Open Access Article
Open Access ArticleOdoribacter splanchnicus lipooligosaccharide: an uncommon structure with weak immunostimulatory activity
Marta
Tiemblo-Martin†
a,
Marcello
Mercogliano†
a,
Kaisa
Hiippala†
b,
Luca
De Simone Carone
 acd,
María Asunción
Campanero-Rhodes
acd,
María Asunción
Campanero-Rhodes
 fg,
Maria
Masiello
a,
Alessandro
Cangiano
ae,
Antonio
Molinaro
fg,
Maria
Masiello
a,
Alessandro
Cangiano
ae,
Antonio
Molinaro
 ad,
Luigi
Paduano
ae,
Dolores
Solís
ad,
Luigi
Paduano
ae,
Dolores
Solís
 fg,
Reetta
Satokari
b,
Flaviana
Di Lorenzo
fg,
Reetta
Satokari
b,
Flaviana
Di Lorenzo
 *ad and
Alba
Silipo
*ad and
Alba
Silipo
 *ad
*ad
aDepartment of Chemical Sciences and Task Force for Microbiome Studies, University of Naples Federico II, Via Cinthia 4, Naples, 80126, Italy. E-mail: flaviana.dilorenzo@unina.it; silipo@unina.it
bHuman Microbiome Research Program, Faculty of Medicine, University of Helsinki, Haartmaninkatu 8 (P O Box 63), 00014, Finland
cDepartment of Biology, University of Naples Federico II, Via Cinthia 4, Naples, 80126, Italy
dCEINGE Biotecnologie Avanzate Franco Salvatore, Via Gaetano Salvatore, 486, Naples, 80131, Italy
eCSGI, Center for Colloid and Surface Science, 50019 Sesto Fiorentino, Italy
fInstituto de Química Física Blas Cabrera, CSIC, Serrano 119, 28006 Madrid, Spain
gCIBER de Enfermedades Respiratorias (CIBERES), Avda Monforte de Lemos 3-5, 28029 Madrid, Spain
First published on 27th November 2025
Abstract
Odoribacter splanchnicus, a prominent member of the human gut microbiota, has emerged as a promising next-generation probiotic due to its anti-inflammatory properties and association with intestinal health. While previous studies have demonstrated its ability to attenuate inflammation and protect epithelial integrity, the molecular basis underlying these effects remained elusive. In this study, we provide the first comprehensive structural and functional characterization of the O. splanchnicus lipooligosaccharide (LOS), a major immunomodulatory surface component. NMR spectroscopy and mass spectrometry revealed a heterogeneous LOS structure composed of a mono-phosphorylated tetra- and penta-acylated lipid A backbone and a highly branched, short core oligosaccharide featuring unique residues such as 4-lactylmannose and galactosaminuronic acid. Small-angle X-ray scattering demonstrated that the LOS self-assembles into core–shell spherical micelles, with a large oligosaccharide corona shielding the lipid A, thereby limiting its accessibility to the innate immunity TLR4/MD-2 receptor complex. Accordingly, the O. splanchnicus LOS exhibited minimal TLR4 activation and failed to induce IL-8 secretion in enterocyte cell lines, in stark contrast to proinflammatory Escherichia coli LPS; in addition, it potently antagonized E. coli LPS-induced inflammation. These findings identify the LOS as a key effector of the O. splanchnicus anti-inflammatory phenotype and support its further development as a live biotherapeutic agent.
Introduction
The gut microbiota has emerged as a vital component of host health, with changes in its composition and activity closely linked to the onset of various diseases.1 Among the diverse microbial species inhabiting the human gut, Odoribacter splanchnicus has recently emerged as a promising next-generation probiotic (NGP), owing to its ability to produce short-chain fatty acids, modulate immune responses, and maintain intestinal homeostasis.2,3 Originally isolated from human abdominal abscesses, O. splanchnicus is now recognized as a prevalent and metabolically active member of the gut microbial community, associated with health-promoting effects such as anti-inflammatory activity and reinforcement of the epithelial barrier.4–6 Conversely, a reduction in its presence has been observed in several disease conditions, including inflammatory bowel disease, non-alcoholic fatty liver disease and cystic fibrosis.7O. splanchnicus was also identified as part of a core group of transferable donor-derived strains in fecal microbiota transplantation-treated ulcerative colitis patients, and colonization of mice with this bacterium led to the induction of mucosal regulatory T cells.8,9In vitro studies further demonstrated that cell-free supernatants from O. splanchnicus isolates exert anti-proliferative effects on human colon cancer cells.10 More intriguingly, the outer membrane vesicles (OMVs) it produces do not elicit IL-8 secretion in intestinal epithelial cells, suggesting a remarkably low proinflammatory potential, while at the same time exhibiting protective, anti-inflammatory activity in vitro.5 We also successfully formulated O. splanchnicus strain 57 for oral colon delivery, indicating the isolate's suitability to be developed into a live biotherapeutic agent.11All these findings are especially noteworthy, considering that O. splanchnicus is a Gram-negative bacterium whose outer membrane is characterized by the presence of lipopolysaccharides (LPSs), microbe associated molecular patterns acknowledged as potent inducers of inflammatory responses. LPSs are tripartite molecules composed of a glycolipid moiety (lipid A), an oligosaccharide region (core OS), and a polysaccharide chain (O-antigen). LPS molecules comprising all three regions are referred to as a smooth-type LPS (S-LPS), whereas those lacking the O-antigen are known as a rough-type LPS or lipooligosaccharide (R-LPS or LOS).12 The lipid A portion typically acts as a potent pro-inflammatory stimulus by binding to the innate immune system receptorial complex made up of Toll-Like Receptor 4/Myeloid Differentiation Protein-2 (TLR4/MD-2).13 However, the immunological properties of an LPS are tightly linked to its chemical structure. The LPS from Escherichia coli, for instance, possesses structural features that make it a robust TLR4 agonist and a powerful inducer of inflammation, namely a hexa-acylated lipid A with 4 + 2 symmetry and two phosphate groups on the glucosamine disaccharide backbone.14 In contrast, structural deviations in the lipid A moiety (as well as in the carbohydrate portion) can significantly reduce or even abrogate its pro-inflammatory activity. Therefore, an LPS can behave as a strong, weak, or even inert immunostimulant, depending on its molecular architecture.15
Therefore, understanding whether the LPS contributes to O. splanchnicus anti-inflammatory potential is critical for delineating the role of this gut bacterium as a modulator of host immune homeostasis. In this framework, we present the first structural characterization of the LOS isolated from O. splanchnicus, together with the evaluation of its functional effects on TLR4 signaling and intestinal epithelial cells. The immunomodulatory phenotype observed here confirmed the hypothesis that its O. splanchnicus LOS possesses structural features that deviate from the canonical pro-inflammatory archetype exemplified by E. coli and is linked to weak TLR4 activation.
Results
O. splanchnicus LOS isolation and chemical analysis
The LOS from O. splanchnicus was isolated following hot phenol-water extraction and further purified by enzymatic treatments and gel-filtration chromatography. Monosaccharide analysis indicated the presence of D-mannose (D-Man), D-glucosamine (D-GlcN), D-galactosaminuronic acid (D-GalNA), L-glycero-D-manno-heptose (Hep), and 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo). Moreover, GC-MS analysis of the acetylated methyl glycosides revealed the presence of a hexose substituted with a lactyl moiety, which was subsequently identified by NMR as 4-O-[1-carboxyethyl]-D-mannose. The electron impact mass spectrum of this monosaccharide, analyzed as an acetylated methyl glycoside, displayed diagnostic fragment ions at m/z 375, 347, 289, 272, 213 and 142. Methylation analysis indicated the presence of 2,7-substituted heptose (2,7-Hep), terminal GalNA (t-GalNA), terminal Man4Lac (t-Man4Lac), terminal and 4-substituted Man (t-Man and 4-Man), 6-substituted GlcN (6-GlcN) and 5-substituted Kdo (5-Kdo). Fatty acid compositional analysis revealed the presence of anteiso-pentadecanoic acid (aC15:0), iso-C15:0 (iC15:0) and C15:0, hydroxyhexadecanoic acid [C16:0 (3-OH)], and anteiso-hydroxyheptadecanoic acid [aC17:0 (3-OH)], iC-17:0 (3-OH), and C17:0 (3-OH).Structural determination of O. splanchnicus LOS
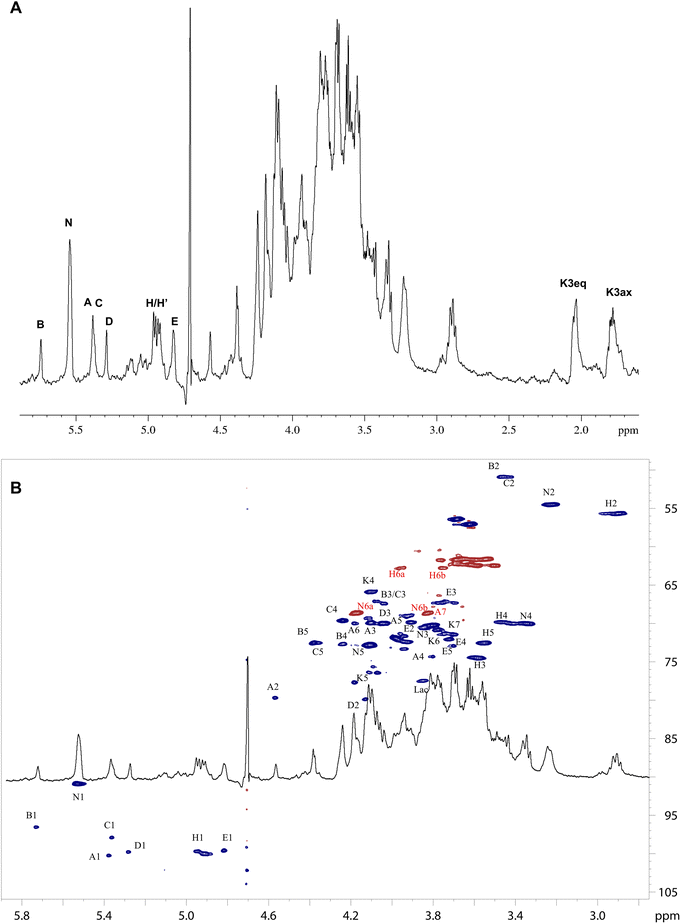
To elucidate the structure of the saccharide portion of the O. splanchnicus LOS, an aliquot was fully de-acylated and further purified by gel-filtration chromatography to get the pure saccharide fraction (OSdeAc), which was extensively studied by NMR spectroscopy, ESI-MS and tandem MS (Fig. 1 and 2). The anomeric configuration of the monosaccharide units was assigned based on 3JH1,H2 and 1JC1,H1 coupling constants, and further confirmed by intra-residue NOE contacts. The relative configuration of sugar residues was elucidated by analyzing vicinal 3JH1,H2 coupling constants and intra-residue NOE contacts. Five spin systems were identified (Fig. 1, S1 and Table S1); all the monosaccharide residues were present as pyranose rings, as indicated by both 13C chemical shift values and the long-range correlations in the HMBC spectrum between positions 1 and 5 (for Kdo between C-2 and H-6).16Spin systems H and N were both gluco configured sugar residues, based on the large 3JH,H ring coupling constants and were identified as GlcpN residues based on the correlation of their H-2 resonance to nitrogen bearing carbon signals (Fig. 1 and Table S1). The intra-residue NOE contact of H-1 with H-3 and H-5, and 3JH1,H2 and 1JCH values were indicative of the β-anomeric configuration of H. Conversely N was identified as an α-GlcN, whose anomeric configuration was determined according to 3JH1,H2 and the sole H1-H2 intra-residual NOE contact. Spin system B was assigned to a galacto configured sugar unit as evident from the low 3JH3,H4 and 3JH4,H5 values (3 Hz and <1 Hz, respectively). Both 3JH1,H2 and 1JCH coupling constants, together with intra-residual NOE contact of H-1 with H-2, were diagnostic for the α-configuration. In addition, and in accordance with the compositional analysis, scalar correlations between H-5 and a carboxylate group, evident from the HMBC spectra, and the correlation of the H-2 resonance with nitrogen-bearing carbon signals allowed this to be identified as a GalNA unit. Spin system E matched with an α-Man, with its C4 bonded to C2 of lactic acid (ether bond). The manno configuration was established by low 3JH1,H2 and 3JH2,H3, both below 2 Hz and diagnostic of the H-2 equatorial orientation. Spin system A matched with an α-heptose, whose manno configuration was established as above. Spin system K was assigned to the Kdo residue based on the diastereotopic methylene proton signals resonating in the high-field region of the NMR spectrum; its α-anomeric configuration was assigned on the basis of the H-3 chemical shift values (Table S1).17 The down-field shifted carbon signals were indicative of glycosylation at position O-6 and O-1 of N, O-6 of H, O-5 and O-4 of K, and O-2 and O-7 of A, while B was identified as a terminal residue, in accordance with the methylation analysis. The analysis of the inter-residual NOE contacts (Fig. S2) helped to define the complete monosaccharide sequence of the LOS from O. splanchnicus. Residues β-(1 → 6) linked H and N composed the lipid A saccharide backbone. Unit H was substituted at position O-6 by the Kdo residue (K), as also confirmed by the weak downfield shift of the signal of C-6 H (Table S1), expected for the α-(2 → 6) ketosidic linkage of Kdo with the β-GlcN of the lipid A. The Kdo unit was, in turn, substituted at O-5 by the heptose residue A, as suggested by the corresponding inter-residue NOE contacts, together with the HMBC correlations A1-K5.
Last, residue A was substituted at position O-2 by the terminal GalNA residue B, and in position O-7 by the terminal Man4Lac residue E. Overall we defined the structure as shown in Scheme 1.
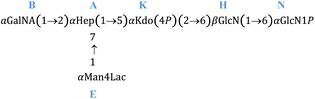 | ||
| Scheme 1 Structure of the OSdeAc from O. splanchnicus obtained from the combination of chemical, NMR and MS investigations. | ||
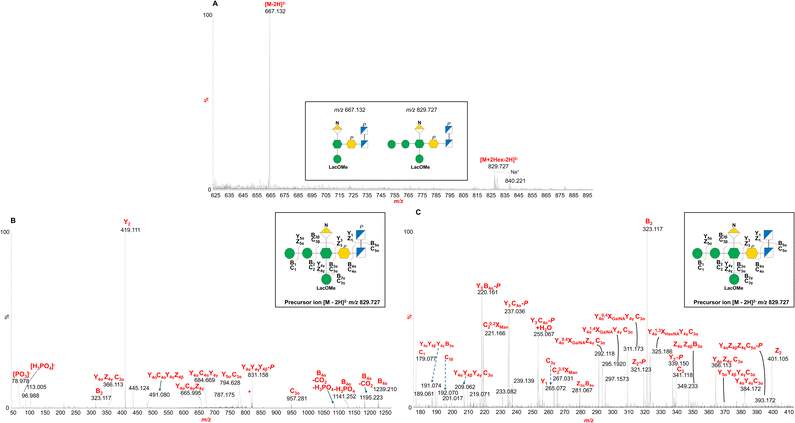
This structural deduction was confirmed and expanded by direct infusion ESI-MS and MS/MS analysis of OSdeAc. As shown in Fig. 2A, the negative-ion ESI-MS spectrum revealed a dominant double charged [M–2H]2− ion at m/z 667.132 consistent with the deacylated LOS structure. The assigned composition includes one Kdo, one heptose (Hep), one hexosaminuronic acid (HexNA), one hexose (Hex) carrying lactic acid (HexLac), one phosphate group, and one methyl group, along with a mono-phosphorylated diglucosamine backbone derived from lipid A (see below). Strikingly, a [M–2H]2− ion at m/z 829.727, consistent with the above-mentioned composition plus two other Hex units (likely mannoses, according to chemical analyses) was also detected. A related sodium adduct was also identified at m/z 840.1221.
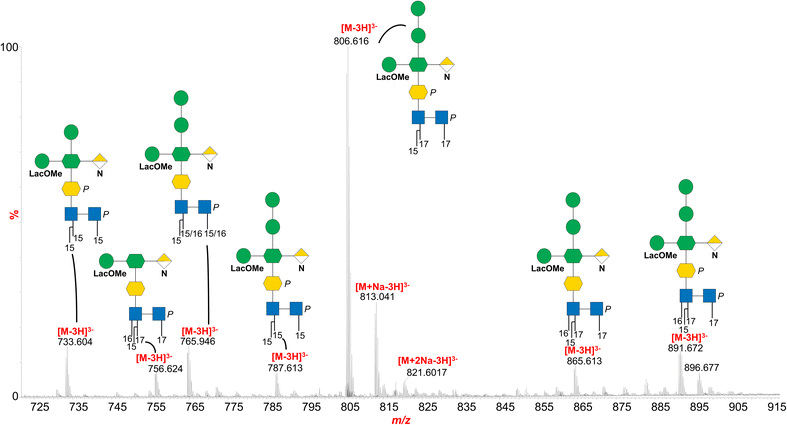
Negative-ion MS/MS analysis of the precursor ion at m/z 829.727 (Fig. 2B and C) provided key insights into the glycan sequence, confirming the structural assignment deduced by NMR and revealing the position of the two additional Hex residues, which could not be resolved by NMR alone. The MS/MS spectrum in fact revealed glycosidic and cross-ring fragments distinctive of the core region in the m/z 50–1250 range. The most intense peak at m/z 419.111 (Fig. 2B) matched with the mono-phosphorylated lipid A diglucosamine backbone (Y2 ion) while the related B4α fragmentation was detected at m/z 1239.210. Peaks attributed to ions originating from phosphate and/or carbon dioxide losses were also identified (Fig. 2B). Of note, the identification of the peak at m/z 265.072 (Fig. 2C) revealed that the methyl group was on the HexLac unit. The sequentiality of the two Hex was demonstrated by the intense B2 fragment at m/z 323.117 (Fig. 2B and C) while their linkage to the Hep by the C3α fragment at m/z 957.281, was in parallel supported by the absence of any peaks attributable to the loss of one (or two) Hex plus HexNA or HexLac. Additional fragments confirmed the peculiar highly branched oligosaccharide structure, such as the peaks at m/z 684.669 and m/z 794.628 that supported the presence of the methylated HexLac and the HexNA on the Hep moiety, in agreement with the NMR analysis. In parallel, the lipid A structure was determined by merging data from compositional and MALDI-TOF MS analyses. In the negative-ion MALDI-TOF MS spectrum (Fig. S3) the existence of different lipid A species was shown, with two main clusters of signals differing in the number of fatty acid chains. Each group of ions exhibited mass differences indicating variation in the fatty acid chain length within the lipid A species. In detail, the most intense cluster of ions in the mass range m/z 1600–1702 was consistent with mono-phosphorylated penta-acylated lipid A species whose fatty acid distribution and composition for the ion peak at m/z 1660.1 was determined as two C17:0 (3-OH) as N-linked acyl chains, two C15:0(3-OH) as primary O-linked fatty acids and C15:0 as a secondary O-acyl substitution, in accordance with fatty acid compositional analysis and previously reported data on Bacteroidetes lipid A.28–31 The cluster of ions visible in the mass range m/z 1391–1434 was attributed to mono-phosphorylated tetra-acylated lipid A species with the main ion peak at m/z 1419.0 assigned to a lipid A species possessing the above-mentioned fatty acid distribution and composition but lacking one primary O-linked C15:0(3-OH). Finally, to gain a more comprehensive structural overview, an aliquot of LOS was hydrolyzed with diluted ammonium hydroxide. This treatment selectively removed O-linked acyl chains, enabling a clearer assessment of structural variability in both the oligosaccharide and lipid A portions across the detected species. The resulting spectrum revealed that the main peaks differed predominantly in the length of the saccharide chain and the number and type of acyl chains on the lipid A moiety (Fig. 3). For instance, the most intense triply charged ion, [M–3H]3− at m/z 806.616, was assigned to a species composed of the complete core OS linked to a lipid A carrying two N-linked 17:0(3-OH) chains and one secondary 15:0 fatty acid. In comparison, the [M–3H]3− ion at m/z 891.672 was attributed to the same structure bearing an additional 16:0(3-OH) unit. Additional species were also detected, featuring lipid A with three or four acyl chains of varying lengths, associated either with the intact core OS or with truncated versions lacking the Hex residues and/or one phosphate group.
Overall, we concluded that the O. splanchnicus LOS is made up of a core region that was predominantly composed of a tetrasaccharide backbone bearing a phosphorylated Kdo unit, which was further substituted at position O-5 by a Hep residue. This heptose was identified as a branched monosaccharide, carrying an α-GalNA unit at position O-2 and an α-Man4Lac at position O-7. The stereochemistry of lactic acid remains to be defined. In addition, MS analysis revealed structural heterogeneity within the LOS species, with the detection of extended species bearing a disaccharide of (1 → 4)-linked hexose residues at the non-reducing end of the core OS. These additional Hex units, likely D-mannoses according to compositional analyses, are attached to position O-4 of the Hep residue, further contributing to the observed microheterogeneity. Consistent with this, integration of MS and linkage analysis data suggested that these extended species represent a minor subpopulation of the LOS, accounting for approximately 10% of the mixture. The presence of the GalNA residue in the LOS is particularly notable, as this monosaccharide is rarely observed in LOS core oligosaccharides, though it has been reported in the O-antigen of Pseudomonas aeruginosa18 and in the core region of Bordetella bronchiseptica, a respiratory pathogen in mammals including humans.19 Remarkably, in B. bronchiseptica, the absence of GalNA correlates with a reduced induction of TNF-α, a key proinflammatory cytokine involved in early host defense.19 This suggests a potential immunomodulatory role for GalNA, either direct or indirect, shaping host responses to bacterial components. Another notable peculiarity of the O. splanchnicus LOS is the presence of the rare sugar residue 4-Lactylmannose (4-O-[1-carboxyethyl]mannose; D-ManpLac). This residue is an uncommon substituent in bacterial polysaccharides, although it has been previously identified in the extracellular polysaccharides Cyanospira capsulate, Mycobacterium lacticolum 121 and the halophilic bacterium Salinivibrio sp. EG9S8QL.20–22 Lactic acid itself has been described as an ether-linked substituent in various bacterial polysaccharides, most notably as an N-linked substituent in N-acetylmuramic acid. However, to the best of our knowledge, this is the first report of D-Man4Lac in a member of the Bacteroidota phylum. Given that the structural specificity of microbial glycans is intimately linked to their biological function, it is tempting to hypothesize that the presence of D-Man4Lac may be involved in bacterial recognition by host immune systems. Interestingly, Cyanobacterium Nostoc commune contains a different derivative, a lactylated uronic acid called “nosturonic acid”,23 and its exopolysaccharides have been extensively studied for their prebiotic functions in selectively promoting beneficial bacterial growth such as bifidobacteria and lactobacilli while inhibiting pathogenic microorganisms. This selective modulation of gut microbiota helps to maintain intestinal barrier integrity and supports host health.24,25 In addition, several studies indicate that mannose derivatives can influence cytokine production and immune cell activation,26 while mannose-containing oligosaccharides improve gut dysbiosis and intestinal barrier damage.27
As for the lipid A portion, this resulted in a complex blend of mono-phosphorylated tetra- and penta-acylated lipid A species carrying linear and branched acyl chains. Analogously, other Bacteroides and Prevotella species, all belonging to the phylum Bacteroidota, like O. splanchnicus, have been found to carry penta-acylated and mono-phosphorylated lipid A (Fig. 4).29,31–35 Genomic analyses suggested that many Bacteroidota lack the enzymatic machinery required for the synthesis of more pro-inflammatory hexa-acylated lipid A species.36 These observations support the notion that O. splanchnicus conforms to a broader structural paradigm of under-acylated, less inflammatory lipid A within the Bacteroidota phylum. In this frame, of particular interest for further investigation is the carbohydrate portion (Fig. 4), whose structural heterogeneity and peculiarities warrant deeper exploration from a biosynthetic standpoint.
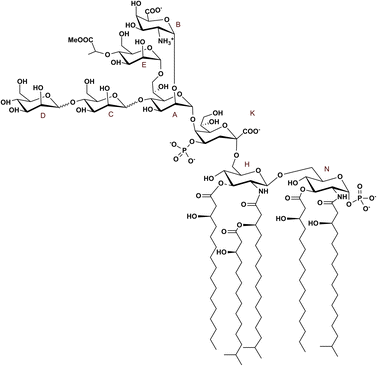 | ||
| Fig. 4 Full structure of the LOS from O. splanchnicus. Branched fatty acids are represented in their iso-form; however they can also be found as linear or anteiso chains. | ||
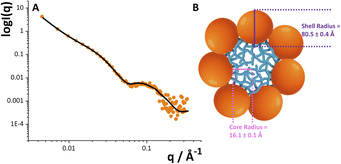
Given the structural peculiarity of this LOS, we then investigated the supramolecular organization of the O. splanchnicus LOS, which is known to critically influence immunogenicity. Small Angle X-ray scattering measurements on the O. splanchnicus LOS unambiguously reveal its propensity to form monodisperse, spherical micelles in aqueous solution, a supramolecular organization that mirrors self-assembly behavior reported for a broad range of bacterial LPSs/LOSs extracted from several bacterial strains.37 As shown in Fig. 5, the experimental data were fitted using a core–shell model (eqn (1)), which effectively describes a spherical micelle composed of a compact hydrophobic core (Lipid A domains) surrounded by a hydrophilic shell.38 The nonlinear least-squares fitting of the model yielded a hydrophobic core radius of 16.1 ± 0.1 Å and a total particle radius of 80.5 ± 0.4 Å. This aggregation state is essential, as it is known to influence how a LPS is recognized by the host immune system.39 In particular, the extended oligosaccharide shell (∼64 Å) sterically shields the lipid A moiety and limits access to the TLR4/MD-2 receptor complex. Such a “structural masking” mechanism has been shown to attenuate LPS immunostimulatory potential.40 Taken together, the unique structural and aggregation properties of the O. splanchnicus LOS ensure its processing in a low-inflammatory manner. This may allow the bacterium to evade immune detection and preserve epithelial integrity, thus favoring the persistence of beneficial commensals in the gut and protecting against colonization by proinflammatory pathobionts.
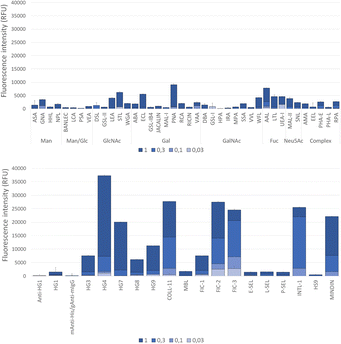
Microarray studies
Lectin recognition of the O. splanchnicus LOS was examined in binding assays to the microarray-printed LOS.41 First, the binding of a panel of 40 model lectins with diverse carbohydrate-binding specificities (Table S2) was tested. As shown in Fig. 6 (upper panel), negligible or very weak signals were detected for most lectins, including those exhibiting specificity for Man-containing structures, although lectin binding to the appropriate control glycoproteins was observed. This behavior indicates that Man residues are not in the appropriate configuration for recognition, as can be anticipated for 4-substituted Man. Significant signals were only detected for a limited number of lectins with nominal specificity for GlcNAc, Gal, or Fuc, which can be ascribed to peculiar features of their fine specificities. In addition, the binding of 17 human innate immune lectins (Table S3) was examined (Fig. 6, lower panel). Intense binding signals were observed for galectin-4, ficolins 2 and 3, intelectin-1, and mindin, as similarly observed for the LPS of Paenalcaligenes hominis.42 The complexity of the O. splanchnicus LOS interactome with innate immune lectins is further expanded by collectin-11, which gave strong binding signals, and the other galectins tested, i.e. galectins 3, 7, 8, and 9, for which weak to moderate signals were detected. Besides calcium-dependent sugar recognition (Table S2), collectin-11 has been shown to bind DNA from diverse origins in a charge-dependent mode and it has been suggested to interact directly with several microbes in a calcium- and charge-dependent manner.43Therefore, it is plausible that charge-mediated interactions could contribute to collectin-11's behavior, possibly owing to a suitable negative charge distribution in the LOS molecule. Regarding galectins, binding of galectin-3 to galacturonic acid in an unconventional orientation, flipped about 180° relative to the canonical binding mode, has been described,44 raising the possibility that it could also accommodate a GalNA residue within the binding site. To the best of our knowledge, no similar studies have been reported for other galectins. These results encourage further in-depth analyses of the peculiar motifs of this LOS and their recognition by the innate immune lectins tested here.
O. splanchnicus LOS-mediated TLR activation
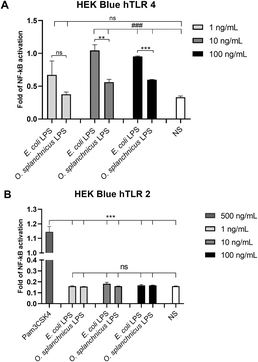
To assess the immunological properties of the O. splanchnicus LOS, different concentrations of the purified LOS were tested in HEK-Blue™ cells transfected with human TLR4, MD-2, and CD14 genes or HEK-Blue™ hTLR2 cells. The O. splanchnicus LOS exhibited a significantly weaker capability to activate TLR4 signaling compared to the pro-inflammatory E. coli LPS used as a positive control (10 and 100 ng, ***p < 0.001, Fig. 7A). In parallel, no significant activation compared to untreated cells was observed in HEK-Blue™ hTLR2, indicating that the O. splanchnicus LOS is unable to activate TLR2 signaling (Fig. 7B).O. splanchnicus LOS impact on enterocytes
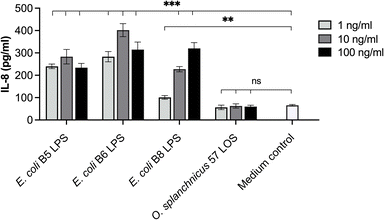
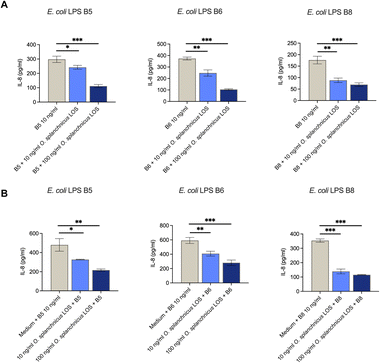
The O. splanchnicus LOS was tested in the enterocyte cell line HT-29 and clearly showed no induction of IL-8 release as the level of the proinflammatory cytokine in the supernatant was similar to the cell medium serving as the background control (Fig. 8).In accordance, the amount of IL-8 released was also significantly lower than IL-8 levels induced by the corresponding concentrations of the different E. coli LPSs used (B5, B6, and B8). In addition, we also evaluated whether the O. splanchnicus LOS was able to compete with proinflammatory E. coli LPSs in the HT-29 cell model. Interestingly, we observed that this peculiar LOS could compete with all three inflammatory E. coli LPSs with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, and significantly decreased IL-8 release compared to cells only induced by E. coli LPSs (Fig. 9A). The effect was even more profound when the ratio of O. splanchnicus LOSs was 10 times higher than proinflammatory LPSs. Finally, in a parallel experiment we first primed HT-29 cells with the O. splanchnicus LOS and, only after its removal, cells were treated with the E. coli LPS. Also in this case, we observed a decrease in the IL-8 release as compared to the control, i.e. attenuation of the E. coli LPS effect (ratios of 1
10, and significantly decreased IL-8 release compared to cells only induced by E. coli LPSs (Fig. 9A). The effect was even more profound when the ratio of O. splanchnicus LOSs was 10 times higher than proinflammatory LPSs. Finally, in a parallel experiment we first primed HT-29 cells with the O. splanchnicus LOS and, only after its removal, cells were treated with the E. coli LPS. Also in this case, we observed a decrease in the IL-8 release as compared to the control, i.e. attenuation of the E. coli LPS effect (ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Fig. 9B). These observations are extremely interesting considering that O. splanchnicus cells and OMVs exert anti-inflammatory effects by attenuating E. coli LPS-induced IL-8 release from enterocytes, although the responsible effector molecules were not identified thus far.4 The present study now points to the LOS of O. splanchnicus as a key mediator of this effect and establishes a link between its anti-inflammatory properties and distinct structural features of both the lipid A and core oligosaccharide domains.
1, Fig. 9B). These observations are extremely interesting considering that O. splanchnicus cells and OMVs exert anti-inflammatory effects by attenuating E. coli LPS-induced IL-8 release from enterocytes, although the responsible effector molecules were not identified thus far.4 The present study now points to the LOS of O. splanchnicus as a key mediator of this effect and establishes a link between its anti-inflammatory properties and distinct structural features of both the lipid A and core oligosaccharide domains.
Conclusions
The use of commensal gut bacteria as “next-generation probiotics” may offer greater benefits compared to traditional probiotic formulations. These benefits may include improved retention in the gastrointestinal environment, enhanced adhesion to intestinal epithelial tissues, potent immunomodulatory effects, and the production of unique beneficial metabolites not typically associated with conventional probiotics.45 Among the gut commensals that meet these criteria is O. splanchnicus, with potential applications in the prevention or management of metabolic and inflammatory diseases.46,47 Indeed, although many aspects of its metabolic pathways and general biological functions remain largely unknown, evidence suggests that elevated levels of O. splanchnicus are positively associated with a healthy gastrointestinal status.9 The results of this study highlight the O. splanchnicus LOS as a structurally and functionally distinct bacterial glycolipid with important implications for host–microbe interactions. Our combined structural, biophysical, and functional analyses reveal a lipid A architecture consistent with the broader Bacteroidota paradigm of hypo-acylated, mono-phosphorylated species with intrinsically reduced proinflammatory potential, in contrast to the canonical hexa-acylated lipid A of enterobacteria. The core OS further introduces rare and immunologically relevant motifs, including GalNA and the unprecedented incorporation of D-Man4Lac in the Bacteroidota phylum, both of which are plausibly linked to unique lectin recognition and to immunomodulation. The observed structural heterogeneity, together with selective binding to innate immune lectins, such as galectin-4, ficolins, and collectin-11, underscore the potential of this LOS to engage host immunity through non-canonical pathways.13 Importantly, the supramolecular organization of the O. splanchnicus LOS into micelles with an extended oligosaccharide shell provides a mechanistic explanation for its weak TLR4 activation and its antagonism of proinflammatory E. coli LPS signaling. These features converge to create a low-inflammatory phenotype that facilitates immune tolerance, epithelial integrity, and the commensal persistence of O. splanchnicus. Although the exact micellar architecture may not be preserved in the complex milieu of the gut, the SAXS data capture the intrinsic self-assembly propensity of this LOS and reveal a structural arrangement in which the oligosaccharide domain effectively limits exposure of the lipid A region. This inherent steric attenuation of lipid A accessibility, together with its hypo-acylated and hypo-phosphorylated nature, are likely the main factors underlying its reduced TLR4 recognition in vivo.By linking distinctive glycan motifs and aggregation states to biological outcomes, this work establishes the LOS of O. splanchnicus as a key determinant of its beneficial immunomodulatory effects, reinforcing the potential of this species as a next-generation probiotic candidate.
Experimental
O. splanchnicus isolation
O. splanchnicus isolate 57 was isolated, purified and identified from the feces of an FMT (fecal microbiota transplant) donor in our previous study.5 The donor provided written informed consent, and the use of the sample was approved by the Ethics Committee of Hospital District of Helsinki and Uusimaa Finland (DnroHUS124/13/03/01/11). The O. splanchnicus isolate was cultivated on Gifu anaerobic medium (GAM; Nissui Pharmaceutical Co., Ltd, Japan) for 48 h under anaerobic conditions at 37 °C in an anaerobic chamber (Whitley 85A anaerobic workstation).36 The isolate was genome sequenced using the MiSeq (Illumina, Inc., San Diego, CA, United States; BioProject PRJNA575760). The bacterium was preserved as frozen stocks in skim milk tubes at −80 °C. For the LOS extraction, O. splanchnicus was propagated from a frozen stock on a GAM agar plate and grown for 48 h anaerobically. After confirming the purity of the isolate, 2 L culture of O. splanchnicus isolate 57 was grown in GAM broth for 48 h followed by pelleting the cells (4700 rpm, 10 min).Isolation and purification of the LOS from O. splanchnicus
The dried bacterial cells (1 g) were extracted with the hot phenol-water protocol.48 The obtained aqueous and phenolic phases were further dialyzed and subjected to enzymatic digestion with DNase (Sigma-Aldrich, Darmstadt, Germany) and RNase (Sigma-Aldrich, Darmstadt, Germany) followed by proteinase K (Sigma-Aldrich, Darmstadt, Germany) (37 and 56 °C). The purity and nature of the extracted material (111 mg) was checked via SDS-PAGE stained with silver nitrate gel (Fig. S1).49Chemical analyses
To determine the monosaccharide composition, the acetylated O-methyl glycoside derivative method was used, and the treatment starts with HCl/MeOH (1.25 M, 358 K, 12 h) followed by acetylation with acetic anhydride in pyridine (358 K, 30 min).50 To determine the absolute configuration of the sugars, O-octylglycoside derivatives were prepared and analyzed as previously described.51 The sugar linkage pattern was determined with an aliquot of LOS suspended in DMSO in the presence of NaOH, stirred at room temperature for 1.5 h. Then, the methylation step was achieved with CH3I, followed by hydrolysis with trifluoroacetic acid (4 M, 373 K, 4 h), a reduction of the carbonyl with NaBD4 and acetylation with pyridine and acetic anhydride. In parallel, in order to analyze the fatty acids present in the sample, another LOS aliquot underwent the previously described methanolysis step and then the mixture was extracted three times with hexane. Hexane which contained the fatty acids was analyzed via GC-MS. All chemical analyses were carried out on a gas-chromatograph Agilent Technologies 6850A (Santa Clara, CA, USA) equipped with a mass selective detector 5973N and a Zebron ZB-5 capillary column (Phenomenex, 30 m × 0.25 mm i.d., 0.25 µm film thickness, flow rate 1 mL min−1, he as the carrier gas). The temperature program 413 K for 5 min, 413 K → 553 K at 10 °C min−1, and 553 K for 10 min was used for lipid analysis, and for sugar analysis the temperature program was 423 K for 5 min, 423 K → 553 K at 3 °C min−1, and 553 K for 5 min.Isolation of the OSdeAc, OS and lipid A fractions
An aliquot of LOS (30 mg) was subjected to O-deacylation with anhydrous hydrazine (1 mL, at 310 K for 90 min), and the suspension created was cooled and transferred into ice-cold acetone to allow precipitation. The precipitate was centrifuged (277 K, 5000×g, 30 min) and washed again with ice-cold acetone, dried, dissolved in water and then freeze-dried. The O-deacylated product obtained underwent N-deacylation with 4 M KOH (393 K, 16 h) resulting in the OSdeAc product. The salts resulting from the reactions were eliminated using gel-filtration chromatography on a Sephadex G-10 column (Pharmacia, 50 × 1.5 cm). In parallel, another aliquot (9 mg) was treated with acetic acid at 1% (373 K, 2 h) in order to separate the lipid A from the core OS portion. The solution was extracted three times with a mixture of CHCl3/MeOH/H20 (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v/v) and then centrifuged (277 K, 7000×g, 15 min). The organic phase contained lipid A and was washed three times with distilled water and freeze-dried to obtain the isolated lipid A product. The aqueous phase contained the product and was also collected and freeze-dried for further analysis. At this point, first, gel filtration chromatography (TSK40) and then high-performance gel permeation chromatography (HPGPC) on a TSKgel G3000 PWXL column were performed.
30, v/v/v) and then centrifuged (277 K, 7000×g, 15 min). The organic phase contained lipid A and was washed three times with distilled water and freeze-dried to obtain the isolated lipid A product. The aqueous phase contained the product and was also collected and freeze-dried for further analysis. At this point, first, gel filtration chromatography (TSK40) and then high-performance gel permeation chromatography (HPGPC) on a TSKgel G3000 PWXL column were performed.
NMR spectroscopy
To assign the structure of the oligosaccharide, the NMR spectra were recorded in D2O at 298 K at pH 7 with a Bruker 600 AVANCE NEO equipped with a cryoprobe. Total correlation spectroscopy (TOCSY) experiments were performed with spinlock times of 100 ms and a data set (t1 × t2) of 4096 × 512 points. The Rotating-frame Overhauser enhancement spectroscopy (ROESY) and nuclear Overhauser enhancement spectroscopy (NOESY) measurements were performed with data sets (t1 × t2) of 4096 × 512 points and mixing times ranging from 100 to 400 ms. Double-quantum-filtered phase-sensitive correlation spectroscopy (DQF-COSY) experiments were conducted using data sets of 4096 × 912 points. To give a 4 K × 2 K point matrix, the data matrix of all homonuclear experiments was filled with zeros in both dimensions. Afterwards, the resolution enhancement was used in the dimensions via a cosinebell function prior to Fourier transformation. To obtain the coupling constant, 2D phase-sensitive DQF-COSY measurements were used. Heteronuclear single-quantum coherence (HSQC) and heteronuclear multiple–bond correlation (HMBC) experiments were carried out in the 1H-detection mode using single-quantum coherence with proton decoupling in the 13C domain using data sets of 2048 × 400 points. Phase-sensitive mode and sensitivity improvements were applied for HSQC, along with echo/antiecho gradient selection and multiplicity editing in the selection step. In the HMBC experiments, an optimization was applied for long-range coupling constants, incorporating a low-pass J filter to eliminate the one-bod correlations, via gradient pulses for selection. Furthermore, a 60 ms delay was used for the evolution of long-range correlations, with an optimization for 6–15 Hz coupling constants. To improve resolution, the data matrix in the heteronuclear experiment was extended to 2048 × 1024 points with forward linear prediction extrapolation.ESI MS and MS/MS analyses
OSdeAc was dissolved in a solution composed of 50% (v/v) methanol and 50% water and directly infused by using a syringe pump (10 µL min−1, at an estimated concentration of 10 µg mL−1) into the source of a Waters Synapt XS mass spectrometer, equipped with an 8 kDa quadrupole with an electrospray ionization (ESI) source. The analysis was conducted with a capillary potential of −2.00 kV, source temperature of 120 °C, sampling cone at 20.0 V, source offset at 0 V, source gas (N2) flow of 0.0 mL min−1, desolvation temperature of 400 °C, cone gas flow of 20 L h−1, desolvation gas flow of 150 L h−1, and nebulizer gas pressure of 2.5 bar. An aliquot of LOS was dissolved in 400 µL of a 1 M aqueous solution of ammonia (NH3). The solution was stirred overnight at 37 °C. Then, the solution was dried under air flow, followed by freeze-drying to obtain the O-deacylated LOS. This product was dissolved in 50% (v/v) 2-propanol and 50% water and directly infused by using a syringe pump (5 µL min−1, at an estimated concentration of 20 µg mL−1). The analysis was carried out with a capillary potential of −1.9 kV, source temperature of 120 °C, sampling cone at 40.0 V, source offset at 10 V, source gas (N2) flow of 0.0 mL min−1, desolvation temperature of 400 °C, cone gas flow of 60 L h−1, desolvation gas flow of 300 L h−1, and nebulizer gas pressure of 2.5 bar. All the analyses were performed in negative polarity and resolution mode with a scan range between m/z 50 and m/z 2000 and a scan time of 1 s. Nitrogen was supplied by a PLINIUS N45-1 nitrogen generator from CLAIND and was used as desolvation and cone gas. MS/MS experiments were performed using ultra-pure argon (SOL SpA) as the collision gas in the trap cell, with a collision energy ramp ranging from 20 to 90 eV. A 2 µg µL−1 solution of NaI in (v/v) 2-propanol/H2O 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 was used for the calibration from 50 to 4000 Da. Data acquisition and MS and MS/MS spectral images were generated by MassLynx™ software (V4.2).52 A reference solution consisting of 100 pg µL−1 Leucine Enkephalin dissolved in a 1
50 was used for the calibration from 50 to 4000 Da. Data acquisition and MS and MS/MS spectral images were generated by MassLynx™ software (V4.2).52 A reference solution consisting of 100 pg µL−1 Leucine Enkephalin dissolved in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (vol/vol) mixture of water/acetonitrile with 0.1% of formic acid was used as lockmass during sample acquisition with a lockspray capillary of −2.5 kV and a scan time of 0.5 s.
1 (vol/vol) mixture of water/acetonitrile with 0.1% of formic acid was used as lockmass during sample acquisition with a lockspray capillary of −2.5 kV and a scan time of 0.5 s.
MALDI-TOF MS analysis
MALDI-TOF MS spectra were obtained using an ABSCIEX TOF/TOFTM 5800 mass spectrometer from Applied Biosystems, equipped with an Nd:YAG laser (wavelength of 349 nm), featuring a pulse width of 3 ns and a maximum repetition rate of 1000 Hz. The lipid A fraction was dissolved in CHCl3/CH3OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) while the matrix was 2,4,6-trihydroxyacetophenone in CH3OH/0.1% trifluoroacetic acid/CH3CN (7
1, v/v) while the matrix was 2,4,6-trihydroxyacetophenone in CH3OH/0.1% trifluoroacetic acid/CH3CN (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2:1, v/v) at a concentration of 75 mg mL−1.53,54 0.5 µL of both the sample and the matrix were deposited on the MALDI plate and allowed to dry at ambient temperature. Each spectrum represented an accumulation of 2000 laser shots, while the MS/MS spectra involved the summation of between 5000 and 7000 shots. Acquisition was performed ensuring random yet uniform sampling across the sample spot.
2:1, v/v) at a concentration of 75 mg mL−1.53,54 0.5 µL of both the sample and the matrix were deposited on the MALDI plate and allowed to dry at ambient temperature. Each spectrum represented an accumulation of 2000 laser shots, while the MS/MS spectra involved the summation of between 5000 and 7000 shots. Acquisition was performed ensuring random yet uniform sampling across the sample spot.
HEK cell culture and stimulation assays
HEK-Blue hTLR4 and HEK-Blue hTLR2 were provided by Invivogen (Invivogen, Toulouse, France) and cultured in Dulbecco's Modified Eagle's Medium (DMEM) 4 g L−1 glucose supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin/streptomycin (pen/strep), 2 mM L-glutamine and 100 µg mL−1 Normocin. To ensure plasmid selection, HEK-Blue selection was added to HEK-Blue cell culture medium according to the manufacturer's instructions. Cell lines were maintained at 37 °C in a humidified atmosphere containing 5% CO2. HEK-Blue hTLR4 and HEK-Blue hTLR2 were seeded into a 96-well plate (3 × 104 cells per well). After an overnight attachment, HEK-Blue cells were stimulated for 18 hours with various concentrations of E. coli O111:B4 LPS (Invivogen, Toulouse, France) or O. splanchnicus LOS. NF-kB activation was measured by Quanti blue assay (Invivogen, Toulouse, France). HEK-Blue hTLR2 cells were also treated with 500 ng mL−1 Pam3CSK4 (Invivogen, Toulouse, France), a positive control for TLR2 activation. The secreted embryonic alkaline phosphatase (SEAP) levels were evaluated in the supernatants to establish NF-kB activation following LPS stimulation by Quanti-Blue assay. For this assay, the absorbance at 620 nm was measured by using a TECAN Infinite M Plex spectrophotometer (Tecan, Grödig, Austria).HT-29 cell line
The human colonic epithelial cell line HT-29 (ACC 299) was purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ). The cell line was grown under an oxic atmosphere with 5% CO2 at 37 °C. HT-29 cells were cultivated in McCoy 5A (Biowest) medium containing 10% heat-inactivated fetal bovine serum (FBS; Gibco) and 100 U mL−1 penicillin/streptomycin solution (PEST). The cells were passaged after reaching 80% confluence (every 3–4 days) using TryplExpress (Gibco) to detach the cells. Passages 6–20 were used in the experiments. For the immunological assays, 12.500 HT-29 cells per well were seeded onto a 96-well microplate and grown for 8 days post-plating under an oxic atmosphere with 5% CO2 at 37 °C. The medium in the wells was changed every 3–4 days and one day before the experiment. The confluence of HT-29 cells in each well was checked before the assay. Three different commercial E. coli LPSs were purchased from Merck. LPSs from strains DSM4779 O55:K59(B5), ATCC 12795 O26(B6) and O127:(B8) were used in the experiments.IL-8 release from HT-29: induction, competition & attenuation
The capacity of the O. splanchnicus 57 LOS to induce IL-8 release from HT-29 cells was measured as previously described.5 Three different commercial E. coli LPSs were used in comparison. In the induction assay, 1 ng mL−1, 10 ng mL−1 and 100 ng mL−1 of LOS were suspended in McCoy 5A medium, added onto 8-day-old HT-29 cells and incubated at 37 °C for 3 h under an oxic atmosphere with 5% CO2. The McCoy 5A medium without LPSs was used as the background control. Supernatants were collected and IL-8 levels were measured by using an ELISA assay (BD OptEIA Set).In the competition assay, 10 ng mL−1 of proinflammatory E. coli LPS was used in the HT-29 cell experiments. Briefly, the O. splanchnicus LOS was mixed with different ratios 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (10 ng mL−1E. coli LPS + 10 ng mL−1O. splanchnicus 57 LOS) or 1
1 (10 ng mL−1E. coli LPS + 10 ng mL−1O. splanchnicus 57 LOS) or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (10 ng mL−1E. coli LPS + 100 ng mL−1O. splanchnicus 57 LOS) and incubated on an 8-day old, fully confluent HT-29 monolayer for 3 h before collecting the supernatants for ELISA IL-8 measurement. E. coli LPS 10 ng mL−1 was used as the baseline control.
10 (10 ng mL−1E. coli LPS + 100 ng mL−1O. splanchnicus 57 LOS) and incubated on an 8-day old, fully confluent HT-29 monolayer for 3 h before collecting the supernatants for ELISA IL-8 measurement. E. coli LPS 10 ng mL−1 was used as the baseline control.
In the attenuation assay, HT-29 cells were first incubated with the O. splanchnicus LOS for 1 h with a concentration of either 10 ng mL−1 or 100 ng mL−1. After removing the O. splanchnicus LOS from the monolayer, the proinflammatory E. coli LPS (10 ng mL−1) was added onto the cells for 4 h followed by collection of the supernatants and IL-8 measurements with ELISA.
Statistical analysis
Significant differences between two groups, for example the sample and control, were calculated using an unpaired t-test. Homoscedasticity testing was carried out with Levene's test to identify equal or unequal variances. All statistical analyses were calculated with GraphPad Prism 10.5.0 (GraphPad Software, United States). A p-value of <0.05 was considered statistically significant.Small Angle X-ray scattering (SAXS)
Small-angle X-ray scattering (SAXS) was employed to probe the self-assembly behavior of lipooligosaccharide (LOS) in an aqueous suspension. Measurements were conducted on the Diamond Light Source B21 beamline (Didcot, UK), operating at an X-ray energy of 13.018 keV and a sample-to-detector distance of 3.7 m. Using this configuration, it was possible to collect data for the scattering vector modulus Q = 4πsin(θ/2) λ between 0.0045 Å−1 and 0.34 Å−1, where θ is the scattering angle.55–57The SAXS experimental data in the present work were fitted with the SASView v 5.0.6 Software (https://www.sasview.org/) using a core–shell sphere model:
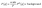 | (1) |
 | (2) |
Microarray binding assays
The O. splanchnicus LOS and control (glyco)proteins (asialofetuin, ribonuclease B, and ribonuclease A) were printed as duplicates at four different concentrations (from 1 to 0.03 mg mL−1) on 16-pad nitrocellulose-coated glass slides (Grace Biolabs ONCYTE NOVA) using a non-contact arrayer (Sprint, Arrayjet Ltd), as described.41 The Cy3 fluorophore (GE Healthcare) was added at 1 µg mL−1 to all the solutions to enable postarray monitoring of the spots58 by using scanning fluorescence signals upon excitation at 532 nm (green laser), using a GenePix 4200-AL scanner (Axon, Molecular Devices). For binding assays, the microarrays were first blocked for 1 h with 0.25% (v/v) Tween-20 in 5 mM sodium phosphate, pH 7.2, 0.2 M NaCl (PBS) or in 10 mM Tris pH 8, 150 mM NaCl (TBS). Then, the arrays were overlaid for 1.5 h with biotin-labelled lectins at 10 µg mL−1 in an appropriate buffer containing 0.1% (v/v) Tween-20. A panel of 40 lectins was assayed (see Table S2 for details on lectins tested, source, and buffer used in each case). Similarly, for testing the binding of human innate immune lectins, the arrays were overlaid with the lectins at 20 µg mL−1 (tags, source of the lectins tested, and buffers used are listed in Table S3). After incubation and subsequent washing steps, pads were incubated for 1 h with either biotinylated rabbit anti-human galectin 1 (Peprotech, working dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) or mouse anti-His antibody pre-complexed with biotinylated goat anti-mouse IgG antibody (Sigma, final working dilutions 1
1000) or mouse anti-His antibody pre-complexed with biotinylated goat anti-mouse IgG antibody (Sigma, final working dilutions 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 and 3
1000 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, respectively). The slides were washed 4 times with PBS and binding was detected by incubating with AlexaFluor-647 (AF647)-labelled streptavidin (invitrogen) at 1 µg mL−1 in PBS or TBS, 0.1% (v/v) Tween-20, for 35 min in the dark, washed again thoroughly with PBS, then with water and finally scanned for AF647 signals (excitation at 635 nm, red laser). Fluorescence intensities were quantified with the GenePix Pro 7 software.
1000, respectively). The slides were washed 4 times with PBS and binding was detected by incubating with AlexaFluor-647 (AF647)-labelled streptavidin (invitrogen) at 1 µg mL−1 in PBS or TBS, 0.1% (v/v) Tween-20, for 35 min in the dark, washed again thoroughly with PBS, then with water and finally scanned for AF647 signals (excitation at 635 nm, red laser). Fluorescence intensities were quantified with the GenePix Pro 7 software.
Author contributions
A. S. conceived the study; M. T. M. executed the LOS isolation purification, and characterization; F. D. L and M. M executed the MRM/MS studies; R. S. and K. H. provided the bacterial strain and studied the impact on enterocytes; L. D. S. C performed HEK cell assays; M. A. C. R. and D. S. executed the microarray binding essays; A. C. and L. P. executed small angle X-ray scattering; funding and resource acquisition: A. S., F. D. L., A. M. and R. S; writing – original draft preparation: M. T. M., M. M., and K. H.; writing – review and editing: A. S., F. D. L., A. M., D. S. and R. S. All authors have read and have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: SI reports Fig. S1–S3 with SDS-PAGE, NMR and MS data, as well as Tables S1–S3 with NMR and microarray data. See DOI: https://doi.org/10.1039/d5sc08335d.Acknowledgements
The authors wish to thank the Diamond Light Source for providing beamtime (reference SM34244-1). This work was supported by the European Research Council (ERC) under the Horizon Europe program under grant agreement no. 101039841 (DEBUGGING LPS) to F. D. L. This work has also received funding from the European Union's Horizon Europe research and innovation programme under grant agreement no. 101095084 for the ENDOTARGET project, which is also supported by the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract number 22.00462. It was also supported by the project “CLariFY” and “GIVING” funded by the Italian Ministry of University and Research, PRIN MUR 2022 (2022SHW3KY) and PRIN MUR PNRR 2022 (P202293ZMC) to F. D. L. MAC-R and DS were funded by grant PID2022-143249OB-I00 of MCIN/AEI/10.13039/501100011033 and “ERDF A way of making Europe”, and by CIBER (Consorcio Centro de Investigación Biomédica en Red (CIBERES), Instituto de Salud Carlos III), CB06/06/0037. This work was also supported in part by the Italian Ministry of Foreign Affairs and International Cooperation (Italy Germany Science and Technology cooperation–Call for joint research proposals for the years 2023–2025). This research was funded under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3—Call for tender no. 341 of 15 March 2022 of Italian Ministry of University and Research funded by the European Union—NextGenerationEU. Project code PE00000003, Concession Decree no. 1550 of 11 October 2022, adopted by the Italian Ministry of University and Research, CUP E63C22002030007, Project title “ON Foods—Research and innovation network on food and nutrition Sustainability, Safety and Security—Working ON Foods”. H2020-MSCA-ITN-2020, contract no. 956758 (AS, MTM). Ministry of Education, Universities and Research, PRIN MUR 2022 (2022ZEZS45) and Ministry of Education, Universities and Research, PRIN MUR PNRR 2022 (P2022M457Z) (AS). PNRR, Missione 4 – Componente 2 – NextGenerationEU – Partenariato Esteso INF-ACT – One Health Basic and Translational Research Actions Addressing Unmet Needs on Emerging Infectious Diseases MUR: PE00000007 (AS).References
- I. Schoultz, M. J. Claesson, M. G. Dominguez-Bello, F. Fåk Hållenius, P. Konturek, K. Korpela, M. F. Laursen, J. Penders, H. Roager, T. Vatanen, L. Öhman and M. C. Jenmalm, J. Intern. Med., 2025, 297, 560–583 CrossRef CAS PubMed.
- M. Göker, S. Gronow, A. Zeytun, M. Nolan, S. Lucas, A. Lapidus, N. Hammon, S. Deshpande, J. F. Cheng, S. Pitluck, K. Liolios, I. Pagani, N. Ivanova, K. Mavromatis, G. Ovchinikova, A. Pati, R. Tapia, C. Han, L. Goodwin, A. Chen, K. Palaniappan, M. Land, L. Hauser, C. D. Jeffries, E. M. Brambilla, M. Rohde, J. C. Detter, T. Woyke, J. Bristow, V. Markowitz, P. Hugenholtz, J. A. Eisen, N. C. Kyrpides and H. P. Klenk, Stand. Genomic Sci., 2011, 4, 200–209 CrossRef.
- J. Li, J. Xu, X. Guo, H. Xu, C. Huang, Y. Nie and Y. Zhou, Microorganisms, 2025, 13, 815 CrossRef CAS.
- J. D. Lewis, E. Z. Chen, R. N. Baldassano, A. R. Otley, A. M. Griffiths, D. Lee, K. Bittinger, A. Bailey, E. S. Friedman, C. Hoffmann, L. Albenberg, R. Sinha, C. Compher, E. Gilroy, L. Nessel, A. Grant, C. Chehoud, H. Li, G. D. Wu and F. D. Bushman, Cell Host Microbe, 2015, 18, 489–500 CrossRef CAS.
- K. Hiippala, G. Barreto, C. Burrello, A. Diaz-Basabe, M. Suutarinen, V. Kainulainen, J. R. Bowers, D. Lemmer, D. M. Engelthaler, K. K. Eklund, F. Facciotti and R. Satokari, Front. Microbiol., 2020, 11, 575455 CrossRef.
- J. Zhuang, Z. Zhuang, B. Chen, Y. Yang, H. Chen and G. Guan, J. Mol. Med., 2025, 31, 1–14 Search PubMed.
- D. G. Burke, F. Fouhy, M. J. Harrison, M. C. Rea, P. D. Cotter, O. O'Sullivan, C. Stanton, C. Hill, F. Shanahan, B. J. Plant and R. P. Ross, BMC Microbiol., 2017, 17, 1–11 CrossRef.
- Q. Chen, Y. Fan, B. Zhang, C. Yan, Q. Zhang, Y. Ke, Z. Chen, L. Wang, H. Shi, Y. Hu, Q. Huang, J. Su, C. Xie, X. Zhang, L. Zhou, J. Ren and H. Xu, Microbiol. Spectr., 2023, 11, e04152 Search PubMed.
- S. F. Lima, L. Gogokhia, M. Viladomiu, L. Chou, G. Putzel, W. B. Jin, S. Pires, C. J. Guo, Y. Gerardin, C. V. Crawford, V. Jacob, E. Scherl, S. E. Brown, J. Hambor and R. S. Longman, Gastroenterology, 2022, 162, 166–178 CrossRef CAS PubMed.
- B. S. Oh, W. J. Choi, J. S. Kim, S. W. Ryu, S. Y. Yu, J. S. Lee, S. H. Park, S. W. Kang, J. Lee, W. Y. Jung, Y. M. Kim, J. H. Jeong and J. H. Lee, Front. Microbiol., 2021, 12, 736343 CrossRef PubMed.
- B. Bosch, S. Moutaharrik, A. Gazzaniga, K. Hiippala, H. A. Santos, A. Maroni and R. Satokari, Eur. J. Pharm. Biopharm., 2023, 190, 73–80 CrossRef CAS.
- F. Di Lorenzo, K. A. Duda, R. Lanzetta, A. Silipo, C. De Castro and A. Molinaro, Chem. Rev., 2021, 122, 15767–15821 CrossRef PubMed.
- S. De Chiara, L. De Simone Carone, R. Cirella, E. Andretta, A. Silipo, A. Molinaro, M. Mercogliano and F. Di Lorenzo, ChemMedChem, 2025, 20, e202400780 CrossRef CAS PubMed.
- B. S. Park and J. O. Lee, Exp. Mol. Med., 2013, 45, e66 CrossRef.
- F. Bäckhed, S. Normark, E. K. H. Schweda, S. Oscarson and A. Richter-Dahlfors, Microb. Infect., 2003, 5, 1057–1063 CrossRef.
- G. I. Birnbaum, R. Rene, J. R. Brisson and H. J. Jennings, J. Carbohydr. Chem., 1987, 6, 17–39 CrossRef CAS.
- R. Marchetti, R. E. Forgione, F. N. Fabregat, C. Di Carluccio, A. Molinaro and A. Silipo, Curr. Opin. Struct. Biol., 2021, 68, 74–83 CrossRef CAS PubMed.
- Y. A. Knirel, O. V. Bystrova, N. A. Kocharova, U. Zähringer and G. B. Pier, J. Endotoxin Res., 2006, 12, 324–336 CAS.
- F. Sisti, J. Fernández, A. Cordero, A. Casabuono, A. Couto and D. Hozbor, Bioorg. Med. Chem. Lett., 2017, 27, 432–436 CrossRef CAS PubMed.
- D. Garozzo, G. Impallomeni, E. Spina, L. Sturiale, A. Cesàro and P. Cescutti, Carbohydr. Res., 1995, 270, 97–106 CrossRef CAS.
- N. K. Kochetkov, A. F. Sviridov, K. A. Arifkhodzhaev, O. S. Chizhov and A. S. Shashkov, Carbohydr. Res., 1979, 71, 193–203 CrossRef CAS.
- E. N. Sigida, I. M. Ibrahim, M. S. Kokoulin, H. H. Abulreesh, K. Elbanna, S. A. Konnova and Y. P. Fedonenko, Mar. Drugs, 2021, 19, 508 CrossRef CAS PubMed.
- R. F. Helm, Z. Huang, D. Edwards, H. Leeson, W. Peery and M. Potts, J. Bacteriol., 2000, 182, 974–982 CrossRef CAS.
- H. Li, S. Liu, Y. Liu, W. Li, A. Niu, P. Ren, Y. Liu, C. Jiang, M. Inam and L. Guan, Carbohydr. Polym., 2022, 281, 119055 CrossRef CAS.
- Y. Yang, P. Ren, Y. Sun, J. Li, X. Zhou, H. Zhang, C. He, H. Dai and L. Guan, Int. J. Biol. Macromol., 2024, 283, 137435 CrossRef PubMed.
- M. Dhanalakshmi, D. Sruthi, K. R. Jinuraj, K. Das, S. Dave, N. M. Andal and J. Das, Med. Chem. Res., 2023, 32, 391–408 CrossRef CAS PubMed.
- Y. Du, Y. Fan, X. Li and F. Chen, npj Biofilms Microbiomes, 2025, 11, 1–22 CrossRef.
- M. Tiemblo Martín, M. Coccimiglio, E. Andretta, L. De Simone Carone, A. Bell, T. Guerpe Amor, C. Di Carluccio, A. Molinaro, Y. van Kooyk, N. Juge, F. Chiodo, F. Di Lorenzo and A. Silipo, Carbohydr. Polym., 2025, 348, 122833 CrossRef.
- M. D. Pither, A. Illiano, C. Pagliuca, A. Jacobson, G. Mantova, A. Stornaiuolo, R. Colicchio, M. Vitiello, G. Pinto, A. Silipo, M. A. Fischbach, P. Salvatore, A. Amoresano, A. Molinaro and F. Di Lorenzo, Carbohydr. Polym., 2022, 297, 120040 CrossRef CAS PubMed.
- F. Di Lorenzo, M. D. Pither, M. Martufi, I. Scarinci, J. Guzmán-Caldentey, E. Łakomiec, W. Jachymek, S. C. M. Bruijns, S. M. Santamaría, J. S. Frick, Y. Van Kooyk, F. Chiodo, A. Silipo, M. L. Bernardini and A. Molinaro, ACS Cent. Sci., 2020, 6, 1602–1616 CrossRef CAS.
- M. Hashimoto, Y. Asai, R. Tamai, T. Jinno, K. Umatani and T. Ogawa, FEBS Lett., 2003, 543, 98–102 CrossRef CAS.
- T. Vatanen, A. D. Kostic, E. D'Hennezel, H. Siljander, E. A. Franzosa, M. Yassour, R. Kolde, H. Vlamakis, T. D. Arthur, A. M. Hämäläinen, A. Peet, V. Tillmann, R. Uibo, S. Mokurov, N. Dorshakova, J. Ilonen, S. M. Virtanen, S. J. Szabo, J. A. Porter, H. Lähdesmäki, C. Huttenhower, D. Gevers, T. W. Cullen, M. Knip and R. J. Xavier, Cell, 2016, 165, 842–853 CrossRef CAS.
- S. Elizabeth, PhD thesis, University of North Carolina at Chapel Hill, 2013, pp. 1965–1985.
- A. Weintraub, U. Zähringer, H. -W Wollenweber, U. Seydel and E. T. Rietschel, Eur. J. Biochem., 1989, 183, 425–431 CrossRef CAS PubMed.
- F. Di Lorenzo, A. Silipo, T. Matier, A. Hanuszkiewicz, J. S. Elborn, R. Lanzetta, L. Sturiale, A. Scamporrino, D. Garozzo, M. A. Valvano, M. M. Tunney and A. Molinaro, Eur. J. Org Chem., 2016, 1732–1738 CrossRef CAS.
- K. Hiippala, V. Kainulainen, M. Suutarinen, T. Heini, J. R. Bowers, D. Jasso-Selles, D. Lemmer, M. Valentine, R. Barnes, D. M. Engelthaler and R. Satokari, Nutrients, 2020, 12, 935 CrossRef CAS PubMed.
- J. Pazol, T. M. Weiss, C. D. Martínez, O. Quesada and E. Nicolau, JCIS Open, 2022, 7, 100058 CrossRef PubMed.
- M. Vaccaro, A. Accardo, D. Tesauro, G. Mangiapia, D. Löf, K. Schillén, O. Söderman, G. Morelli and L. Paduano, Langmuir, 2006, 22, 6635–6643 CrossRef CAS.
- A. E. Mohr, M. Crawford, P. Jasbi, S. Fessler and K. L. Sweazea, FEBS Lett., 2022, 596, 849–875 CrossRef CAS PubMed.
- L. Hong, M. Gontsarik, H. Amenitsch and S. Salentinig, Small, 2022, 18, 2104211 CrossRef CAS.
- M. A. Campanero-Rhodes, I. Kalograiaki, B. Euba, E. Llobet, A. Ardá, J. Jiménez-Barbero, J. Garmendia and D. Solís, Biomolecules, 2021, 11, 595 CrossRef CAS.
- F. Nieto-Fabregat, M. Mercogliano, A. Cangiano, G. Vitiello, E. Andretta, L. A. Clifton, A. Vanacore, L. Buono, M. A. Campanero-Rhodes, D. Solís, C. Di Carluccio, G. Pecoraro, A. Molinaro, G. Smaldone, J. K. Kim, D. H. Kim, L. Paduano, F. Di Lorenzo and A. Silipo, JACS Au, 2025, 5, 3311–3327 CrossRef PubMed.
- M. L. Henriksen, J. Brandt, S. S. C. Iyer, N. M. Thielens and S. Hansen, Mol. Immunol., 2013, 56, 757–767 CrossRef CAS PubMed.
- Y. Zheng, J. Su, M. C. Miller, J. Geng, X. Xu, T. Zhang, M. Mayzel, Y. Zhou, K. H. Mayo and G. Tai, Glycobiology, 2021, 31, 341–350 CrossRef CAS PubMed.
- M. E. Abouelela and Y. A. Helmy, Microorganisms, 2024, 12, 430 CrossRef CAS PubMed.
- N. J. Foegeding and M. X. Byndloss, Cell Host Microbe, 2021, 29, 851–853 CrossRef CAS PubMed.
- C. Xing, M. Wang, A. A. Ajibade, P. Tan, C. Fu, L. Chen, M. Zhu, Z. Z. Hao, J. Chu, X. Yu, B. Yin, J. Zhu, W. J. Shen, T. Duan, H. Y. Wang and R. F. Wang, Cell Host Microbe, 2021, 29, 959–974 CrossRef CAS.
- O. Westphal, O. Lüderitz, F. Bister and B. Naturforsch, J. Chem. Sci., 1965, 7, 148–155 Search PubMed.
- M. D. Pither, A. Silipo, A. Molinaro and F. Di Lorenzo, Methods Mol. Biol., 2023, 2613, 153–179 CrossRef CAS.
- F. D. Lorenzo, L. Sturiale, A. Palmigiano, L. Lembo-Fazio, I. Paciello, C. P. Coutinho, I. Sá-Correia, M. L. Bernardini, R. Lanzetta, D. Garozzo, A. Silipo and A. Molinaro, ChemBioChem, 2013, 14, 1105–1115 CrossRef.
- F. D. Lorenzo, A. Palmigiano, I. Paciello, M. Pallach, D. Garozzo, M. L. Bernardini, V. Cono, M. M. Yakimov, A. Molinaro and A. Silipo, Mar. Drugs, 2017, 15(7), 201 CrossRef.
- L. De Simone Carone, G. Barra, R. Cirella, M. Ziaco, M. Mercogliano, F. Olmeo, E. Andretta, V. Mazziotti, C. Fusco, G. D'Ippolito, K. A. Duda, F. M. Farquharson, P. Louis, A. Fontana, A. Silipo, A. Molinaro, F. Chiodo and F. Di Lorenzo, Angew. Chem., Int. Ed., 2025, e202512947 CAS.
- F. Di Lorenzo, Antonie van Leeuwenhoek, 2017, 110, 1401–1412 CrossRef CAS PubMed.
- J. K. Kim, H. A. Jang, M. S. Kim, J. H. Cho, J. Lee, F. Di Lorenzo, L. Sturiale, A. Silipo, A. Molinaro and B. L. Lee, J. Biol. Chem., 2017, 292, 19226–19237 CrossRef CAS.
- A. Cangiano, N. Gallucci, A. Giarra, J. Ali, L. Chiappisi, N. P. Cowieson and L. Paduano, Nanoscale, 2025, 17, 15829–15840 RSC.
- N. P. Cowieson, C. J. C. Edwards-Gayle, K. Inoue, N. S. Khunti, J. Doutch, E. Williams, S. Daniels, G. Preece, N. A. Krumpa, J. P. Sutter, A. D. Tully, N. J. Terrill and R. P. Rambo, J. Synchrotron Radiat., 2020, 27, 1438–1446 CrossRef CAS.
- N. Gallucci, M. S. Appavou, N. Cowieson, G. D'Errico, R. Di Girolamo, S. Lettieri, F. Sica, G. Vitiello and L. Paduano, J. Colloid Interface Sci., 2024, 659, 926–935 CrossRef CAS PubMed.
- M. A. Campanero-Rhodes, R. A. Childs, M. Kiso, S. Komba, C. Le Narvor, J. Warren, D. Otto, P. R. Crocker and T. Feizi, Biochem. Biophys. Res. Commun., 2006, 344, 1141–1146 CrossRef CAS PubMed.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2026 |