Open Access Article
Open Access ArticleDecoding polyethylene formation in Cr/PNP catalyzed ethylene oligomerization via experimentally guided machine learning
Youcai Zhu ,
Yue Mu,
Xiaoke Shi,
Long Chen,
Shu Yang,
Li Sun
,
Yue Mu,
Xiaoke Shi,
Long Chen,
Shu Yang,
Li Sun and
Zhen Liu
and
Zhen Liu *
*
School of Chemical Engineering, East China University of Science and Technology, Shanghai 200237, China. E-mail: liuzhen@ecust.edu.cn
First published on 19th January 2026
Abstract
Polyethylene formation remains a critical side reaction in ethylene selective oligomerization, lowering α-olefin yields and lacking reliable predictive strategies. Here, an automated workflow incorporating structural parsing and indexing was developed to construct a Cr/PNP catalysts library, extract comprehensive molecular descriptors, and integrate them with machine learning models trained on experimentally measured PE values. A combined classification-regression strategy enabled accurate identification of low-PE catalysts and quantitative prediction of PE contents, establishing a broadly applicable framework for catalyst design and side-product control in homogeneous catalysis. Analysis of SHAP feature importance and DFT calculations revealed that the intrinsic properties of the ligand primarily determine whether it produces low or high PE. In contrast, optimizing experimental conditions plays a pivotal role in further reducing PE in low-PE ligands.
1. Introduction
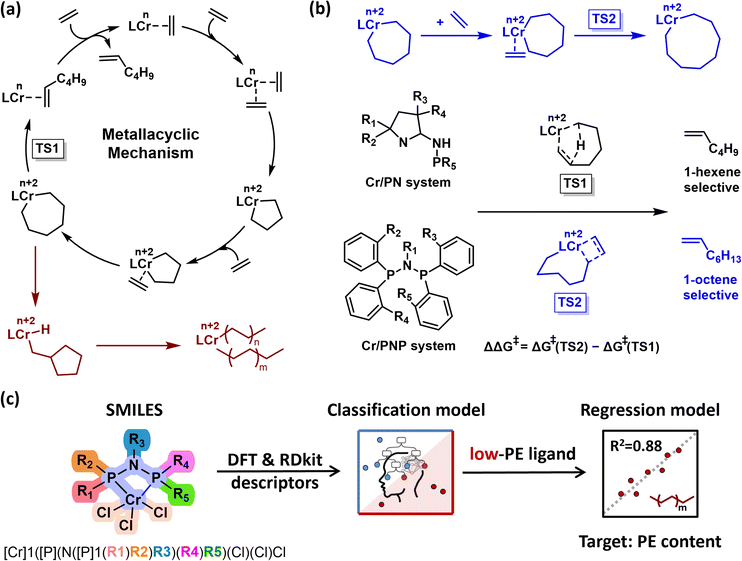
Linear α-olefins are important commodity chemicals widely used in applications such as ethylene copolymerization and the production of plasticizers, detergents, surfactants, and lubricants.1–4 A significant portion of the light fraction (C4–C8) serves as comonomers in the copolymerization with ethylene to produce linear low-density polyethylene (PE).5 Among them, 1-hexene and 1-octene are particularly valuable for enhancing the mechanical performance of the resulting polymers, especially in terms of tear resistance.6 To enable the efficient and selective production of these valuable α-olefins, significant attention has been directed toward the development of transition metal catalysts. Chromium-based catalysts3,7,8 have been most extensively studied for selective ethylene oligomerization, especially in comparison to those based on tantalum,9,10 titanium,11,12 and other metals.13,14Selective ethylene trimerization typically follows a metallacyclic mechanism, where the relative stability of metallacyclic intermediates dictates selectivity toward specific α-olefins (Fig. 1a, black line).3,15,16 Although crossover studies have experimentally supported this mechanism,3 differentiating it from classical Cossee–Arlman pathways, several mechanistic uncertainties remain unresolved, including the oxidation states of key intermediates,17 the reaction's kinetic order in ethylene,18 and the elementary steps involved in α-olefin product release.19 Another particularly challenging issue is the mechanism responsible for the formation of PE byproducts — an undesirable and costly side reaction.20 This side reaction not only reduces the yield of target α-olefins, but also leads to the accumulation of high-molecular-weight PE, which can cause reactor fouling and operational inefficiencies. The most widely accepted explanation for PE formation is a catalyst degradation pathway, where an off-cycle intermediate produces PE through a Cossee-type chain growth pathway (Fig. 1a, red line).20 However, alternative pathways have also been demonstrated, Gibson showed that chain transfer to aluminum mediated by Me3Al can also generate PE.16 These findings suggest that mitigating PE byproduct formation solely through mechanistic understanding is highly challenging, and thus alternative approaches are required.
Given these mechanistic ambiguities, a common strategy is to model transition states computationally using quantum chemical methods. However, such approaches often suffer from intrinsic errors on the order of 2–3 kcal mol−1.21 In the context of ethylene oligomerization, the energy barrier difference between the key transition states, β-hydrogen transfer (TS1) and ethylene insertion (TS2), can be as small as a few tenths of a kcal mol−1. Such narrow ΔΔG‡ windows render the predicted product selectivity highly sensitive to computational uncertainties.22 Errors stemming from functional choice, solvation models, or inaccurate conformer sampling may lead to misassignment of the favored product pathway (e.g., erroneously favoring 1-hexene over 1-octene). In such scenarios, a hybrid catalyst design framework that integrates quantum–mechanical transition state modeling with machine learning techniques offers a promising solution. Reaction-relevant descriptors were extracted from quantum calculations and integrated into data-driven models.23,24 This approach enables the systematic identification of critical features governing catalyst performance, which are often difficult to capture through conventional analysis, and facilitates their use in rational catalyst screening.
In recent years, data-driven approaches have demonstrated increasing effectiveness across a variety of chromium-based catalytic systems for ethylene trimerization and tetramerization. These methods offer promising alternatives or complement to traditional transition state modeling, particularly in addressing cases where minute energy differences between competing pathways challenge the reliability of quantum chemical predictions. Ess combined DFT-derived transition state models with machine learning to predict the 1-hexene/1-octene selectivity of Cr/PN catalysts based on the computed energy difference (ΔΔG‡) between competing transition states (Fig. 1b).25 Their model identified key structural descriptors (e.g., Cr–N distance and Cr distance out of pocket) and enabled the design of new Cr/PN ligands predicted to exhibit >95% selectivity for 1-octene. This approach was further extended to Cr/PNP catalytic systems, where we identified key factors correlating with the ΔΔG‡ across 240 transition state structures and established a standardized feature selection protocol that yielded 20 descriptors, including 13 multidimensional and 7 Mordred descriptors.26 A combined DFT-ANN framework was also developed for Cr/PCCP catalysts, where steric and electronic descriptors extracted from DFT-optimized structures enabled accurate modeling and screening of novel ligands. Experimental 1-octene selectivity and activity of a newly synthesized ligand closely matched the ANN predictions, underscoring the reliability of this approach for guiding catalyst design.27
While significant progress has been made in modulating 1-hexene/1-octene selectivity, the issue of PE formation remains relatively underexplored. As Sydora28 aptly stated, “any discussion of ethylene tri-/tetramerization catalysts would not be complete without mentioning PE formation.” The unclear mechanism underlying PE byproduct formation remains a major obstacle to the industrial application of Cr-catalyzed ethylene oligomerization. Therefore, we chose the Cr/PNP catalytic system as the model platform, given its well-established high selectivity in ethylene trimerization and tetramerization.5,29,30 Based on experimental data, a diverse set of descriptors was extracted from DFT-optimized structures, encompassing electronic, steric, and geometric features of the catalysts. Classification and regression models were then integrated to systematically investigate the key factors governing PE formation (Fig. 1c).
2. Computational details
2.1 DFT calculations
The DFT calculations, including geometry optimizations and frequency calculations, were performed using the Gaussian 09 program package.31 Lowest energy conformers and protomers were searched using automated conformer-rotamer ensemble sampling tool (CREST).32 Subsequently, full optimization of all structures was conducted employing the B3LYP-D3 functional in conjunction with the def2-SVP basis set and the SMD solvation model, with toluene employed as the model solvent.33–37 Harmonic frequency calculations were employed throughout to assure that the structures were adequately optimized. The nature of the lowest energy stationary points was then confirmed by frequency calculations to ensure that they were characterized as local minima with no imaginary frequency or transition states with only one appropriate imaginary frequency. The larger triple-zeta basis set def2-TZVP in combination with the functional M06-L was adopted to calculate the single-point energy (E).38–40 Thermal correction of free energy (Gtherm) is performed by thermochemical analysis. The SMD solvation model for toluene was used to calculate the solvation free energy (Gsol). The free energy required to change the standard state of 1 atm/1 M (Gstd) is included. Therefore, the relative Gibbs free energy (ΔG) is defined as ΔG = ΔGtherm + ΔGsol + ΔGstd + ΔE.2.2 Model training
The machine learning models were trained using Jupyter Notebook and the Scikit-learn library, with all code written in Python.41 We employed both classification and regression models. For classification, we used Logistic Regression, Decision Trees, and Random Forest.42–44 Six models were applied for regression: Linear Regression, Ridge Regression, Random Forest (RF) Regressor, Support Vector Regression (SVR), XGBoost Regressor, and Lasso Regression.44–46 Cross-validation was performed by randomly splitting the data into 20% test and 80% training sets, using 5-fold cross-validation. Hyperparameter optimization was conducted via Grid Search.473. Result and discussion
3.1 Database construction
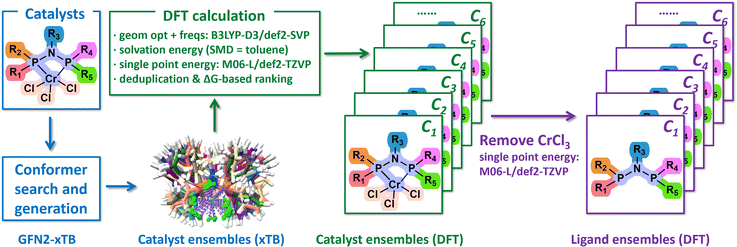
As the catalyst optimization relies on experimentally determined PE contents, we constructed a comprehensive descriptor library incorporating PNP ligands evaluated in ethylene oligomerization reactions. The dataset encompasses most of the ligands reported to date, with more than 200 experimental entries including variations in temperature, pressure, and other reaction conditions. It should be emphasized that the PE contents used as targets in our models are overall experimental observables, which may include contributions from multiple active species and degradation pathways, rather than purely from a single idealized catalytic cycle. The aim was to construct a PNP descriptor library applicable to reactions with varying ligand coordination environments. The resulting parameters needed to be comparable across PNP skeleton with different P- or N-substituents, so that disparate PNP ligands could be meaningfully compared.One key challenge in establishing the computational workflow for PNP ligands is the representation of their conformational space, the relative conformer energies, and the resulting contribution to ligand properties. This is particularly critical for steric descriptors, which vary substantially with conformation. While no single model system (e.g., free ligand or specific reference complexes) can fully capture the conformational space accessible to PNP ligands in catalytically relevant environments, it is possible to define reasonable limits for attainable geometries and properties. Probing these ranges allows us to assess ligand behavior in Cr-catalyzed ethylene oligomerization and to predict catalytic performance.
Reliable geometries were obtained using GFN2-xTB combined with CREST-based workflows to generate conformer ensembles (Fig. 2). The conformational space was explored in two reference states: (i) coordinated species represented by the (PNP)CrCl3 model complex and (ii) the corresponding free ligands. Free ligands typically occupy more space around the nitrogen lone pair or phosphine substituents, making them appear sterically more demanding than their coordinated counterparts, yet both states are essential for accurately describing catalytic behavior and potential side reactions such as ligand dissociation. Specifically, the conformer ensembles generated by CREST were further refined using DFT calculations, and the resulting structures were ranked by their Gibbs free energies to identify the lowest-energy conformer set. To extract ligand-level information, single-point energy calculations were subsequently performed on geometries derived from this lowest-energy catalyst conformer set, in which the Cr and Cl atoms were removed while preserving the ligand geometry. This strategy ensured that the ligand descriptors were obtained in a manner consistent with the catalytically relevant conformations.
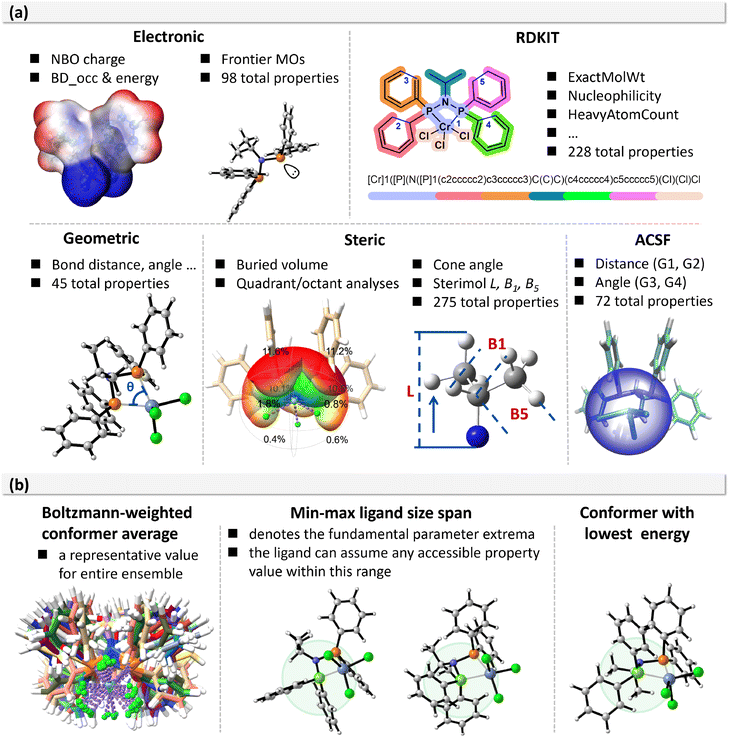
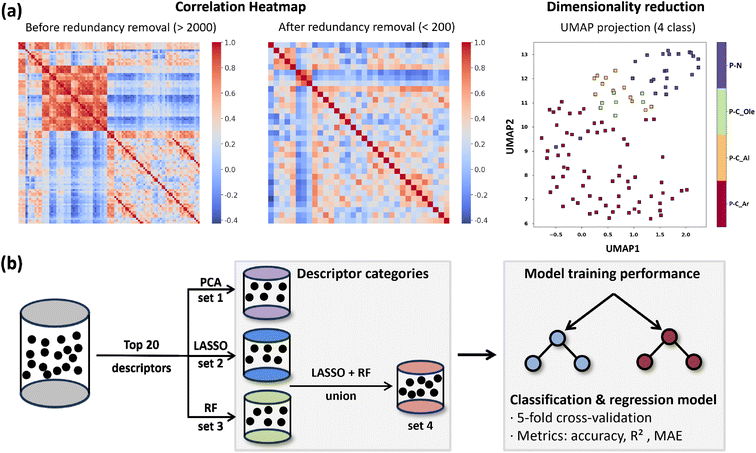
To comprehensively capture the factors relevant to transition-metal catalysis, both catalysts and ligands were characterized using five categories of descriptors: electronic, RDKit, geometric, steric, and atom-centered symmetry functions (ACSF). The descriptor set includes global catalyst-level properties (e.g., molecular electrostatic potential, percent buried volume, and bite angle) together with ligand-specific features, such as the lone-pair environment of the phosphorus donor atoms and sterimol parameters of the nitrogen substituents (Fig. 3a). Of particular importance, the steric descriptors were designed to reflect quadrant-specific contributions for ligands with different symmetry elements. For example, in the case of a C2v symmetric ligand (Section 2 of SI), the percent buried volume of the northwestern quadrant illustrates how optimized geometries often deviate from ideal symmetry, leading to quadrant-specific values that are not equivalent.
To account for this, we report the minimum, maximum, and average values of symmetry-equivalent quadrants, thereby capturing subtle directional steric effects that traditional global parameters cannot reflect. In total, more than 700 descriptors were evaluated. For each descriptor, four variants were generated: Boltzmann-weighted average, maximum, minimum, and the value of the lowest-energy conformer. This strategy ensured both conformational diversity and statistical representativeness (Fig. 3b).
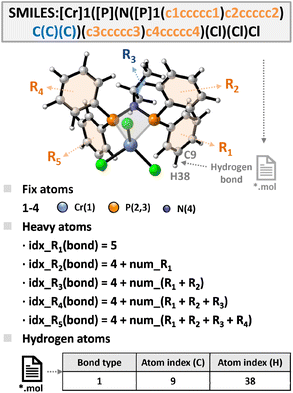
To enable efficient and scalable descriptor extraction, we developed a fully automated structural parsing and indexing workflow (Fig. 4). This approach integrates SMILES strings with information from generated MOL files, and through grouping, decomposition, and heavy-atom tracking, it precisely identifies key atomic relationships. Based on the core scaffold and predefined SMILES rules, the structures and atom ordering of substituents R1–R5 are determined. Because tools such as RDKit place heavy atoms before hydrogens, the indices of substituents can be directly derived from SMILES, while bond connectivity in MOL files provides the corresponding hydrogen indices. These atom indices then serve as anchors for computing geometric and electronic descriptors in a reproducible way. The fully automated process eliminates manual intervention, improves efficiency and consistency, and ensures the comparability of descriptors across large ligand libraries.
3.2 Feature engineering and model development
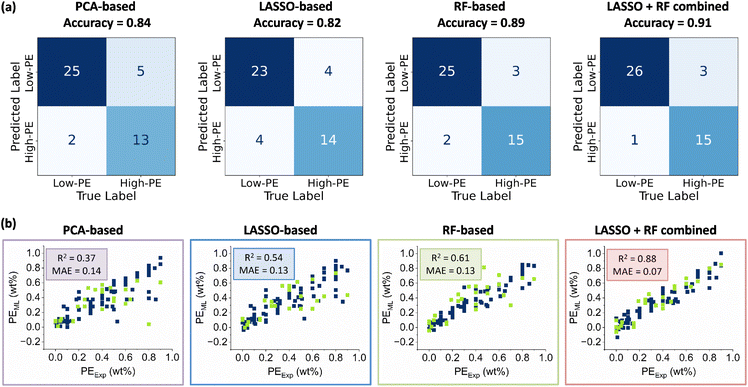
To reduce redundancy and highlight the most informative descriptors, we first eliminated highly correlated variables, reducing the descriptor space from more than 2000 to fewer than 200 non-redundant descriptors (Fig. 5a). UMAP-based dimensionality reduction further confirmed the clustering of ligands into the four major classes present in the database (P–N, P–C_Ole, P–C_Al, and P–C_Ar). This indicates that the reduced descriptor set preserves sufficient chemical information to represent the structural diversity of phosphorus substituents, thereby validating the effectiveness of the selected descriptors. Subsequently, three complementary feature selection strategies were applied to identify the most relevant descriptors: principal component analysis (PCA), linear LASSO regression with cross-validation (LASSO-CV), and nonlinear random forest (RF) ranking (Fig. 5b). For each method, the top 20 descriptors were retained, and the union of LASSO and RF selections was constructed to capture both linear and nonlinear effects. These refined descriptor subsets were then used to train classification and regression models, with model performance evaluated by 5-fold cross-validation using accuracy (for classification) and R2/MAE (for regression) as metrics.Direct regression of the full dataset yielded poor predictive performance due to the highly imbalanced distribution of PE values (Section 3 of SI). To address this challenge, a classification-regression strategy was implemented. First, classification models were constructed to distinguish between catalysts with low PE (<1 wt%) and high PE (≥1 wt%) using four different descriptor sets (Fig. 6a). Among them, the combined LASSO + RF descriptor set achieved the best performance under the decision tree model, with an accuracy of 0.91. Notably, 26 out of 27 low-PE cases were correctly predicted, demonstrating an excellent precision for the low-PE class. Since PE values below 1 wt% are often considered industrially acceptable, this classification framework provides a robust basis for practical catalyst screening.
Regression models were then applied to the low-PE catalysts, which are particularly favorable for ethylene oligomerization, to predict their quantitative values. Six machine learning algorithms were evaluated, including linear regression, ridge regression, RF, SVR, XGBoost, and LASSO (Section 3 of SI). Among them, XGBoost consistently delivered the highest predictive accuracy. Across the four descriptor sets, the combined LASSO + RF descriptors again yielded the best performance, with an R2 of 0.88 and a MAE of 0.07 (Fig. 6b). These results demonstrate that the proposed descriptor strategy effectively captures the structural and electronic features governing catalyst performance, enabling accurate, quantitative predictions in the low-PE regime.
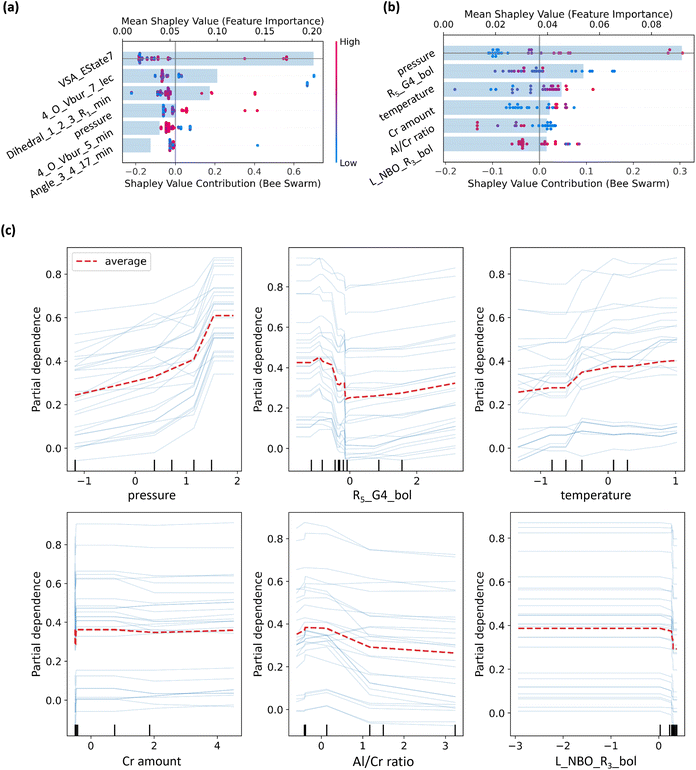
The SHAP summary plots for both the Decision Tree Regressor and XGBoost models provide significant insights into the contributions of individual features to the predictions (Fig. 7).48,49 Detailed interpretations of the descriptors corresponding to the SHAP analysis are provided in Section 4 of the SI. In the Decision Tree Regressor model, VSA_Estate7 and 4_O_Vbur_7_lec stand out as the most important features (Fig. 7a). Apart from pressure, the top six most important features are all descriptors related to the structure of catalyst, indicating that the inherent properties of catalyst primarily determine whether the PE values are high or low (Table S12).
By contrast, pressure emerges as the most important feature in the XGBoost model when used to describe the experimental conditions, with a clear positive correlation with PE content (Fig. 7b). A similar trend is observed for temperature, where higher temperatures are associated with higher PE content. In contrast, R5_G4_bol shows a negative correlation with PE content. Notably, four out of the top five most important features are related to experimental conditions, emphasizing their significant role in influencing PE content predictions. Consistent with this, a quantitative SHAP analysis shows that the experimental conditions collectively account for over 70% of the total importance of the features in the regression model (Table S13). Therefore, feature importance is primarily determined by ligand-specific properties in the classification model, which play a critical role in distinguishing between materials with high (PE > 1 wt%) and low (PE < 1 wt%) PE. However, once a ligand has been classified as a low PE material, optimizing the experimental conditions becomes crucial. In this instance, factors such as pressure, temperature, the amount of Cr, and the Al/Cr ratio play a much larger role in controlling PE formation.
The partial dependence plots (PDPs) illustrate the impact of individual features on the predicted PE values in the regression model (Fig. 7c). Pressure and temperature have a significant positive effect on the predicted PE, with increasing values leading to higher PE predictions. R5_G4_bol exhibits clear threshold effects, with substantial changes in predicted PE when their values cross certain points. The L_NBO_R3_bol shows minimal influence on the PE prediction. These results highlight the importance of experimental conditions, such as pressure and temperature, in the regression model for low PE ligands. Optimizing these conditions can further reduce the PE content, emphasizing their critical role in predicting and improving low PE outcomes.
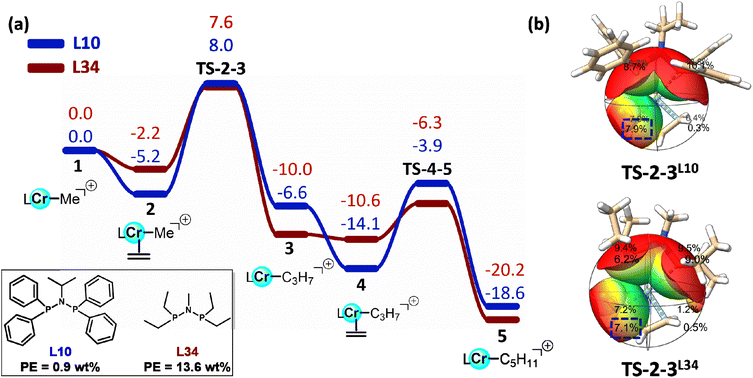
To further elucidate the physical meaning of the selected descriptors and their relationship to PE formation, we carried out DFT studies on representative PNP ligands, L10 and L34. Cr/L10 system has been experimentally identified as one of the most selective catalysts for 1-octene formation, exhibiting a relatively low PE content (0.9 wt%), whereas Cr/L34 system leads to significantly higher PE formation (13.6 wt%).5 Although the detailed mechanism of PE formation remains unclear, chain growth via insertion of ethylene into a metal-hydride or metal–methyl bond is well established. Therefore, our computational study focused on the Cossee–Arlman mechanism.50,51 At the same time, experimental evidence from Duchateau indicates a non-redox Cr(II) pathway for ethylene polymerization.52 Accordingly, the [LCr(II)–Me]+ complex was adopted as the catalytic model to explore the chain propagation step responsible for PE formation.
For both Cr/L10 and Cr/L34 catalytic systems, the reaction initiates from the Cr–Me intermediate 1, and subsequent ethylene coordination affords intermediate 2, with both processes being exergonic. Subsequently, ethylene insertion into the Cr–C bond leads to the formation of a new C–C bond, requiring activation barriers of 13.2 kcal mol−1 for Cr/L10 system and 9.8 kcal mol−1 for Cr/L34 system, respectively (Fig. 8a). Further coordination and insertion of an additional ethylene molecule proceed with lower energy barriers, indicating a gradual facilitation of chain propagation. From a kinetic perspective, the lower barrier for Cr/L34 system suggests a higher propensity toward PE formation, consistent with the experimental observations. To rationalize these differences, we analysed the buried volume parameters of the two systems at the transition state (TS-2-3) corresponding to the rate-determining ethylene insertion step (Fig. 8b). In particular, the buried volume for octant 7 with a radius of 4 Å, corresponding to the ethylene insertion site, was found to be 7.9% for Cr/L10 system, larger than 7.1% for Cr/L34 system. This indicates that the increased local steric hindrance effectively suppresses chain growth, thereby reducing PE formation. This observation aligns well with the SHAP analysis, confirming that the selected structural descriptors not only correlate strongly with catalytic performance but also provide physically interpretable insight into the steric effects governing polymerization behaviour.
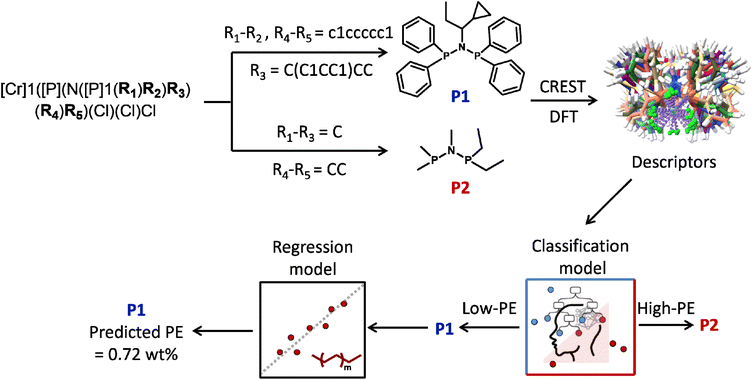
To translate the above models into practical ligand design, we finally outline a simple workflow for screening new PNP ligands (Fig. 9). We encode the generic PNP skeleton as [Cr]1([P](N([P]1(R1)R2)R3)(R4)R5)(Cl)(Cl)Cl, where R1–R5 can be any user-defined substituents. For any proposed set of R1–R5, the corresponding ligand is first written as a SMILES string and converted into a 3D structure (e.g., via RDKit). Starting from this structure, CREST is used to generate conformational ensembles, which are refined at the DFT level; the same set of geometrical, steric and electronic descriptors as in the training stage is then extracted and fed into the pretrained PE models.
As shown in Fig. 9, this allows us to compare, for example, two novel representative ligands generated from the same scaffold: (i) P1, obtained when R3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C1CC1)CC and R1–R2, R4–R5 = c1ccccc1, and (ii) P2, obtained when R1–R3
C(C1CC1)CC and R1–R2, R4–R5 = c1ccccc1, and (ii) P2, obtained when R1–R3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and R4–R5
C and R4–R5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC. After CREST/DFT refinement and descriptor calculation, the classification model predicts P2 to fall in the high-PE region, whereas P1 is classified as a low-PE ligand. Applying the regression model to P1 then yields a quantitative prediction of the polymer content (predicted PE = 0.72 wt%), indicating that P1 is expected to generate a relatively low amount of PE under the reference conditions. From an industrial perspective, such low-PE ligands are especially attractive because they are less likely to cause reactor fouling or plugging and can be further optimized to deliver high overall selectivity to the prime products 1-hexene and 1-octene.
CC. After CREST/DFT refinement and descriptor calculation, the classification model predicts P2 to fall in the high-PE region, whereas P1 is classified as a low-PE ligand. Applying the regression model to P1 then yields a quantitative prediction of the polymer content (predicted PE = 0.72 wt%), indicating that P1 is expected to generate a relatively low amount of PE under the reference conditions. From an industrial perspective, such low-PE ligands are especially attractive because they are less likely to cause reactor fouling or plugging and can be further optimized to deliver high overall selectivity to the prime products 1-hexene and 1-octene.
4. Conclusions
This work presents an integrated computational and data-driven framework to investigate PE byproduct formation in Cr/PNP catalyzed ethylene oligomerization. A comprehensive virtual ligand library was constructed, and conformational ensembles were systematically explored using GFN2-xTB/CREST sampling and DFT refinement to derive catalyst- and ligand-level descriptors capturing key steric, electronic, geometric and ACSF features. A fully automated structural parsing and indexing workflow ensured reproducible descriptor extraction across diverse PNP ligands, greatly facilitating the systematic acquisition of descriptors for transition metal catalysts. This workflow is readily extendable to other reactions, enabling the extraction of additional descriptors and providing a powerful tool for transition metal catalyst development.To address the challenge of predicting PE formation, a classification-regression strategy was implemented. Classification models effectively distinguished low-PE from high-PE catalysts, with the combined LASSO + RF descriptor set achieving an accuracy of 0.91 and excellent recall in the industrially relevant low-PE regime. For catalysts with PE < 1 wt%, regression analysis showed that XGBoost combined with the LASSO + RF descriptor set delivered the best performance, with R2 = 0.88 and MAE = 0.07. Through SHAP feature importance analysis, it was found that the ligand's inherent properties determine whether it will generate low or high PE. DFT calculations consistently confirm that the steric features of the ligands directly influence the reaction barriers associated with the formation of PE. Meanwhile, the optimization of experimental conditions is crucial for further reducing PE in low-PE ligands.
From a broader design perspective, the current PE models are intended to serve as the first screening layer in a multi-objective workflow for Cr/PNP oligomerization catalysts. New ligands can first be evaluated by the classification and regression models developed here to identify candidates that are intrinsically disfavored toward PE formation. These low-PE candidates can then be subjected to further optimization of 1-hexene/1-octene selectivity using existing machine-learning models for 1-hexene/1-octene selectivity in Cr/PNP systems. In this way, the risk of excessive PE formation is addressed upfront, and prime-product selectivity is fine-tuned within a ligand space that has already been filtered for robustness against polymer formation. In future work, we envisage using this workflow to guide the synthesis and catalytic testing of new model-predicted low-PE ligands under experimentally optimized conditions, thereby enabling prospective validation and further refinement of the models.
Author contributions
Youcai Zhu: software, investigation, writing – original draft. Yue Mu & Xiaoke Shi: software, writing – review & editing. Shu Yang & Li Sun: methodology. Zhen Liu: project administration, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
All computational scripts, trained machine learning models, and raw descriptor data used in this study are openly available on GitHub at: https://github.com/youcaizhu668/EthyleneOligomerization_Cr-PNP_ML Additional data supporting the findings of this work are provided in the supporting information (SI). Supplementary information: full details of the ligand database construction, the definition of steric, electronic, geometric, and other descriptors, complete machine learning model development, and a benchmark of the computational methods used in this work. See DOI: https://doi.org/10.1039/d5sc07894f.Acknowledgements
We thank the National Natural Science Foundation of China (22171084). We acknowledge the supercomputer at East China University of Science and Technology for generous computing resources.References
- L. H. N. G. R. Lappin, J. D. Sauer and J. D. Wagner, Higher Olefins, Higher Olefins, Wiley & Sons, Inc, 2005 Search PubMed.
- T. Agapie, J. A. Labinger and J. E. Bercaw, J. Am. Chem. Soc., 2007, 129, 14281–14295 CrossRef CAS PubMed.
- T. Agapie, S. J. Schofer, J. A. Labinger and J. E. Bercaw, J. Am. Chem. Soc., 2004, 126, 1304–1305 CrossRef CAS PubMed.
- B. Yang, A. J. Schaefer, B. L. Small, J. A. Leseberg, S. M. Bischof, M. S. Webster-Gardiner and D. H. Ess, Chem. Sci., 2024, 15, 18355–18363 RSC.
- A. Bollmann, K. Blann, J. T. Dixon, F. M. Hess, E. Killian, H. Maumela, D. S. McGuinness, D. H. Morgan, A. Neveling, S. Otto, M. Overett, A. M. Slawin, P. Wasserscheid and S. Kuhlmann, J. Am. Chem. Soc., 2004, 126, 14712–14713 CrossRef CAS.
- Y. V. Kissin, Polymers of Higher Olefins, Wiley & Sons, Inc, 2005 Search PubMed.
- R. D. Köhn, M. Haufe, G. Kociok-Köhn, S. Grimm, P. Wasserscheid and W. Keim, Angew. Chem., Int. Ed., 2000, 39, 4337–4339 Search PubMed.
- Y. Zhu, Y. Mu, X. Shi, S. Yang, L. Sun and Z. Liu, J. Catal., 2026, 454, 116613 CrossRef CAS.
- C. Andes, S. B. Harkins, S. Murtuza, K. Oyler and A. Sen, J. Am. Chem. Soc., 2001, 123, 7423–7424 CrossRef CAS.
- R. Arteaga-Muller, H. Tsurugi, T. Saito, M. Yanagawa, S. Oda and K. Mashima, J. Am. Chem. Soc., 2009, 131, 5370–5371 CrossRef CAS PubMed.
- P. J. W. Deckers, B. Hessen and J. H. Teuben, Angew. Chem., Int. Ed., 2001, 40, 2516–2519 CrossRef CAS.
- E. Otten, A. A. Batinas, A. Meetsma and B. Hessen, J. Am. Chem. Soc., 2009, 131, 5298–5312 CrossRef CAS.
- F. Kong, P. Rios, C. Hauck, F. J. Fernandez-de-Cordova, D. A. Dickie, L. G. Habgood, A. Rodriguez and T. B. Gunnoe, J. Am. Chem. Soc., 2023, 145, 179–193 CrossRef CAS.
- B. L. Small and M. Brookhart, J. Am. Chem. Soc., 1998, 120, 7143–7144 CrossRef CAS.
- G. J. P. Britovsek, D. S. McGuinness, T. S. Wierenga and C. T. Young, ACS Catal., 2015, 5, 4152–4166 CrossRef CAS.
- A. K. Tomov, J. J. Chirinos, D. J. Jones, R. J. Long and V. C. Gibson, J. Am. Chem. Soc., 2005, 127, 10166–10167 CrossRef CAS PubMed.
- S. Tang, Z. Liu, X. Yan, N. Li, R. Cheng, X. He and B. Liu, Appl. Catal., A, 2014, 481, 39–48 CrossRef CAS.
- R. Walsh, D. H. Morgan, A. Bollmann and J. T. Dixon, Appl. Catal., A, 2006, 306, 184–191 CrossRef CAS.
- Y. Qi, Q. Dong, L. Zhong, Z. Liu, P. Qiu, R. Cheng, X. He, J. Vanderbilt and B. Liu, Organometallics, 2010, 29, 1588–1602 CrossRef CAS.
- T. Gunasekara, J. Kim, A. Preston, D. K. Steelman, G. A. Medvedev, W. N. Delgass, O. L. Sydora, J. M. Caruthers and M. M. Abu-Omar, ACS Catal., 2018, 8, 6810–6819 Search PubMed.
- M. Bursch, J. M. Mewes, A. Hansen and S. Grimme, Angew. Chem., Int. Ed., 2022, 61, e202205735 CrossRef CAS PubMed.
- D.-H. Kwon, J. T. Fuller, U. J. Kilgore, O. L. Sydora, S. M. Bischof and D. H. Ess, ACS Catal., 2018, 8, 1138–1142 Search PubMed.
- J. J. Dotson, L. van Dijk, J. C. Timmerman, S. Grosslight, R. C. Walroth, F. Gosselin, K. Puntener, K. A. Mack and M. S. Sigman, J. Am. Chem. Soc., 2023, 145, 110–121 CrossRef CAS PubMed.
- T. Gensch, G. Dos Passos Gomes, P. Friederich, E. Peters, T. Gaudin, R. Pollice, K. Jorner, A. Nigam, M. Lindner-D'Addario, M. S. Sigman and A. Aspuru-Guzik, J. Am. Chem. Soc., 2022, 144, 1205–1217 CrossRef CAS.
- S. M. Maley, D. H. Kwon, N. Rollins, J. C. Stanley, O. L. Sydora, S. M. Bischof and D. H. Ess, Chem. Sci., 2020, 11, 9665–9674 Search PubMed.
- Z. Luo, J. Peng, Y. Mu, L. Sun, Z. Zhu and Z. Liu, J. Catal., 2023, 428, 115127 CrossRef CAS.
- H. Fan, Y. Zhang, F. Alam, J. Ma, B. Hao, Y. Chen, Y. Wang, J. Huang and T. Jiang, J. Catal., 2024, 429, 115237 Search PubMed.
- O. L. Sydora, Organometallics, 2019, 38, 997–1010 CrossRef CAS.
- M. J. Overett, K. Blann, A. Bollmann, J. T. Dixon, D. Haasbroek, E. Killian, H. Maumela, D. S. McGuinness and D. H. Morgan, J. Am. Chem. Soc., 2005, 127, 10723–10730 CrossRef CAS.
- J. Rabeah, M. Bauer, W. Baumann, A. E. C. McConnell, W. F. Gabrielli, P. B. Webb, D. Selent and A. Brückner, ACS Catal., 2012, 3, 95–102 CrossRef.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision E.01, Gaussian, Inc., Wallingford CT, 2009 Search PubMed.
- P. Pracht, F. Bohle and S. Grimme, Phys. Chem. Chem. Phys., 2020, 22, 7169–7192 RSC.
- A. D. Becke, J. Chem. Phys., 1993, 98, 1372–1377 Search PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter, 1988, 37, 785–789 CrossRef CAS.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS.
- L. Xu and M. L. Coote, J. Phys. Chem. A, 2019, 123, 7430–7438 Search PubMed.
- P. Macchi, D. M. Proserpio and A. Sironi, J. Am. Chem. Soc., 1998, 120, 1447–1455 CrossRef CAS.
- Y. Zhao and D. G. Truhlar, J. Phys. Chem. A, 2008, 112, 6794–6799 CrossRef CAS PubMed.
- Y. Zhu, X. Guo, X. Ding, L. Sun, M. Zhang and Z. Liu, Mol. Catal., 2022, 518, 112108 CAS.
- Y. Zhu, L. Sun, Z. Zeng and Z. Liu, ACS Catal., 2024, 14, 13684–13696 CrossRef CAS.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- S. Sperandei, Biochem. Med., 2014, 24, 12–18 Search PubMed.
- J. R. Quinlan, IEEE Trans. Syst. Man Cybern., 1990, 20, 339–346 CrossRef.
- L. Breiman, Mach. Learn., 2001, 45, 5–32 CrossRef.
- C. G. Tianqi Chen, In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, 785–794 Search PubMed.
- R. Tibshirani, J. Roy. Stat. Soc. B Stat. Methodol., 1996, 58, 267–288 CrossRef.
- Y. B. James Bergstra, J. Mach. Learn. Res., 2012, 13, 281–305 Search PubMed.
- A. V. Ponce-Bobadilla, V. Schmitt, C. S. Maier, S. Mensing and S. Stodtmann, Clin. Transl. Sci., 2024, 17, e70056 CrossRef.
- S. Tan, R. Wang, G. Song, S. Qi, K. Zhang, Z. Zhao and Q. Yin, Fuel, 2024, 355 CAS.
- D. S. McGuinness, Chem. Rev., 2011, 111, 2321–2341 CrossRef CAS.
- E. Arlman, J. Catal., 1964, 3, 99–104 CrossRef CAS.
- A. Jabri, C. B. Mason, Y. Sim, S. Gambarotta, T. J. Burchell and R. Duchateau, Angew. Chem., Int. Ed., 2008, 47, 9717–9721 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |