Open Access Article
Open Access ArticleDSSA-PPI: enhancing binding affinity change prediction upon protein mutations using disentangled structure–sequence aware attention
Juhong Wu a,
Jiehui Suna,
Tian Lianga,
Yongqi Zhanga,
Han Zhanga,
Tianyi Zhangd,
Xianmin Feng
a,
Jiehui Suna,
Tian Lianga,
Yongqi Zhanga,
Han Zhanga,
Tianyi Zhangd,
Xianmin Feng d,
Ping Gaoa,
Peng Xu*b and
Jinyu Li
d,
Ping Gaoa,
Peng Xu*b and
Jinyu Li *ace
*ace
aCollege of Chemistry, Fuzhou University, Fuzhou 350116, Fujian, China. E-mail: j.li@fzu.edu.cn
bCollege of Biological Science and Engineering, Fuzhou University, Fuzhou 350116, Fujian, China. E-mail: pengxu@fzu.edu.cn
cCollege of Biological and Pharmaceutical Engineering, Jilin Agricultural Science and Technology University, Jilin 132101, China
dJilin Medical University, Jilin 132013, China
eFujian Provincial Key Laboratory of Theoretical and Computational Chemistry, Xiamen 361005, Fujian, China
First published on 23rd December 2025
Abstract
Accurate assessment of the effects of mutations on protein–protein interactions (PPIs) is crucial for understanding disease pathogenesis and the development of targeted therapies. Here, we present DSSA-PPI, a hybrid deep learning framework that enhances the prediction of mutation-induced binding affinity changes (ΔΔG) by leveraging structural and sequence information through a disentangled attention mechanism. Building upon the complementary strengths of a geometric equivariant graph neural network PPIFormer and a protein language model ESM-2, our framework employs a novel representation learning strategy that integrates sequence- and structure-specific contributions, thereby improving the precision of PPI ΔΔG predictions. DSSA-PPI demonstrates robust performance across diverse mutational contexts on the standard protein binding affinity SKEMPI v2 dataset and outperforms existing methods on multiple benchmarks under identical cross-validation. In a case study of the SARS-CoV-2 receptor-binding motif (RBM) interaction with angiotensin-converting enzyme 2 (ACE2), our model accurately identifies top-ranking mutations that enhance binding affinity. Additionally, it guided the optimization of a peptide inhibitor, improving its inhibitory activity against activated factor XI (FXIa) by over 40-fold. These results highlight DSSA-PPI as a versatile and reliable tool for predicting mutation-induced perturbations in PPIs.
1 Introduction
Protein–protein interactions (PPIs) govern a wide range of biological processes, including signal transduction,1,2 immune responses,3,4 and metabolic regulation.5,6 Mutations that disrupt or modulate these interactions can lead to dysregulated cellular functions and are implicated in numerous diseases, including cancer,7,8 autoimmune disorders,9,10 and neurodegenerative diseases.11,12 Understanding how amino acid substitutions influence PPIs is critical for elucidating disease mechanisms and developing targeted therapeutic strategies. In protein engineering—particularly in antibody design and binder development—optimizing PPIs often requires exploring vast mutational landscapes, where combinatorial mutations can generate thousands to millions of potential variants. While high-throughput experimental techniques like deep mutational scanning (DMS) enable the large-scale profiling of PPIs,13 these methods remain resource-intensive and impractical for exhaustively assessing expansive mutational spaces. Consequently, there is an urgent need for accurate and scalable computational approaches to predict the effects of mutations on binding affinity.Over the years, numerous computational methods have been developed to predict changes in protein–protein binding affinity upon mutations.11–16 Traditional physics-based approaches, such as FoldX,14 Flex ddG,15 meta-dynamics,16 and molecular mechanics/generalized born surface area (MM/GBSA),17 rely on rigorous free-energy calculations but suffer from high computational costs due to extensive conformational sampling requirements. In contrast, machine learning (ML)-based methods offer a more efficient, data-driven alternative. These approaches can be broadly categorized into sequence-based and structure-based models, depending on their input features.
Sequence-based models leverage evolutionary information and physicochemical properties derived from amino acid sequences to infer binding affinity changes. For instance, SAAMBE-SEQ employs hand-crafted sequence features, including position-specific scoring matrices (PSSMs) and physicochemical descriptors, in conjunction with a gradient-boosted tree (GBT) predictor.18 Similarly, ISLAND integrates sequence-based features with kernel-based similarity measures to enhance prediction accuracy.19 However, these methods fail to account for the structural context essential for accurately modeling binding interactions.
Structure-based models, by contrast, utilize geometric representations of protein complexes to encode topological and spatial features, often achieving superior predictive performance. These models can be broadly divided into two categories: featurization-based methods and end-to-end methods. Featurization-based methods rely on hand-crafted or algorithmically derived features to encode the structural and physicochemical properties of protein. A prominent line of work in this category leverages algebraic topology to simplify complex 3D structures while preserving essential biological features, has shown promising potential in the field of mutational effect on protein binding affinity changes prediction. For example, TopNetTree employs element- and site-specific persistent homology to extract topological invariants from protein–protein complexes, effectively capturing their structural complexity for mutation effect prediction.20 Among the most advanced models in this category, MT-TopLap integrates persistent Laplacian features with physicochemical descriptors and transformer-based protein language model embeddings, achieving superior performance in predicting binding affinity changes.21 In contrast, end-to-end methods directly learn structural representations from raw input without manual feature engineering. GeoPPI, for example, employs a graph attention network to learn structural representations through a self-supervised side-chain reconstruction task, followed by a GBT predictor to estimate mutational effects.22 MpbPPI extends this paradigm with an equivariant graph neural network (GNN) and multi-task pre-training to further enhance structure-aware learning.23 However, both models rely on GBT predictors, which do not explicitly model the physical mechanisms underlying PPI perturbations. Recent advances in geometric deep learning have led to frameworks such as MuToN, which employs a geometric transformer to capture local structural perturbations and receptor–ligand interactions.24 Despite their promise, these methods remain constrained by their dependence on high-quality structural inputs for mutant complexes. While tools like AlphaFold25 and RoseTTAFold26 can predict mutant structures, their computational cost becomes prohibitive when applied to large mutational libraries.27,28 Moreover, selecting the most plausible conformations from multiple predicted structures introduces additional challenges.
To address these limitations, PPIFormer adopts a pre-training strategy inspired by masked language modeling, reconstructing masked structural elements to predict binding affinity changes without requiring explicit mutant structures.29 By computing the log-odds of probability differences between wild-type and mutant residues, PPIFormer enables predictions based solely on a single input structure. Nevertheless, existing approaches—whether sequence- or structure-based—have yet to fully exploit the complementary strengths of both modalities. For example, ProAffiMuSeq iteratively combines sequence and structural descriptors but lacks a mechanism to model their interdependencies;30 while ProBASS concatenates embeddings from ESM-2 and ESM-IF1 without explicitly capturing their interplay.31
Here, we present DSSA-PPI, a hybrid deep learning framework that integrates sequence and structural information via a cross-attention mechanism to predict mutation-induced changes in protein–protein binding affinity. Building upon PPIFormer, our model retains its advantage of bypassing explicit mutant structures while enhancing predictive accuracy through the incorporation of ESM-2-derived sequence representations. DSSA-PPI demonstrates robust performance across diverse mutational contexts in the SKEMPI v2 dataset and outperforms existing methods on standard benchmarks. Stratified analyses further demonstrate its reliability across mutations of different physicochemical types. We further validate its utility through a case study on SARS-CoV-2 spike protein variants binding to angiotensin-converting enzyme 2 (ACE2), where DSSA-PPI accurately identifies affinity-enhancing mutations and captures key mutational signatures of variants of concern (VOCs). Furthermore, DSSA-PPI successfully guides the engineering of a cyclic peptide inhibitor's binding affinity to activated factor XI (FXIa), validating its practical application. Collectively, these results establish DSSA-PPI as a powerful and versatile tool for predicting mutation-induced perturbations in PPIs.
2 Results and discussion
2.1 Overview of DSSA-PPI
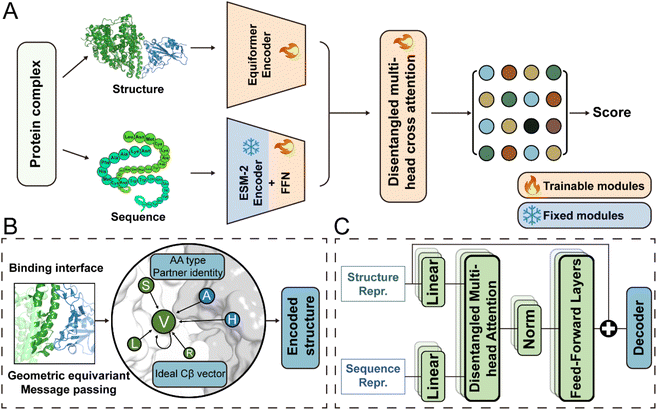
The global framework of DSSA-PPI is illustrated in Fig. 1A, with detailed architectural components presented in Fig. S1. It comprises three primary modules: a structure encoder, a sequence encoder, and a transformer-based fusion block equipped with disentangled attention mechanisms for both structural and sequence information.2.2 Performance for protein binding affinity prediction
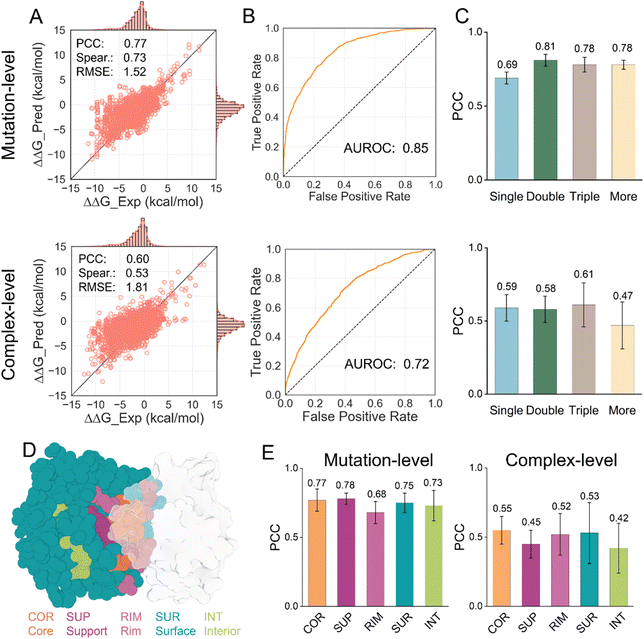
We conducted a comprehensive evaluation of the predictive performance of DSSA-PPI using the SKEMPI v2 benchmark dataset, which contains 7085 experimentally measured mutation effects (including both single and multiple amino acid substitutions) across 345 distinct protein–protein interactions. Following established conventions, we employed a leave-one-complex-out (LOCO) cross-validation and a 10-fold cross-validation strategy, utilizing two distinct splitting methodologies: mutation-level splitting and complex-level splitting. Multiple evaluation metrics were employed to comprehensively evaluate model performance, with Pearson's correlation coefficient (PCC) serving as the primary evaluation criterion.Under the LOCO split, we limited training to 3 epochs per fold due to the high computational cost of end-to-end training across multiple folds. Despite this constraint, DSSA-PPI maintained competitive performance relative to existing methods (Table S1). For mutation-level splitting, our model achieved an overall PCC of 0.77, a Spearman's correlation of 0.73, and a root-mean-square error (RMSE) of 1.52 for ΔΔG prediction (Fig. 2A). For binary classification of binding affinity changes, the model recorded an area under the receiver operating characteristic curve (AUROC) of 0.85 (Fig. 2B). Notably, the model exhibited consistent performance across folds in mutation-level cross-validation. In contrast, larger variability was observed in complex-level evaluation, suggesting that mutation-level splitting may overestimate performance due to potential information leakage among mutations within the same complex (Table S2).
We further evaluated the predictive performance of DSSA-PPI across varying mutational distances. While the model demonstrated consistent and robust performance on mutation-level splitting, its performance declined on the more challenging complex-level splitting, which involves testing on unseen protein complex structures (Fig. 2C). Specifically, the model exhibited reduced accuracy at higher mutational distances (mutational distance > 3). Such cases often involve extensive or highly disruptive alterations that may induce complex changes in protein–protein interactions, including significant conformational rearrangements. These dynamic structural perturbations present a fundamental challenge, as the current model implicitly assumes moderate local shifts upon mutation and does not explicitly model large-scale conformational changes. The impact of protein dynamics is a well-recognized limitation in this field.33–35 Future extensions incorporating global dynamic features, such as those captured by the Gaussian Network Model (GNM)36 and the Anisotropic Network Model (ANM),37 may help improve predictive performance in these challenging scenarios. With the rapid advancement of deep learning frameworks capable of modeling protein conformational ensembles,38–40 fully integrating dynamic information into ΔΔG prediction is expected to become increasingly feasible.
Finally, we investigated the predictive ability of DSSA-PPI across different structural regions of protein complexes. Using the SKEMPI v2 annotations, the dataset was categorized into core (COR), support (SUP), rim (RIM), surface (SUR), and interior (INT) regions based on mutation locations (Fig. 2D). Notably, despite being trained exclusively on binding interface residues, DSSA-PPI maintained consistent predictive accuracy across all structural areas (Fig. 2E).
2.3 Ablation analysis of model components
To elucidate the contributions of key components in our model architecture, we conducted a series of ablation experiments. Specifically, we examined the effects of (i) removing the sequence encoder and (ii) replacing the disentangled attention fusion module with a simple concatenation operation. As presented in Table 1, both modifications resulted in notable performance degradation, underscoring the importance of these components. These results indicate that: (i) evolutionary context captured by the sequence encoder is essential for accurate prediction of binding affinity changes, and (ii) the disentangled attention mechanism facilitates a more effective integration of structural and sequence-based features than naïve concatenation.| PCC ↑ | Spearman ↑ | RMSE ↓ | AUROC ↑ | |
|---|---|---|---|---|
| a The term “– w/o R2R” denotes the removal of structure-to-structure attention term, similar to other terms. | ||||
| DSSA-PPI | 0.77 | 0.73 | 1.52 | 0.85 |
| – w/o seq. enc. | 0.74 | 0.68 | 1.72 | 0.80 |
| – w/o cross-attn. | 0.72 | 0.65 | 1.80 | 0.77 |
| – w/o R2Ra | 0.75 | 0.70 | 1.65 | 0.80 |
| – w/o R2S | 0.76 | 0.71 | 1.59 | 0.83 |
| – w/o R2Spartner | 0.77 | 0.71 | 1.54 | 0.84 |
| – w/o S2R | 0.76 | 0.71 | 1.53 | 0.83 |
| – w/o Spartner2R | 0.76 | 0.70 | 1.53 | 0.85 |
To further assess the contributions of individual attention terms in the disentangled attention module, we performed ablations by removing one term at a time: including structure-to-structure (R2R), structure-to-sequence (R2S), structure-to-partner sequence (R2Spartner), sequence-to-structure (S2R), partner sequence-to-structure (Spartner2R). These ablation results reveal that all five terms are necessary to achieve the best overall performance. Notably, the removal of R2R and R2S results in drops in overall performance, with the PCC decreasing from 0.77 to 0.75 and 0.76, respectively. This underscored the important role of structure information and structural representation to attend to its own sequence context. Removing of Spartner2R have least impact, suggesting that binding partner sequence attention to structure is relatively less critical than other items.
Collectively, these findings validate our architectural design and emphasize the critical role of synergistically combining structural and sequence information, as well as the necessity of each individual attention term, for robust and accurate prediction of binding affinity changes.
2.4 Benchmarking against existing methods under identical complex-level cross-validation
To establish the predictive capabilities of DSSA-PPI, we conducted comprehensive benchmarking against the very recent approaches using three widely adopted datasets: S1131, S4169, and M1707. As a baseline comparison, we first performed random 10-fold mutation-level cross-validation. As shown in Table S3, all machine learning-based methods, including DSSA-PPI, exhibited strong performance across benchmarks under this setting.However, recent studies have raised significant concerns that random mutation-level splits can lead to overly optimistic performance estimates,41,42 particularly when mutations from the same protein complex appear in both training and test sets. Consistent with these observations, we also found that performance metrics under mutation-level splitting were substantially higher than those obtained under complex-level cross-validation (Fig. 2A, B and Table S2).
To address this, we adopted a more rigorous evaluation protocol based on complex-level splitting, following the protocol of MpbPPI, which ensures that mutations from the same protein complex do not appear across different folds. We adopted predefined five-fold complex-level cross-validation splits for S1131 and S4169 from MpbPPI. For M1707, which lacks predefined partitions, we created analogous complex-level splits based on PDB identifiers to ensure methodological consistency. To ensure robust performance estimation, we repeated five-fold cross-validation five times using independent random seeds for each dataset. The same data splits were applied consistently across all baseline methods to ensure fair comparisons.
Across all benchmarks, DSSA-PPI generally outperformed baseline models on both regression (PCC, RMSE) and classification (AUROC) tasks (Tables 2 and S4). On the S1131 dataset, which consists of non-redundant single-point mutations localized at protein–protein interfaces. DSSA-PPI achieved a PCC of 0.71 and an AUROC of 0.77. This performance exceeded that of PPIFormer (0.68/ 0.72), MT-TopLap (0.62/0.72), ESM-2 with a three-layer feedforward network regressor (ESM-2-FFN) (0.65/0.74), ProBASS (0.65/0.80), and SAAMBE-SEQ (0.49/0.70). On the S4169 dataset, which includes a more diverse mutation landscape, DSSA-PPI maintained strong performance with a PCC of 0.58 and AUROC of 0.70. For the challenging M1707 dataset—characterized by up to nine mutations across multiple chains within a single complex—DSSA-PPI achieved a PCC of 0.42 and AUROC of 0.72.
| Method | S1131 | S4169 | M1707 | |||
|---|---|---|---|---|---|---|
| PCC ↑ | AUROC ↑ | PCC ↑ | AUROC ↑ | PCC ↑ | AUROC ↑ | |
| a Data are represented as mean ± SD (n = 5).b Bold values indicate the best results. The dash sign indicates the results of the corresponding methods are not available.c ESM-2-FFN refers to the ESM-2 model followed by a three-layer feed-forward network, which shares the same architecture as the sequence encoder used in DSSA-PPI.d Indicates statistically significant difference compared to DSSA-PPI (p < 0.05), determined by a one-tailed unpaired t-test. | ||||||
| DSSA-PPI | 0.71 ± 0.01a,b | 0.77 ± 0.02 | 0.58 ± 0.01 | 0.70 ± 0.02 | 0.42 ± 0.02 | 0.72 ± 0.02 |
| PPIFormer | 0.68 ± 0.01d | 0.72 ± 0.02d | 0.54 ± 0.01d | 0.60 ± 0.01d | 0.31 ± 0.03d | 0.61 ± 0.02d |
| ESM-2-FFNc | 0.65 ± 0.02d | 0.72 ± 0.02d | 0.45 ± 0.02d | 0.62 ± 0.02d | 0.25 ± 0.05 | 0.63 ± 0.02d |
| MT-TopLap | 0.62 ± 0.03d | 0.74 ± 0.01d | 0.50 ± 0.03d | 0.64 ± 0.01d | — | — |
| MuToN | 0.70 ± 0.01 | 0.74 ± 0.01 | 0.57 ± 0.01 | 0.64 ± 0.01d | — | — |
| MpbPPI | 0.48 ± 0.04d | 0.75 ± 0.02d | 0.43 ± 0.03d | 0.67 ± 0.01d | 0.34 ± 0.02d | 0.64 ± 0.02d |
| ProBASS | 0.65 ± 0.01d | 0.80 ± 0.01 | 0.51 ± 0.01d | 0.68 ± 0.03 | 0.20 ± 0.04d | 0.59 ± 0.02d |
| SAAMBE-SEQ | 0.49 ± 0.01d | 0.70 ± 0.02d | 0.29 ± 0.01d | 0.59 ± 0.01d | — | — |
| FoldX | 0.44 | 0.70 | 0.30 | 0.62 | 0.30 | 0.59 |
Compared with physics-based approaches, DSSA-PPI also showed superior performance across S1131, S4169, and M1707 (Table S5). Under current benchmarking protocols, structure-based methods, including physics-based tools, are typically evaluated on single static structures because generating conformational ensembles through molecular dynamics is computationally demanding. This practice does not reflect the full capabilities of physics-based methods, which generally achieve higher accuracy when conformational sampling is incorporated.17,43 In contrast, ML-based models have been shown to exhibit greater robustness to structural variability, as reported in several studies.23,24 A systematic benchmarking framework that evaluates both physics-based and machine learning based methods using dynamically sampled conformations would provide a more balanced and rigorous comparison. Although such an assessment is beyond the scope of the present study, it represents an important direction for future work.
Beyond numerical performance, we conducted an in-depth architectural analysis to elucidate the sources of the predictive advantage of DSSA-PPI. We benchmarked it against five representative ML-based baseline models: PPIFormer (structure-based), ESM-2-FFN (sequence-based), ProBASS (structure–sequence hybrid), MT-TopLap (topology-based hybrid), and SAAMBE-SEQ (biophysics-informed), as summarized in Table S6.
Our comparative analysis revealed several key insights. First, end-to-end deep learning models generally outperformed methods based on handcrafted features, such as SAAMBE-SEQ, across all datasets. Interestingly, PPIFormer outperformed ESM-2-FFN in regression tasks, while ESM-2-FFN generally showed superior performance in classification (Table 2). This divergence may suggest that structural and sequence representations encode distinct but complementary aspects of protein binding. ProBASS attempts to leverage this by fusing multimodal embeddings from ESM-IF and ESM-2 via simple feature concatenation. While this yielded modest improvements over ESM-2-FFN on S4169 datasets (PCC of 0.51 vs. 0.45), it failed to generalize well to more complex cases such as M1707, where performance decreased compared to the ESM-2 backbone (PCC: 0.20 for ProBASS vs. 0.25 for ESM-2-FFN). A similar pattern was observed with MT-TopLap, which combines ESM-2 embeddings with handcrafted topological features. Despite this multimodal feature fusion, MT-TopLap did not outperform ESM-2-FFN, indicating that naive feature concatenation may not be sufficient to capture the complex interplay between structure and sequence representations.
By contrast, DSSA-PPI builds upon PPIFormer and ESM-2, but employs a disentangled structure–sequence attention module specifically designed to model inter-modal interactions. This architecture consistently outperforms both backbone models across benchmark datasets. To further assess the generality of the disentangled attention design, we evaluated DSSA-PPI-PF, a variant where the structure encoder is initialized with PPIFormer pre-trained weights and the pretraining is omitted. Despite lacking co-adaptive structure–sequence pretraining, DSSA-PPI-PF still outperformed PPIFormer and achieved performance comparable to the fully end-to-end DSSA-PPI model (Table S7). This finding highlights the plug-and-play robustness and effectiveness of the disentangled attention module.
In summary, our architectural investigation indicates that the performance gains of DSSA-PPI stem from the disentangled attention mechanism, which facilitates synergistic integration of structural and sequential representations.
2.5 Stratified performance analysis based on chemical characteristics mutation types
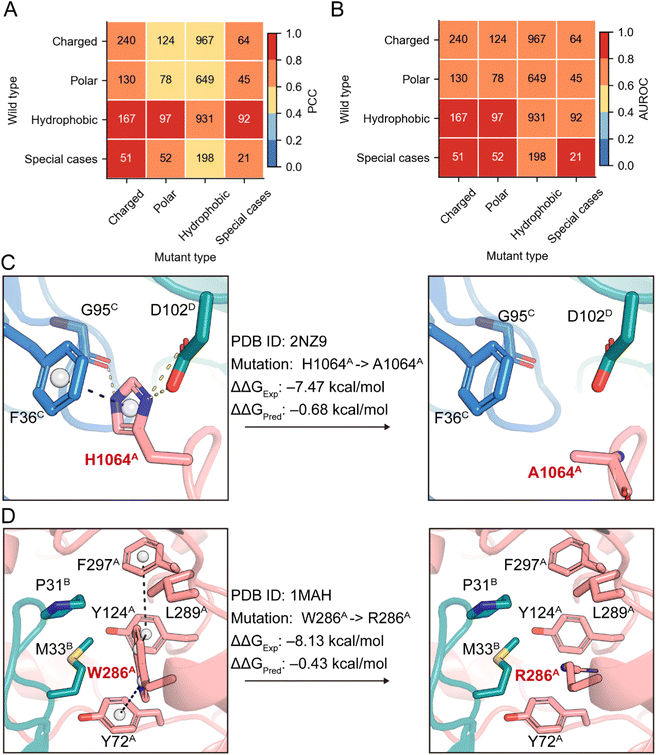
Having established the overall predictive superiority of DSSA-PPI on benchmark datasets, we next sought to examine its predictive performance in a more fine-grained manner. To this end, we conducted a systematic error analysis stratified by amino acid chemical properties. This analysis leveraged the S4169 dataset, which comprises all single-point mutations from the SKEMPI v2.0 database. Predictive performance was assessed using PCC and AUROC, averaged over five repetitions of five-fold cross-validation.We first categorized mutations based on the chemical characteristics of the substituted residues: charged, polar, hydrophobic, and special cases (Gly, Cys, Pro). As shown in Fig. 3A and B, DSSA-PPI demonstrated consistent performance across most categories. For regression, all groups achieved a PCC greater than 0.4, with four categories exceeding 0.8. In the classification setting, DSSA-PPI also performed reliably, with AUROC scores above 0.6 across all categories. These results suggest that DSSA-PPI captures the general physicochemical behavior of amino acid substitutions, enabling generalization across diverse mutation types. Due to the severe class imbalance in the S4169 dataset, we did not perform further breakdown analysis at the level of individual wild-type to mutant amino acid pairs (Fig. S2).
To further investigate specific failure modes, we manually inspected the top 10 mutations with the largest absolute prediction errors (|ΔΔGExp − ΔΔGPred|). These outlier mutations often involved wild-type residues participating in intricate multichain interactions or exhibiting extreme physicochemical shifts. For instance, in PDB complex 2NZ9 (Fig. 3C), the wild-type H1064A participates in hydrogen bonding with D102D and G95C, and engages in π–π stacking with F36C. Upon mutation to alanine, DSSA-PPI correctly predicted a decrease in binding affinity but substantially underestimated the magnitude of ΔΔG, likely due to the disruption of multichain cooperative interactions that are difficult to model and may not be fully captured by the coarse-grained, residue-level representations employed in DSSA-PPI. Another failure case involves a drastic substitution from tryptophan to arginine in 1MAH. The wild-type tryptophan resides in a tightly packed hydrophobic pocket at the interface (Fig. 3D). Introducing a charged arginine residue alters the local electrostatic environment and induces desolvation penalties. DSSA-PPI failed to fully capture this physicochemical disruption, underestimating the binding destabilization.
These findings suggest that while DSSA-PPI demonstrates strong overall performance, it faces challenges in specific edge cases involving complex interaction networks. In such cases, the perturbations introduced by certain substitutions may not be fully captured by the coarse-grained, residue-level representations used in DSSA-PPI. Future improvements could include incorporating explicit side-chain modeling or surface-based physicochemical representations to better characterize the local interaction environment and enhance prediction accuracy in structurally intricate contexts.
2.6 Evaluation on SARS-CoV-2 RBM mutations affecting ACE2 binding
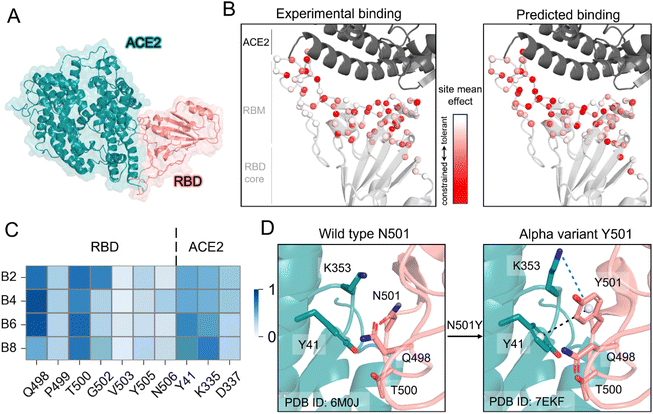
To further demonstrate the predictive power of our model, we evaluated its performance on the fitness of binding affinity changes induced by mutations in the receptor binding motif (RBM) of the receptor binding domain (RBD) of the SARS-CoV-2 spike protein, which directly interacts with the human ACE2 receptor (Fig. 4A). We employed an ensemble model trained via 10-fold cross-validation on SKEMPI v2, with predictions averaged for robustness. All comparative models were evaluated under identical conditions to ensure fair benchmarking. As the MT-TopLap was pretrained directly on the SARS-CoV-2 RBD/ACE2 deep mutational scanning (DMS) dataset,21,44 it was excluded from this case study to maintain an unbiased comparison.We conducted saturation single-point mutational analysis on approximately 1700 RBM mutations, with predictions completed within seconds for DSSA-PPI, demonstrating high computational efficiency. Compared to DMS experimental data,45 DSSA-PPI consistently outperformed all baseline methods across multiple evaluation metrics (Tables 3 and S8). Notably, DSSA-PPI predictions captured the overall mutational impact patterns observed in DMS data at both the site level (Fig. 4B) and the mutation level (Fig. S3). Additionally, we employed a hit-rate metric to assess the ability of models to prioritize top-ranking mutations. Among the top 10 high-affinity mutations identified by DMS experiments, DSSA-PPI successfully identified 4 within the top 1% (18 mutations) and 6 within the top 5% (86 mutations) of its predictions. In comparison, PPIFormer, ESM-2-FFN, and MuToN identified 2, 2, and 1 mutation(s) within the top 5%, respectively. MpbPPI, ProBASS, SAAMBE-SEQ, and FoldX failed to rank any of these mutations within the thresholds.
| Method | Spearman ↑ | Top10Hit@1% ↑ | Top10Hit@5% ↑ |
|---|---|---|---|
| DSSA-PPI | 0.31 | 40% | 60% |
| PPIFormer | 0.19 | 0% | 20% |
| ESM-2-FFN | −0.04 | 0% | 20% |
| MuToN | −0.01 | 0% | 10% |
| MpbPPI | 0.08 | 0% | 0% |
| ProBASS | −0.01 | 0% | 0% |
| SAAMBE-SEQ | −0.20 | 0% | 0% |
| FoldX | −0.65 | 0% | 0% |
Further, we investigated hot-spot mutations in the RBM region that are associated with early SARS-CoV-2 variants of concern (VOCs), including N501Y, E484K, T478K, K417N, L452R, and Q493R. We compared the ranking and predicted binding fitness scores from DSSA-PPI with experimental data from DMS relative to the wild-type spike protein (Table 4). In general, DSSA-PPI ranked these hot-spot mutations in a manner consistent with experimental data, except for E484K. Interestingly, while DMS reported T478K and Q493R as mildly destabilizing mutations, DSSA-PPI predicted both to be stabilizing, aligning with their known roles in enhancing ACE2 binding in the Delta and Omicron variants.46,47 These discrepancies suggest that DSSA-PPI may capture co-evolutionary and contextual information beyond the wild-type sequence, potentially enabling the identification of future adaptive mutations. Additionally, DSSA-PPI ranked N501Y as a top affinity-enhancing mutation, consistent with its well-established role in strengthening ACE2 binding. To probe the model's underlying reasoning, we visualized the attention scores associated with residue N501. The attention map revealed that N501 predominantly attends to Q498 and T500 within the RBD, as well as Y42 and K353 on ACE2 (Fig. 4C). Notably, the inter-chain attention patterns align with structural evidence: N501Y forms a π–π stacking interaction with Y42 and a cation–π interaction with K353 (Fig. 4D), both of which are known to enhance ACE2 binding. These results underscore the efficiency and predictive accuracy of DSSA-PPI, and suggest its ability to identify mutations of high biological relevance.
| Mutation | N501Y | E484K | T478K | K417N | L452R | Q493R |
|---|---|---|---|---|---|---|
| a Experimental rankings (%) represent the percentile of each mutation's binding fitness relative to all ∼1700 single-point RBM mutations in the DMS dataset. A lower percentile indicates higher binding affinity. | ||||||
| Exp. Rank (%)a | 0.24% | 4.88% | 22.03% | 52.23% | 4.11% | 29.61% |
| Pred. Rank (%) | 0.06% | 70.56% | 13.69% | 52.35% | 12.93% | 2.29% |
| Exp. Score | 1.07 | 0.09 | −0.13 | −0.89 | 0.13 | −0.28 |
| Pred. Score | 1.85 | −1.24 | 0.13 | −0.72 | 0.16 | 0.74 |
2.7 DSSA-PPI-guided reengineering of a peptide inhibitor against activated factor XI (FXIa)
Given the superior performance of DSSA-PPI in predicting and ranking mutation-induced changes in binding affinity, we next sought to assess its applicability in the design of peptide inhibitors of serine proteases. As a model system, we re-engineered a cyclic peptide inhibitor of murine urokinase-type plasminogen activator (muPA), mupain-1 (CPAYSRYLDC), to improve its affinity toward FXIa (Fig. 5A), an emerging target for low-hemorrhagic antithrombotic therapy.48,49 Mupain-1 was originally identified through phage display as a selective inhibitor of muPA,50 but has negligible inhibitory activity against FXIa (Ki > 1000 µM),51 making it an ideal starting point for DSSA-PPI-guided optimization.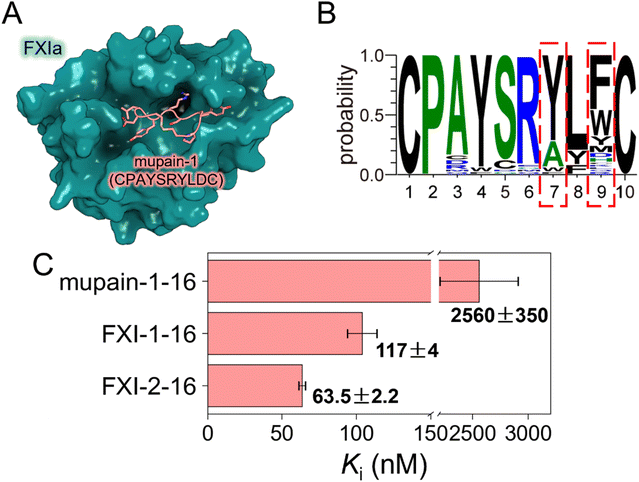 | ||
| Fig. 5 Reengineering of mupain-1 against FXIa guided by DSSA-PPI. (A) Structural model of the FXIa–mupain-1 complex. (B) Preferred amino acid frequencies at each position among the top 50 DSSA-PPI-predicted variants, highlighting mutation hotspots at Y7 and D9. (C) Binding affinities (Ki, nM) of the peptides toward human FXIa are shown as mean ± SD (n = 3). Data for mupain-1–16 are from Xu et al.51 | ||
To adapt DSSA-PPI for peptide–protease interactions, we fine-tuned the model on the SKEMPI v2.0 dataset, which comprises experimentally validated protein–peptide interaction data. Binding affinity measurements of mupain-1 variants targeting plasma kallikrein (PK)51—a serine protease homologous to FXIa—were used as an independent validation set to select the refined model exhibiting the highest Spearman correlation. The optimized DSSA-PPI was subsequently employed to predict and prioritize mutations that could enhance mupain-1 binding to FXIa.
A systematic single-point mutation scan revealed that hydrophobic substitutions at D9 were most likely to improve FXIa affinity (Fig. S4). However, given the intrinsically weak binding of the parental peptide (Ki > 1000 µM), we reasoned that single-site substitutions might not achieve sufficient enhancement. We therefore performed an exhaustive two-site combinatorial mutation scan, in which all possible double mutants were generated and ranked according to their predicted ΔΔG values. Analysis of the top 50 predicted variants (Fig. 5B) revealed that the cyclic architecture was strictly conserved, with cysteine residues at positions 1 and 10 retained, consistent with their established critical role in maintaining peptide binding affinity.50 Hydrophobic substitutions at D9 were strongly favored, consistent with the single-point scan, while substitutions of Y7 with alanine occurred at relatively high frequency. Guided by these predictions, two candidate peptides were designed: CPAYSRALFC (FXI-1) and CPAYSRALWC (FXI-2). To further enhance FXIa binding, the arginine residue at position 6 was replaced with a non-natural analogue (L-3-(N-amidino-4-piperidyl)alanine), previously shown to non-specifically improve affinity towards multiple serine proteases.51,52 Experimental validation confirmed that both designed peptides exhibited markedly enhanced FXIa binding affinity, with Ki values of 117 ± 4 nM for FXI-1-16 and 63.5 ± 2.2 nM for FXI-2-16 (Fig. 5C and S5), compared with 2560 ± 350 nM for the parental mupain-1–16.51 These engineered peptides therefore provide promising starting points for further optimization toward potent FXIa inhibitors for anticoagulant development.
Collectively, these results demonstrate the practical utility and transferability of DSSA-PPI in guiding peptide optimization and protein engineering, enabling rapid identification of high-affinity variants.
3 Conclusions
In this study, we introduced DSSA-PPI, a deep learning framework for predicting changes in protein–protein binding affinity induced by amino acid mutations. DSSA-PPI overcomes the limitations of structure-based models, which require precise mutant structures, by achieving superior or competitive performance using only a single wild-type structure. Through the synergistic integration of structural and sequence information, our model demonstrated strong predictive accuracy across diverse benchmark datasets and outperformed existing approaches in affinity prediction. Stratified analyses further demonstrated its reliability across mutations of different physicochemical types. The practical utility of DSSA-PPI was shown in two case studies. First, the model accurately identified high-affinity mutations in the SARS-CoV-2 RBM interacting with ACE2. Second, it successfully guided the reengineering of a peptide inhibitor targeting FXIa. These results highlight its potential for real-world protein engineering applications. However, like other current approaches, DSSA-PPI exhibits reduced predictive accuracy for highly disruptive mutations (i.e., mutational distance > 3) that may induce large-scale conformational changes. This limitation underscores the need for future work to incorporate structural dynamics for improved prediction. In summary, DSSA-PPI provides an efficient tool for protein–protein interaction engineering, offering valuable insights for rational protein design and therapeutic development.4 Methods
4.1 Overview of DSSA-PPI architecture
DSSA-PPI is composed of three main components: (i) structure encoder, (ii) sequence encoder, and (iii) disentangled attention-based fusion module. The framework is initially pre-trained on the PPIRef50K dataset, followed by fine-tuning for downstream protein–protein binding affinity prediction to evaluate its performance.4.2 Constructing attributed graph for interface
We first used PDBFixer v1.8 for each protein–protein complex to model the residue geometry coordinates if they are missing in PDB structures. Then, we represented the protein complexes as an attributed graph for their interface at the residue level. Interface residues are identified based on the Euclidean distance between their heavy atoms. A residue is considered part of the interface if the distance to the nearest residue from the binding partner is within a specific distance cutoff (10 Å for the pre-training dataset and 15 Å for SKEMPI v2 datasets). The attributed graph was defined as G = (V,E), where V is the set of residues in the protein complex interface (each residue node is represented by its Cα atom) and E is the set of edges between them. For each residue node vi ∈ VNi=1, where N is the number of residues, its feature is represented as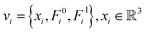 is the Cα coordinate of the ith residue. F0i,Fi1 are two kinds of feature: type-0 scalars (3D-invariant)
is the Cα coordinate of the ith residue. F0i,Fi1 are two kinds of feature: type-0 scalars (3D-invariant) 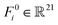 , and type-1 vectors (3D-equivariant)
, and type-1 vectors (3D-equivariant) 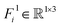 . Feature F0i contains the one-hot encoded representations of the interface residue type (dimension of 20) and a semantic binary value indicating its location in a binding partner. Feature Fi1 represents virtual beta-carbon orientations, calculated using backbone atom positions (Cα, N, C atoms) based on ideal bond angles and lengths.53 To capture the interactive relationships between residues, we define E as a {0, 1}N×N matrix, where 1 indicates an edge between two residue nodes if the Euclidean distance between them is less than 7 Å; otherwise, it is 0.
. Feature F0i contains the one-hot encoded representations of the interface residue type (dimension of 20) and a semantic binary value indicating its location in a binding partner. Feature Fi1 represents virtual beta-carbon orientations, calculated using backbone atom positions (Cα, N, C atoms) based on ideal bond angles and lengths.53 To capture the interactive relationships between residues, we define E as a {0, 1}N×N matrix, where 1 indicates an edge between two residue nodes if the Euclidean distance between them is less than 7 Å; otherwise, it is 0.
4.3 Geometric structure encoder
Geometric properties like translational invariance and rotational equivariance are essential in protein-related representation learning. To incorporate these inductive biases, we build our structure encoder on Equiformer,32 a transformer-based SE(3)-equivariant graph attention network. Equiformer enables the model to learn geometric representations that are equivariant to 3D rotations and translations by operating on features expressed as irreducible representations of the SE(3) group. These features can correspond to different rotation orders: type-0 (scalars), type-1 (vectors), and higher-order tensors.In our framework, following the design of PPIFormer,29 we construct our geometric structure encoder using eight stacked SE(3)-Equiformer blocks (Fig. S1A). Each block performs message passing over the protein interface graph and updates type-specific features via an attention mechanism that respects SE(3) symmetry. The input to each block l is the graph G, with node coordinates 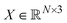 and connection matrix E, along with node feature matrices
and connection matrix E, along with node feature matrices 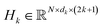 of different types k ∈ {0, 1}. The layer-wise update process is defined as follows:
of different types k ∈ {0, 1}. The layer-wise update process is defined as follows:
| h0(1),h1(1) = f(G(X,E,F0,F1)) | (1) |
| h0(l),h1(l) = f(G(X,E,H0(l−1),H1(l−1))), l = 2,…,8 | (2) |
4.4 Sequence encoder
To model the sequence-level information of protein–protein complexes, we employ ESM-2, a state-of-the-art pretrained protein language model. ESM-2 is trained on over 250 million protein sequences using a masked language modeling (MLM) objective, enabling it to capture rich contextual and evolutionary signals directly from sequence data. For each protein–protein complex. We extract the amino acid sequences of both binding partners. These sequences are either retrieved from the PDB database using the complex's PDB code or directly parsed from the input PDB structure file. Each sequence of the protein complex is then encoded into a 1280-dimensional vector, obtained from the 33rd layer embedding of ESM-2. These sequence embeddings are then processed by a parameter-shared feed-forward network (FFN) comprising three hidden layers, each with a hidden dimension of 256. The FFN maps the 1280-dimensional input down to a 128-dimensional output vector (Fig. S1B). These embeddings are subsequently fused with the structural representations derived from the geometric encoder for downstream binding affinity change prediction.4.5 Disentangled attention-based fusion module
To fully exploit a given protein complex's structural and sequence information, we employ a structure-sequence-aware disentangled multi-head attention mechanism (Fig. S1C). This approach has been effectively used in several protein language models and has benefited from integrated multimodal information.54–56 Specifically, let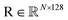 denote the structure representation of a protein complex interface (with N residues), and let
denote the structure representation of a protein complex interface (with N residues), and let 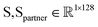 be the sequence representation of the protein and its binding partner, respectively. To model cross-modality and cross-partner interactions between structure (r) and sequence (s) and binding partner sequence (sp), we apply learnable linear projections to compute queries and keys as follows:
be the sequence representation of the protein and its binding partner, respectively. To model cross-modality and cross-partner interactions between structure (r) and sequence (s) and binding partner sequence (sp), we apply learnable linear projections to compute queries and keys as follows:
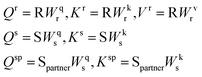 | (3) |
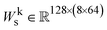 . These projections produce 8 attention heads, each with a dimensionality of 64. The projected tensors are then reshaped into the multi-head attention format:
. These projections produce 8 attention heads, each with a dimensionality of 64. The projected tensors are then reshaped into the multi-head attention format:
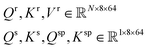 | (4) |
The cross-attention score  for each residue i is computed as the sum of five directional attention terms:
for each residue i is computed as the sum of five directional attention terms:
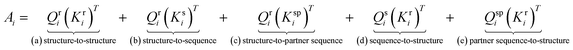 | (5) |
This design enables the structural representation to integrate information not only from itself, but also from both the intra-protein sequence and the binding partner's sequence, allowing comprehensive modeling of intra- and inter-protein interactions across both structural and sequence modalities. To stabilize training, the combined attention score is normalized by 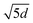 , where d = 64 is the head dimension. The final hidden state for residue i is:
, where d = 64 is the head dimension. The final hidden state for residue i is:
 | (6) |
This updated hidden state is passed through a fully connected layer (size: 128), followed by another softmax layer, yielding a probability distribution for the 20 amino acid types for each residue in the protein complex (Fig. 1A).
Author contributions
Conceptualization: J. W. and J. L.; methodology: J. W.; validation: J. S., Y. Z., and H. Z.; formal Analysis: J. W. and T. L.; investigation: J. W., Y. Z., H. Z., and P. G.; data curation: J. W. and J. S.; writing – original draft: J. W.; writing – review & editing: T. Z., X. F., P. X. and J. L.; supervision: J. L.; project administration: J. L.; funding acquisition: J. L.Conflicts of interest
The authors declare no conflict of interest.Data availability
The pretrained DSSA-PPI model weights and preprocessed model input data can be found in https://doi.org/10.5281/zenodo.15088340. The data and codes that support the findings of this manuscript are openly available GitHub repository https://github.com/Agitw/DSSA-PPI.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5sc08898d.
Acknowledgements
We gratefully acknowledge financial support from the Natural Science Foundation of China (22573016).References
- H. Lu, Q. Zhou, J. He, Z. Jiang, C. Peng, R. Tong and J. Shi, Recent Advances in the Development of Protein–Protein Interactions Modulators: Mechanisms and Clinical Trials, Signal Transduct. Targeted Ther., 2020, 5, 213 Search PubMed.
- X. Qian, X. Zhao, L. Yu, Y. Yin, X.-D. Zhang, L. Wang, J.-X. Li, Q. Zhu and J.-L. Luo, Current Status of GABA Receptor Subtypes in Analgesia, Biomed. Pharmacother., 2023, 168, 115800 Search PubMed.
- T. Tuller, S. Atar, E. Ruppin, M. Gurevich and A. Achiron, Common and Specific Signatures of Gene Expression and Protein–Protein Interactions in Autoimmune Diseases, Gene Immun., 2013, 14, 67–82 Search PubMed.
- H. Hong, J. Su, Y. Zhang, G. Xu, C. Huang, G. Bao and Z. Cui, A Novel Role of Lactate: Promotion of Akt-Dependent Elongation of Microglial Process, Int. Immunopharmacol., 2023, 119, 110136 Search PubMed.
- S. J. Humphrey, D. E. James and M. Mann, Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation, Trends Endocrinol. Metab., 2015, 26, 676–687 Search PubMed.
- J. Wu, Y. Shi, M. Zhou, M. Chen, S. Ji, X. Liu, M. Zhou, R. Xia, X. Zheng and W. Wang, Nutrient Vitamins Enabled Metabolic Regulation of Ferroptosis via Reactive Oxygen Species Biology, Front. Pharmacol, 2024, 15, 1434088 Search PubMed.
- M. Petrosino, L. Novak, A. Pasquo, R. Chiaraluce, P. Turina, E. Capriotti and V. Consalvi, Analysis and Interpretation of the Impact of Missense Variants in Cancer, Int. J. Mol. Sci., 2021, 22, 5416 Search PubMed.
- F. Song, C.-L. Lu, C.-G. Wang, C.-W. Hu, Y. Zhang, T.-L. Wang, L. Han and Z. Chen, Uncovering the Mechanism of Kang-Ai Injection for Treating Intrahepatic Cholangiocarcinoma Based on Network Pharmacology, Molecular Docking, and in Vitro Validation, Front. Pharmacol, 2023, 14, 1129709 Search PubMed.
- J. Zhao, J. Ma, Y. Deng, J. A. Kelly, K. Kim, S.-Y. Bang, H.-S. Lee, Q.-Z. Li, E. K. Wakeland and R. Qiu, A Missense Variant in NCF1 Is Associated with Susceptibility to Multiple Autoimmune Diseases, Nat. Genet., 2017, 49, 433–437 Search PubMed.
- Y. Qin, L. Chen, Q. Fei, X. Shao, W. Lv, J. Yang, F. Xu and J. Shi, Upregulation of CD226 on Subsets of T Cells and NK Cells Is Associated with Upregulated Adhesion Molecules and Cytotoxic Factors in Patients with Tuberculosis, Int. Immunopharmacol., 2023, 120, 110360 Search PubMed.
- M. R. Geisheker, G. Heymann, T. Wang, B. P. Coe, T. N. Turner, H. A. Stessman, K. Hoekzema, M. Kvarnung, M. Shaw and K. Friend, Hotspots of Missense Mutation Identify Neurodevelopmental Disorder Genes and Functional Domains, Nat. Neurosci., 2017, 20, 1043–1051 Search PubMed.
- Z. Hu, Y. Gu, M. Ye, Y. Ma, Y. Wang, S. Pan, C. Huang and X. Lu, Innate Immune Stimulation Prevents Chronic Stress-Induced Depressive and Anxiogenic-like Behaviors in Female Mice, Int. Immunopharmacol., 2022, 111, 109126 Search PubMed.
- D. M. Fowler and S. Fields, Deep Mutational Scanning: A New Style of Protein Science, Nat. Methods, 2014, 11, 801–807 Search PubMed.
- J. Schymkowitz, J. Borg, F. Stricher, R. Nys, F. Rousseau and L. Serrano, The FoldX Web Server: An Online Force Field, Nucleic Acids Res., 2005, 33, W382–W388 Search PubMed.
- K. A. Barlow, S. Ó Conchúir, S. Thompson, P. Suresh, J. E. Lucas, M. Heinonen and T. Kortemme, Flex ddG: Rosetta Ensemble-Based Estimation of Changes in Protein–Protein Binding Affinity upon Mutation, J. Phys. Chem. B, 2018, 122, 5389–5399 Search PubMed.
- S. Xie, Y. Zhou, H. Zhu, X. Xu, H. Zhang, C. Yuan, M. Huang, P. Xu, J. Li and Y. Liu, Interface-Driven Structural Evolution on Diltiazem as Novel uPAR Inhibitors: From in Silico Design to in Vitro Evaluation, Mol. Divers., 2025, 29, 1261–1274 Search PubMed.
- E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Z. H. Zhang and T. Hou, End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design, Chem. Rev., 2019, 119, 9478–9508 Search PubMed.
- G. Li, S. Pahari, A. K. Murthy, S. Liang, R. Fragoza, H. Yu and E. Alexov, SAAMBE-SEQ: A Sequence-Based Method for Predicting Mutation Effect on Protein–Protein Binding Affinity, Bioinformatics, 2021, 37, 992–999 Search PubMed.
- W. A. Abbasi, A. Yaseen, F. U. Hassan, S. Andleeb and F. U. A. A. Minhas, ISLAND: In-Silico Proteins Binding Affinity Prediction Using Sequence Information, BioData Min., 2020, 13, 1–13 Search PubMed.
- M. Wang, Z. Cang and G.-W. Wei, A Topology-Based Network Tree for the Prediction of Protein–Protein Binding Affinity Changes Following Mutation, Nat. Mach. Intell., 2020, 2, 116–123 Search PubMed.
- J. Wee and G.-W. Wei, Evaluation of AlphaFold 3's Protein–Protein Complexes for Predicting Binding Free Energy Changes upon Mutation, J. Chem. Inf. Model., 2024, 64, 6676–6683 Search PubMed.
- X. Liu, Y. Luo, P. Li, S. Song and J. Peng, Deep Geometric Representations for Modeling Effects of Mutations on Protein-Protein Binding Affinity, PLoS Comput. Biol., 2021, 17, e1009284 Search PubMed.
- Y. Yue, S. Li, L. Wang, H. Liu, H. H. Y. Tong and S. He, MpbPPI: A Multi-Task Pre-Training-Based Equivariant Approach for the Prediction of the Effect of Amino Acid Mutations on Protein–Protein Interactions, Brief. Bioinform., 2023, 24, bbad310 Search PubMed.
- P. Li and Z.-P. Liu, MuToN Quantifies Binding Affinity Changes upon Protein Mutations by Geometric Deep Learning, Adv. Sci., 2024, 11, 2402918 Search PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek and A. Potapenko, Highly Accurate Protein Structure Prediction with AlphaFold, Nature, 2021, 596, 583–589 Search PubMed.
- M. Baek, F. DiMaio, I. Anishchenko, J. Dauparas, S. Ovchinnikov, G. R. Lee, J. Wang, Q. Cong, L. N. Kinch and R. D. Schaeffer, Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network, Science, 2021, 373, 871–876 Search PubMed.
- N. Raouraoua, C. Mirabello, T. Véry, C. Blanchet, B. Wallner, M. F. Lensink and G. Brysbaert, MassiveFold: Unveiling AlphaFold's Hidden Potential with Optimized and Parallelized Massive Sampling, Nat. Comput. Sci., 2024, 1–5 Search PubMed.
- O. Kilim, A. Mentes, B. Pál, I. Csabai and Á. Gellért, SARS-CoV-2 Receptor-Binding Domain Deep Mutational AlphaFold2 Structures, Sci. Data, 2023, 10, 134 Search PubMed.
- A. Bushuiev, R. Bushuiev, P. Kouba, A. Filkin, M. Gabrielova, M. Gabriel, J. Sedlar, T. Pluskal, J. Damborsky, S. Mazurenko and J. Sivic, Learning to Design Protein-Protein Interactions with Enhanced Generalization, ICLR, 2024, https://openreview.net/forum?id=xcMmebCT7s Search PubMed.
- S. Jemimah, M. Sekijima and M. M. Gromiha, ProAffiMuSeq: Sequence-Based Method to Predict the Binding Free Energy Change of Protein–Protein Complexes upon Mutation Using Functional Classification, Bioinformatics, 2020, 36, 1725–1730 Search PubMed.
- S. N. S. Gurusinghe, Y. Wu, W. DeGrado and J. M. Shifman, ProBASS—a Language Model with Sequence and Structural Features for Predicting the Effect of Mutations on Binding Affinity, Bioinformatics, 2025, 41, btaf270 Search PubMed.
- Y.-L. Liao and T. Smidt, Equiformer, Equivariant Graph Attention Transformer for 3D Atomistic Graphs, ICLR, 2023, https://openreview.net/forum?id=KwmPfARgOTD Search PubMed.
- E. H. Kellogg, A. Leaver-Fay and D. Baker, Role of Conformational Sampling in Computing Mutation-Induced Changes in Protein Structure and Stability, Protein Struct. Funct. Bioinform., 2011, 79, 830–838 Search PubMed.
- A. Manson, S. T. Whitten, J. C. Ferreon, R. O. Fox and V. J. Hilser, Characterizing the Role of Ensemble Modulation in Mutation-Induced Changes in Binding Affinity, J. Am. Chem. Soc., 2009, 131, 6785–6793 Search PubMed.
- G. Verkhivker, S. Agajanian, D. Oztas and G. Gupta, Dynamic Profiling of Binding and Allosteric Propensities of the SARS-CoV-2 Spike Protein with Different Classes of Antibodies: Mutational and Perturbation-Based Scanning Reveals the Allosteric Duality of Functionally Adaptable Hotspots, J. Chem. Theory Comput., 2021, 17, 4578–4598 Search PubMed.
- I. Bahar and R. L. Jernigan, Vibrational dynamics of transfer RNAs: comparison of the free and synthetase-bound forms., J. Mol. Biol., 1998, 281(5), 871–884 Search PubMed 9719641.
- A. R. Atilgan, S. R. Durell, R. L. Jernigan, M. C. Demirel, O. Keskin and I. Bahar, Anisotropy of Fluctuation Dynamics of Proteins with an Elastic Network Model, Biophys. J., 2001, 80, 505–515 Search PubMed.
- S. Lewis, T. Hempel, J. Jiménez-Luna, M. Gastegger, Y. Xie, A. Y. K. Foong, V. G. Satorras, O. Abdin, B. S. Veeling, I. Zaporozhets, Y. Chen, S. Yang, A. E. Foster, A. Schneuing, J. Nigam, F. Barbero, V. Stimper, A. Campbell, J. Yim, M. Lienen, Y. Shi, S. Zheng, H. Schulz, U. Munir, R. Sordillo, R. Tomioka, C. Clementi and F. Noé, Scalable Emulation of Protein Equilibrium Ensembles with Generative Deep Learning, Science, 2025, 389, eadv9817 Search PubMed.
- N. E. Charron, K. Bonneau, A. S. Pasos-Trejo, A. Guljas, Y. Chen, F. Musil, J. Venturin, D. Gusew, I. Zaporozhets, A. Krämer, C. Templeton, A. Kelkar, A. E. P. Durumeric, S. Olsson, A. Pérez, M. Majewski, B. E. Husic, A. Patel, G. De Fabritiis, F. Noé and C. Clementi, Navigating Protein Landscapes with a Machine-Learned Transferable Coarse-Grained Model, Nat. Chem., 2025, 17, 1284–1292 Search PubMed.
- Y. Jin, Q. Huang, Z. Song, M. Zheng, D. Teng and Q. Shi, P2DFlow: A Protein Ensemble Generative Model with SE(3) Flow Matching, J. Chem. Theory Comput., 2025, 21, 3288–3296 Search PubMed.
- M. Tsishyn, F. Pucci and M. Rooman, Quantification of Biases in Predictions of Protein–Protein Binding Affinity Changes upon Mutations, Brief. Bioinform., 2024, 25, bbad491 Search PubMed.
- R. Zhang, N. Chen, F. Zhou and X. Gao, Navigating Trustworthiness of Deep Learning in ΔΔG Prediction : Addressing Data Bias, Model Evaluation, and Interpretation, in ICML 2024 Workshop on Machine Learning for Life and Material Science, 2024, https://openreview.net/forum?id=XU1ACMwTlr Search PubMed.
- L. A. Chi, J. E. Barnes, J. S. Patel and F. M. Ytreberg, Exploring the Ability of the MD+FoldX Method to Predict SARS-CoV-2 Antibody Escape Mutations Using Large-Scale Data, Sci. Rep., 2024, 14, 23122 Search PubMed.
- J. Wee, J. Chen, K. Xia and G.-W. Wei, Integration of Persistent Laplacian and Pre-Trained Transformer for Protein Solubility Changes upon Mutation, Comput. Biol. Med., 2024, 169, 107918 Search PubMed.
- T. N. Starr, A. J. Greaney, S. K. Hilton, D. Ellis, K. H. D. Crawford, A. S. Dingens, M. J. Navarro, J. E. Bowen, M. A. Tortorici, A. C. Walls, N. P. King, D. Veesler and J. D. Bloom, Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding, Cell, 2020, 182, 1295–1310 Search PubMed.
- R. A. A. Pondé, Physicochemical Effect of the N501Y, E484K/Q, K417N/T, L452R and T478K Mutations on the SARS-CoV-2 Spike Protein RBD and Its Influence on Agent Fitness and on Attributes Developed by Emerging Variants of Concern, Virology, 2022, 572, 44–54 Search PubMed.
- J. Lan, X. He, Y. Ren, Z. Wang, H. Zhou, S. Fan, C. Zhu, D. Liu, B. Shao, T.-Y. Liu, Q. Wang, L. Zhang, J. Ge, T. Wang and X. Wang, Structural Insights into the SARS-CoV-2 Omicron RBD-ACE2 Interaction, Cell Res., 2022, 32, 593–595 Search PubMed.
- Y. Zhou, D. Wang, J. Wu, Y. Qi, M. Song, H. Yao, C. K. Liao, H. Lin, M. Huang, D. Zhuo, L. Jiang, C. Yuan, Y. Chen, M. Huang, J. Li and P. Xu, Discovery of the Low-Hemorrhagic Antithrombotic Effect of Montelukast by Targeting FXIa in Mice, Arterioscler. Thromb. Vasc. Biol., 2025, 45, e150–e162 Search PubMed.
- I. Koulas and A. C. Spyropoulos, A Review of FXIa Inhibition as a Novel Target for Anticoagulation, Hämostaseologie, 2023, 43, 028–036 Search PubMed.
- L. M. Andersen, T. Wind, H. D. Hansen and P. A. Andreasen, A Cyclic Peptidylic Inhibitor of Murine Urokinase-Type Plasminogen Activator: Changing Species Specificity by Substitution of a Single Residue, Biochem. J., 2008, 412, 447–457 Search PubMed.
- P. Xu, M. Xu, L. Jiang, Q. Yang, Z. Luo, Z. Dauter, M. Huang and P. A. Andreasen, Design of Specific Serine Protease Inhibitors Based on a Versatile Peptide Scaffold: Conversion of a Urokinase Inhibitor to a Plasma Kallikrein Inhibitor, J. Med. Chem., 2015, 58, 8868–8876 Search PubMed.
- B. Zhao, P. Xu, L. Jiang, B. Paaske, T. Kromann-Hansen, J. K. Jensen, H. P. Sørensen, Z. Liu, J. T. Nielsen, A. Christensen, M. Hosseini, K. K. Sørensen, N. C. Nielsen, K. J. Jensen, M. Huang and P. A. Andreasen, A Cyclic Peptidic Serine Protease Inhibitor: Increasing Affinity by Increasing Peptide Flexibility, PLoS One, 2014, 9, e115872 Search PubMed.
- J. Dauparas, I. Anishchenko, N. Bennett, H. Bai, R. J. Ragotte, L. F. Milles, B. I. M. Wicky, A. Courbet, R. J. de Haas, N. Bethel, P. J. Y. Leung, T. F. Huddy, S. Pellock, D. Tischer, F. Chan, B. Koepnick, H. Nguyen, A. Kang, B. Sankaran, A. K. Bera, N. P. King and D. Baker, Robust Deep Learning–Based Protein Sequence Design Using ProteinMPNN, Science, 2022, 378, 49–56 Search PubMed.
- Y. Tan, R. Wang, B. Wu, L. Hong and B. Zhou, Retrieval-Enhanced Mutation Mastery: Augmenting Zero-Shot Prediction of Protein Language Model, arXiv, 2024, preprint, arXiv:2410.21127, DOI:10.48550/arXiv.2410.21127.
- D. Wang, M. Pourmirzaei, U. L. Abbas, S. Zeng, N. Manshour, F. Esmaili, B. Poudel, Y. Jiang, Q. Shao, J. Chen and D. Xu, S-PLM: Structure-Aware Protein Language Model via Contrastive Learning Between Sequence and Structure, Adv. Sci., 2025, 12, 2404212 Search PubMed.
- M. Li, Y. Tan, X. Ma, B. Zhong, H. Yu, Z. Zhou, W. Ouyang, B. Zhou, P. Tan and L. Hong, ProSST: Protein Language Modeling with Quantized Structure and Disentangled Attention, NeurIPS, 2024, DOI:10.52202/079017-1126.
| This journal is © The Royal Society of Chemistry 2026 |