIF) values for each descriptor were less than 5, indicating the absence of multicollinearity between the descriptors. The p-values for all descriptors less than 0.05, demonstrating statistically significant contributions to the predictive power of model (4). The fitting results between the predicted and calculated values of the model (4) were good, as shown in Table S5 and Fig. S4A. Additionally, Fig. S4B, illustrates the AD characterisation of model (4), revealing |δ| < 3, which indicates no outliers (Table 2).
A comparison of the four models indicated that model (1), which ustilised ELUMO as a descriptor, demonstrated the best overall performance with the highest Radj2 (0.89) and Qext2 (0.89) values and lowest RMSEext (0.49) value. This finding suggests that electronic properties, specifically ELUMO, play a crucial role in the base-catalyzed hydrolysis mechanism of PCDDs. In practical applications, the choice of model should balance complexity, accuracy, and generalizability.33,34 Therefore, model (1) serves as a robust and reliable tool for the mechanism analysis of PCDDs, which is essential for understanding their environmental behaviour and potential impact.
3.2 Mechanism interpretation
Electrophilicity is related to the ELUMO of an electrophile, where the LUMO represents the innermost orbital with available positions to accept electrons.35,36 A lower ELUMO value implies that a molecule is more inclined to accept electrons,37,38 meaning that PCDDs act as electrophiles, while OH− ions serve as nucleophiles during the base-catalyzed hydrolysis of PCDDs. PCDDs primarily undergo hydrolysis via two pathways: dioxin ring-opening and hydrolytic dichlorination.13 In the ring-opening pathway, the electrophilic C–O bond, particularly in PCDDs with lower ELUMO values, is more susceptible to nucleophilic attack by OH−, resulting in the cleavage of the dioxin ring and the formation of open-ring products (chlorinated hydroxydiphenyl ethers). In the hydrolytic dechlorination pathway, lower ELUMO values enhance the electrophilicity of C–Cl bonds, facilitating the substitution of chlorine atoms with hydroxyl groups to yield hydroxylated PCDDs.Molecular polarizability is defined as the ratio of the induced dipole moment to the electric field that produces this dipole moment.39 Molecules with high polarizability possess electrons that can move relatively easily compared to those with low polarizability.40 Consequently, increased molecular polarizability enhances the likelihood of spatial electron distribution changes, which in turn increases reactivity towards nucleophiles or electrophiles.41,42 In base-catalyzed hydrolysis reactions, the electron clouds in PCDDs with higher α can move more easily. This phenomenon may contribute to a reduction in the stability of reactive sites, thereby decreasing the Gibbs free energies (ΔG‡) of reaction required for bond cleavage and ultimately enhancing the kOH. For instance, 1,2-D2CDD exhibits a polarizability of 233.387 a.u. and a ΔG‡ of 104.599 kJ mol−1, while 1,2,6-T3CDD has a polarizability of 248.523 a.u. and a ΔG‡ of 89.92 kJ mol−1. Thus, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH is positively correlated with α.
kOH is positively correlated with α.
According to the molecular structure of PCDDs presented in Fig. S1, Vm was positively correlated with the number of chlorine atoms in PCDD congeners (Vm = 10.559NCl + 109.414, R2 = 0.983). Therefore, a higher Vm value indicated a greater number of chlorine atoms, as the Vm value of PCDD congeners depends solely on the number of chlorine atoms.43 Due to their strong electronegativity, the electron density at the reaction site will decrease with an increasing number of chlorine atoms, resulting in a faster attack by OH− ions.42,44 Consequently, an increase in the Vm value is associated with an increase in the kOH value for PCDDs.
The QSAR model (4) in Table 1 demonstrated the effects of different degrees of chlorination and chlorine positions on the base-catalyzed hydrolysis rate of PCDDs. For examples, the electrostatic potential (ESP) distributions on the molecular surfaces of 2,7-D2CDD, 1,2,3,4-T4CDD, 1,2,3,6-T4CDD, 1,2,3,7,8,9-H6CDD, 1,2,4,6,8,9-H6CDD and 1,2,3,4,6,7,8,9-O8CDD were calculated to evaluate the relationship between constitutional descriptors (NCl, Nβ and Nm) and kOH of PCDDs. The results are presented in Fig. 2, with the blue regions indicating positive ESP (low electronic density), which OH− ions generally prefer to attack. According to Fig. 2, positive ESP values of PCDDs were primarily distributed at the Cγ position, increasing with the number of chlorine atoms. This finding aligned with the positive correlation between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kOH and NCl in model (4), where a greater number of chlorine atoms enhanced the kOH value. For the PCDD congeners with the same NCl and Nm values, such as 1,2,3,7,8,9-H6CDD and 1,2,4,6,8,9-H6CDD, the former with a higher Nβ value exhibited a greater positive ESP than the latter with a lower Nβ value. For PCDD congeners with the same NCl and Nβ values, those with higher Nm values had a lower positive ESP, as seen with 1,2,3,4-T4CDD and 1,2,3,6-T4CDD. This finding indicated that variations in the number of Cβ position and paired meta-position chlorine atoms significantly affected the electron distribution of PCDDs with the same NCl values, leading to notable changes in their hydrolysis rates. Therefore, the size of the blue region was closely related to the number and position of chlorine atoms in PCDDs. This observation supported the relationship described in the model and explained the effects of the number of chlorine atoms, Cβ position chlorine atoms and paired meta-position chlorine atoms on the base-catalyzed hydrolysis rate of PCDDs.
kOH and NCl in model (4), where a greater number of chlorine atoms enhanced the kOH value. For the PCDD congeners with the same NCl and Nm values, such as 1,2,3,7,8,9-H6CDD and 1,2,4,6,8,9-H6CDD, the former with a higher Nβ value exhibited a greater positive ESP than the latter with a lower Nβ value. For PCDD congeners with the same NCl and Nβ values, those with higher Nm values had a lower positive ESP, as seen with 1,2,3,4-T4CDD and 1,2,3,6-T4CDD. This finding indicated that variations in the number of Cβ position and paired meta-position chlorine atoms significantly affected the electron distribution of PCDDs with the same NCl values, leading to notable changes in their hydrolysis rates. Therefore, the size of the blue region was closely related to the number and position of chlorine atoms in PCDDs. This observation supported the relationship described in the model and explained the effects of the number of chlorine atoms, Cβ position chlorine atoms and paired meta-position chlorine atoms on the base-catalyzed hydrolysis rate of PCDDs.
 | ||
| Fig. 2 Electrostatic potential (ESP) distribution of PCDDs calculated at the M062X/6−31+ G(d,p)/SMD level. | ||
4 Model comparison
This study is the first to develop models for evaluating the hydrolysis reaction of PCDDs, addressing a gap in the literature, as no previous models have been reported in this area. To assess the accuracy and interpretability of these models, they were compared with two types of QSAR models: one for predicting the reaction of PCDDs with hydroxyl radicals and another for predicting the base-catalyzed hydrolysis rate of various organic compounds.Previous researchers have developed QSAR models that investigate how structural and quantum chemical descriptors influence the reactivity of PCDDs with hydroxyl radicals. Yan et al. utilised partial least squares (PLS) regression to develop polyparameter linea free energy models that concentrate on the rate constants for gas-phase reactions of hydroxyl radicals with PCDDs and dibenzofurans (PCDD/Fs).45 Their findings indicated that electron-donating capacity, represented by descriptors such as EHOMO and qH+, was the primary factor affecting reaction rates. Likewise, Qi et al. employed MLR and found that EHOMO significantly influenced the reaction rate constant, while NCl played a critical role, independent of chlorine positions.46 Luo et al. uesd MLR and reported that the position of chlorination on PCDDs is a key determinant in the kinetics of hydroxyl radical oxidation kinetics.47 Furthermore, Chen et al. developed a model using PLS regression with structural descriptors to predict the photolysis rate constants of PCDD/Fs on cherry leaf wax layers.48 They discovered that PCDD/Fs with higher NCl and α values, along with lower ELUMO, exhibited faster photodegradation, which aligns with our hydrolysis model findings and highlights the significance of these descriptors in predicting the reactivity and environmental persistence of PCDDs. Therefore, our models build upon these findings, further emphasising the critical role of key descriptors such as NCl and α values in the hydrolysis reaction.
As illustrated in Table 3, we conducted a comparison of the models developed in this study with QSAR models created through MLR for hydrolysis rates of various organic compounds.49–52 All models were built using rigorous statistical algorithms and well-defined descriptors, consistent with our findings in Model (1). Berger et al. observed that the acid-catalyzed hydrolysis rate constants of sulfonylureas decreased significantly with increasing ELUMO values.49 Similarly, Xu et al. reported that phthalate esters with higher α values displayed a significant increase in the kOH of one side chain, corroborating with the findings of our model (2).51 Compared to these models, the current model offers more accessible descriptors, enhanced clarity and a larger dataset of 75 samples, thereby improving reliability and mitigating the risk of overfitting. This provides a simpler and more interpretable alternative to more complex models. Consequently, our model presents a more robust and interpretable framework for assessing the hydrolysis mechanism of PCDDs, utilising key descriptors while ensuring high predictive accuracy and simplicity. This makes it a valuable tool for evaluating the environmental persistence of PCDDs under base-catalyzed conditions.
| Model napb | Training set | Validation set | ||||||
|---|---|---|---|---|---|---|---|---|
| R adj 2 | RMSEtra | Q LOO 2 | R ext 2 | RMSEext | Q ext 2 | |||
| a n represents the number of chemicals in the data set. b p represents the total number of predictor variables. | ||||||||
| Bernhard et al. (1995) | 6 | 1 | 0.838 | — | — | — | — | — |
| Wang et al. (2018) | 40 | 8 | 0.822 | 1.472 | — | — | — | — |
| Xu et al. (2019) | 23 | 3 | 0.865 | 0.389 | 0.801 | 0.925 | 0.311 | 0.840 |
| Xu et al. (2019) | 5 | 1 | 0.975 | 0.276 | 0.914 | — | — | — |
| Xu et al. (2021) | 24 | 3 | 0.842 | — | 0.729 | 0.919 | — | 0.843 |
5 Conclusion
As of 2025, the Chemical Abstracts Service Registry encompasses over 290 million unique chemical substances (Chemical Abstracts Service, 2025).53 This extensive database relies exclusively on experimental measurements for risk assessment, underscoring the necessity for alternative approaches, such as QSAR models, to effectively evaluate the risks associated with both existing and newly identified chemicals. In this study, four QSAR models were developed for 75 PCDDs utilising molecular descriptors with the Gaussian 16 software package. Statistical diagnostics confirmed the quality of these models, while both internal and external validation demonstrated their robustness and predictive capability. The QSAR models derived from quantum chemical (ELUMO and α), geometric (Vm), and constitutional (NCl, Nβ, and Nm) descriptors exhibited a strong correlation with kOH and effectively portrayed the base-catalyzed hydrolysis of PCDDs. The developed models facilitate the exploration of the mechanisms responsible for the base-catalyzed hydrolysis of PCDDs. Additionally, these models provide molecular descriptors that can be utilised in the development of future machine learning and artificial intelligence models for the hydrolysis of aromatic heterocyclic compounds.Author contributions
Kun Xie: writing – original draft, investigation, conceptualization. Haiqin Zhang: writing – review & editing, methodology, conceptualization.Conflicts of interest
The authors have no conflict of interest to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: details about QSAR model development, values of the descriptors used in the QSAR model, Williams plots, Fig. S1−S4 and Tables S1−S5. See DOI: https://doi.org/10.1039/d5va00397k.Acknowledgements
We acknowledge funding from the Key Lab of Eco-restoration of Regional Contaminated Environment (Shenyang University), Ministry of Education (KF-23-03).References
- R. Lei, Z. Xu, Y. Xing, W. Liu, X. Wu, T. Jia, S. Sun and Y. He, J. Hazard. Mater., 2021, 418, 126265 CrossRef CAS PubMed.
- C. L. Fletcher and W. A. McKay, Chemosphere, 1993, 26, 1041–1069 CrossRef CAS.
- P. S. Kulkarni, J. G. Crespo and C. A. M. Afonso, Environ. Int., 2008, 34, 139–153 CrossRef CAS PubMed.
- E. Q. S. Dossier , Polychlorinated dibenzo-p-dioxins (PCDDs), polychlorinated dibenzofurans (PCDFs), and dioxin-like polychlorinated biphenyls (DL-PCBs), Sub-group on Review of the Priority Substances List, Working Group E, Common Implementation Strategy for the Water Framework Directive, 2011, p. 35 Search PubMed.
- C.-M. Chen, Chemosphere, 2004, 54, 1413–1420 CrossRef CAS PubMed.
- S. Sidhu and P. Edwards, Int. J. Chem. Kinet., 2002, 34, 531–541 CrossRef CAS.
- U. Quaß, M. Fermann and G. Bröker, Chemosphere, 2004, 54, 1319–1327 CrossRef PubMed.
- M. Kim, L. Y. Li and J. R. Grace, Environ. Pollut., 2016, 213, 99–111 CrossRef CAS PubMed.
- G. McKay, Chem.–Eng. J., 2002, 86, 343–368 CrossRef CAS.
- K. J. Friesen and G. R. B. Webster, Environ. Sci. Technol., 1990, 24, 97–101 CrossRef CAS.
- M. Lippmann and G. D. Leikauf, Environmental Toxicants: Human Exposures and Their Health Effects, Wiley & Sons, Limited, John, 2020 Search PubMed.
- W. Mabey and T. Mill, J. Phys. Chem. Ref. Data, 1978, 7, 383–415 CrossRef CAS.
- H. Zhang, K. Xie, Q. Luo, J. Tang and Y. Zhang, Environ. Sci. Technol., 2024, 58, 5483–5490 CrossRef CAS PubMed.
- Y. Liu, P. Peng, X. Li, S. Zhang and M. Ren, J. Hazard. Mater., 2008, 152, 40–47 CrossRef CAS PubMed.
- Q. Zhang, L. Gao, M. Zheng, L. Liu and C. Li, Bull. Environ. Contam. Toxicol., 2014, 92, 585–589 CrossRef CAS PubMed.
- P. Kumar, A. Kumar and D. Singh, Environ. Toxicol. Pharmacol., 2022, 103893 CrossRef CAS PubMed.
- F. Li, X. Li, X. Liu, L. Zhang, L. You, J. Zhao and H. Wu, Environ. Toxicol. Pharmacol., 2011, 32, 478–485 CrossRef CAS PubMed.
- P. de Voogt, D. C. G. Muir, G. R. B. Webster and H. Govers, Chemosphere, 1990, 21, 1385–1396 CrossRef CAS.
- Q. Li, H. Yang, N. Hao, M. Du, Y. Zhao, Y. Li and X. Li, J. Environ. Manag., 2023, 345, 118898 CrossRef CAS PubMed.
- OECD, Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models, Organization for Economic Cooperation and Development, 2014 Search PubMed.
- Y. Gao, J. Zhang, S. Cui, Y. Wu, M. Huang and S. Zhuang, Elsevier eBooks, 2023, 89–99 Search PubMed.
- R. Todeschini and V. Consonni, Methods and Principles in Medicinal Chemistry, 2000 Search PubMed.
- K. Roy, S. Kar and R. N. Das, Understanding the Basics of QSAR for Applications in. Pharmaceutical Sciences and Risk Assessment, 2015, 47–80 Search PubMed.
- A. R. Katritzky, V. S. Lobanov and M. Karelson, Chem. Soc. Rev., 1995, 24, 279 RSC.
- M. J. Frisch, G. W. Trucks and H. B. Schlegelet al., Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- F. P. Pineda, J. Ortega-Castro, J. Raúl Álvarez-Idaboy, J. Frau, B. M. Cabrera, J. C. Ramírez, J. Donoso and F. Muñoz, J. Phys. Chem. A, 2011, 115, 2359–2366 CrossRef CAS PubMed.
- T. Lu and F. Chen, J. Comput. Chem., 2011, 33, 580–592 CrossRef PubMed.
- T. I. Netzeva, A. P. Worth, T. Aldenberg, R. Benigni, M. T. D. Cronin, P. Gramatica, J. S. Jaworska, S. Kahn, G. Klopman, C. A. Marchant, G. Myatt, N. Nikolova-Jeliazkova, G. Y. Patlewicz, R. Perkins, D. W. Roberts, T. W. Schultz, D. T. Stanton, J. J. M. van de Sandt, W. Tong and G. Veith, Altern. Lab. Anim., 2005, 33, 155–173 CrossRef CAS PubMed.
- P. Gramatica, E. Giani and E. Papa, J. Mol. Graph. Model., 2007, 25, 755–766 CrossRef CAS PubMed.
- A. Golbraikh and A. Tropsha, J. Mol. Graph. Model., 2002, 20, 269–276 CrossRef CAS PubMed.
- W. Y. Shiu, W. Doucette, F. A. P. C. Gobas, A. Andren and D. Mackay, Environ. Sci. Technol., 1988, 22, 651–658 CrossRef CAS.
- T. N. G. Borhani, M. Saniedanesh, M. Bagheri and J. S. Lim, Water Res., 2016, 98, 344–353 CrossRef CAS PubMed.
- M. Taghi Goodarzi, B. Dejaegher and Y. Vander Heyden, J. AOAC Int., 2012, 95, 636–651 CrossRef PubMed.
- P. Gramatica, Int. J. Quant. Struct.-Prop. Relat., 2020, 5, 61–97 Search PubMed.
- L.-G. Zhuo, W. Liao and Z.-X. Yu, Asian J. Org. Chem., 2012, 1, 336–345 CrossRef CAS.
- A. Bendjeddou, T. Abbaz, A. Gouasmia and D. Villemin, Int. Res. J. Pure Appl. Chem., 2016, 12, 1–9 CrossRef CAS.
- K. F. Khaled, Appl. Surf. Sci., 2008, 255, 1811–1818 CrossRef CAS.
- J. Attarki, M. Khnifira, W. Boumya, H. Hajjaoui, A. Mahsoune, M. Sadiq, M. Achak, N. Barka and M. Abdennouri, Surfaces, 2023, 6, 450–465 CrossRef CAS.
- Y. Wang, J. Chen, W. Tang, D. Xia, Y. Liang and X. Li, Chemosphere, 2019, 214, 79–84 CrossRef CAS PubMed.
- J. Chen, X. Quan, K.-W. Schramm, A. Kettrup and F. Yang, Chemosphere, 2001, 45, 151–159 CrossRef CAS PubMed.
- J. E. Edwards and R. G. Pearson, J. Am. Chem. Soc., 1962, 84, 16–24 CrossRef CAS.
- Z. Yang, S. Luo, S.-H. Ho, T. Ye, Z. Wei, D. Chen and M. Jin, Environ. Pollut., 2016, 211, 157–164 CrossRef CAS PubMed.
- K. R. Cox and W. G. Chapman, J. Am. Chem. Soc., 2001, 123, 6745 CrossRef CAS.
- M. B. Smith, March’s Advanced Organic Chemistry : Reactions, Mechanisms, and Structure, WILEY, 2019 Search PubMed.
- C. Yan, J. Chen, L. Huang, G. Ding and X. Huang, Chemosphere, 2005, 61, 1523–1528 CrossRef CAS PubMed.
- C. Qi, C. Zhang and X. Sun, Int. J. Mol. Sci., 2015, 16, 18812–18824 CrossRef CAS PubMed.
- S. Luo, S.-H. Ho, Z. Wei, Z. Yang, L. Chai and M. Jin, Chemosphere, 2017, 172, 333–340 CrossRef CAS PubMed.
- J. Chen, X. Quan, F. Yang and W. J. G. M. Peijnenburg, Sci. Total Environ., 2001, 269, 163–170 CrossRef CAS PubMed.
- B. M. Berger and N. Lee Wolfe, Environ. Toxicol. Chem., 1996, 15, 1500–1507 CAS.
- L. Wang, B. Chen and T. Zhang, Chem. Eng. J., 2018, 342, 372–385 CrossRef CAS.
- T. Xu, J. Chen, Z. Wang, W. Tang, D. Xia, Z. Fu and H. Xie, Environ. Sci. Technol., 2019, 53, 5828–5837 CrossRef CAS PubMed.
- T. Xu, J. Chen, X. Chen, H. Xie, Z. Wang, D. Xia, W. Tang and H. Xie, Environ. Sci. Technol., 2021, 55, 6022–6031 CrossRef CAS PubMed.
- Chemical Abstracts Service, 2025. https://www.cas.org/cas-data.
| This journal is © The Royal Society of Chemistry 2026 |

