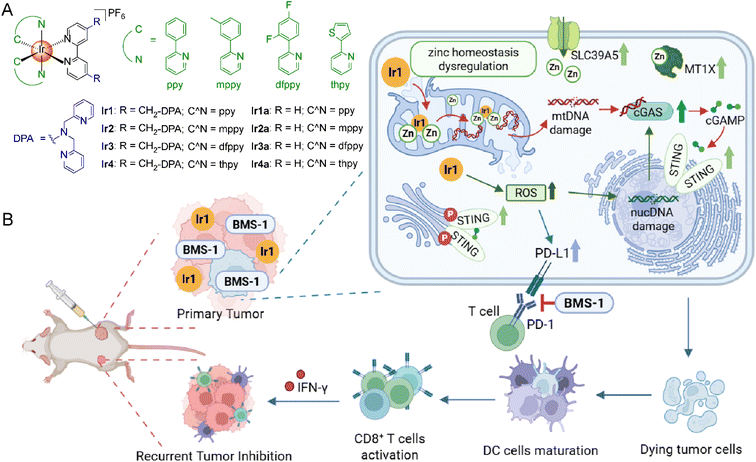
Open Access Article
Open Access ArticlePhosphorescent iridium complexes activated by endogenous zinc as a mitochondrial DNA nuclease for stimulation of the cGAS-STING pathway
Zhi-Yuan
Li†
ac,
Long-Bo
Yu†
ac,
Qing-Hua
Shen
ac,
Liang
Hao
abc,
Peng
Wang
ac,
Xiao-Xiao
Chen
ac,
Yu-Yi
Ling
*abc and
Cai-Ping
Tan
 *acd
*acd
aMOE Key Laboratory of Bioinorganic and Synthetic Chemistry, School of Chemistry, Sun Yat-Sen University, Guangzhou 510006, China. E-mail: lingyy@gdmu.edu.cn; tancaip@mail.sysu.edu.cn
bThe Marine Biomedical Research Institute of Guangdong Zhanjiang, School of Ocean and Tropical-Medicine, Guangdong Medical University, Zhanjiang, Guangdong 524023, China
cGuangdong Basic Research Center of Excellence for Functional Molecular Engineering, Guangzhou 510006, China
dState Key Laboratory of Anti-Infective Drug Discovery and Development, School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou 510006, China
First published on 25th November 2025
Abstract
Zinc is a crucial element in cellular processes, and its homeostasis has intricate relationships with the initiation, progression, and therapeutic intervention of cancer. Activation of the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway has been proven to be an effective strategy for cancer immunotherapy. Herein, we report four phosphorescent iridium complexes (Ir1–Ir4) with zinc chelating ligands. Among them, Ir1 can bind and image mitochondrial chelatable zinc ions via phosphorescence-lifetime responses, consequently modulating the expression of zinc-regulatory proteins. Furthermore, the in situ formed heteronuclear metal complex Ir1-Zn2 shows nuclease mimetic activities, capable of hydrolyzing mitochondrial DNA (mtDNA) to release mtDNA fragments for the activation of the cGAS-STING pathway. In conclusion, we designed a mitochondria-targeting phosphorescent Ir(III) complex with dual functions in dysregulation of zinc homeostasis and generation of nuclease in situ, which provides an innovative approach to stimulate the cGAS-STING pathway.
Introduction
Cyclic GMP-AMP synthase (cGAS) is an intracellular sensing molecule that can recognize DNA within cells, usually originating from pathogens, and activates the stimulator of interferon genes (STING) pathway, thereby triggering antitumor immune responses.1,2 The cGAS-STING pathway holds significant promise in cancer treatment, especially in enhancing the immune response and advancing combination immunotherapy.3,4 However, further research and clinical trials are essential to fully harness its potential in cancer treatment.5,6 Specifically, the development of small-molecule activators of the cGAS-STING pathway is still very limited.7 Cyclic dinucleotides (CDNs) and their analogs, such as ADU-S100 (ref. 8) and MK-1454,9,10 have entered clinical trials. A series of non-nucleotide small-molecule STING agonists, such as diABZI and its derivatives,11,12 MSA-2,13 SR-717,14 NVS-STG2,15 and PDIC-NS16 as well as multivalent polymers like PC7A,17,18 were reported to be effective activators of the cGAS-STING pathway. Diverse nanomaterials, e.g., inorganic nanomaterials, organic polymeric materials, lipid-based nanomaterials, and biomimetic nanomaterials have been developed for the encapsulation and delivery of CDNs or STING agonists.19–23 Specifically, intracellular metal ions, e.g., Mn2+ (ref. 24) and Zn2+,25,26 have demonstrated regulatory effects on the cGAS-STING pathway. Additionally, metal complexes, such as platinum,27–31 rhodium,32 iridium,33,34 ruthenium,35,36 manganese,37 gold,38 and osmium complexes,39 have shown potential in STING activation. Moreover, the combination therapy of STING agonists with immune checkpoint inhibitors (ICIs) has been proven to further enhance anti-tumor immune responses.40–43As the second most abundant transition metal element in the human body, zinc not only acts as a cellular signaling mediator to regulate signaling pathways and enzyme activities, but also participates in the regulation of cellular ionic homeostasis and biomolecule metabolism as protein components and enzyme cofactors.44,45 The intracellular concentration of labile zinc ranges from pM to nM,46 and the cellular zinc homeostasis is regulated by complex mechanisms. Important components of this regulation include zinc finger proteins, superoxide dismutase (SOD) and metal-responsive element-binding transcription factor (MTF)-1, and the zinc transporter proteins, including Zrt/Irt-like protein (ZIP, SLC39A) family, Zn2+ transporter protein (ZnTs, SLC30A) family, and metallothioneins (MTs).47–49 Zn2+ has been found to function in various physiological processes, including neuronal transmission and cancer immunity.25 Dysregulation of zinc homeostasis is closely associated with cancer development and progression, with abnormal accumulation of zinc observed in breast cancer patients.50,51 Researchers such as Lippard,52 Guo,46 Nam,53–55 and Nagano56et al. designed numerous imaging probes for detecting changes in and localization of intracellular chelatable zinc.
Cyclometallic iridium complexes have gained widespread use in bioimaging and antitumor therapy attributed to their excellent photophysical and biochemical properties including large Stokes shifts, high photostability, two-photon absorption and long-lifetime phosphorescence emission, adjustable spectral characteristics, efficient cellular uptake with specific subcellular localization, diverse anti-tumor mechanisms and multi-target therapeutic mechanisms.57–59 Ir(III) complexes have been reported as phosphorescent imaging agents for intracellular labile zinc.53,60,61 These phosphorescent metal complexes exhibit strong binding capacity and effective imaging affinity for intracellular labile zinc, while their potential impacts on zinc homeostasis modulation and antitumor mechanisms have yet to be explored.
Based on the above studies, a series of cyclometallic Ir(III) complexes Ir1–Ir4 with ditopic zinc ion-binding moiety 2,2′-dipicolylamine (DPA), are designed as mitochondrial DNA (mtDNA) nucleases activated by endogenous zinc (Scheme 1A). Ir1–Ir4 can bind with 2 equiv. of Zn2+via DPA moieties and exhibit a corresponding phosphorescence response. The most active complex, Ir1, can bind chelatable zinc in mitochondria and enrich intracellular zinc into mitochondria, which breaks the zinc homeostasis by affecting zinc regulatory proteins. Upon binding with Zn2+, the as formed Ir1-Zn2 can bind to DNA more tightly, and act as mtDNA-targeted nucleases. By acting as a nuclease mimic to impair mtDNA and as a ROS producer to damage nuclear DNA (nucDNA), Ir1 can further activate the cGAS-STING pathway. Finally, in vivo experiments demonstrate that the combination therapy of Ir1 and programmed death-ligand 1 (PD-L1) inhibitor BMS-1 can effectively inhibit the growth of primary and recurrent tumors, highlighting its promising potential for antitumor immunotherapy. In summary, our work reports a phosphorescent Ir(III) complex that disrupts intracellular zinc homeostasis as well as utilizes endogenous metals to achieve artificial metalloenzyme simulation, providing a novel strategy for the activation of the cGAS-STING pathway.
Results and discussion
Ir1–Ir4 exhibit Zn2+-responsive phosphorescence
Ir1 was synthesized by the literature method with slight modifications,62 and Ir2–Ir4 were synthesized using a similar method (Scheme S1). Briefly, the corresponding chloro-bridged precursors and the N^N ligand 4,4′-bis(dipicolylaminomethyl)-2,2′-bipyridine (dDPA-bpy) were heated to reflux in CH2Cl2/CH3OH (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for 6 h to obtain the crude product, which was further purified by silica gel column chromatography. Ir1a–Ir4a without the DPA moieties were synthesized as the controls, among which Ir1a, Ir3a and Ir4a were synthesized by the literature methods,63–65 and Ir2a was synthesized by a similar method (Scheme S1). All the complexes were characterized by ESI-MS, 1H NMR and 13C NMR, and their purities were proved by HPLC (Fig. S1–S25).
1, v/v) for 6 h to obtain the crude product, which was further purified by silica gel column chromatography. Ir1a–Ir4a without the DPA moieties were synthesized as the controls, among which Ir1a, Ir3a and Ir4a were synthesized by the literature methods,63–65 and Ir2a was synthesized by a similar method (Scheme S1). All the complexes were characterized by ESI-MS, 1H NMR and 13C NMR, and their purities were proved by HPLC (Fig. S1–S25).
The UV/Vis absorption spectra of Ir1–Ir4 in PBS, CH2Cl2 and CH3CN show the strong intra-ligand charge transfer (ILCT) at 250–320 nm and less intense spin-allowed/spin-forbidden metal-to-ligand charge transfer (1MLCT/3MLCT) at 320–500 nm (Fig. 1A, S26 and Table S1).66 The absorption values of Ir1–Ir4 at ILCT bands are higher than those of Ir1a–Ir4a (Fig. S26), which is caused by the increased energy of ILCT by DPA substitution. Under excitation at 405 nm, all the complexes emitted yellow-to-red emission in PBS, CH2Cl2 and CH3CN, with quantum yields ranging from 0.002 to 0.16 and phosphorescence lifetimes ranging from 36.46 to 1485.53 ns (Fig. 1B, S27 and Table S1). The oil-water partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w) of Ir1–Ir4 range between 1.24 and 3.15, while log
Po/w) of Ir1–Ir4 range between 1.24 and 3.15, while log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w of Ir1a–Ir4a range between −0.76 and −0.02 (Table S2), which indicates that Ir1–Ir4 possess superior lipophilicity and biocompatibility to Ir1a–Ir4a. The UV/Vis absorption spectra exhibit a linear relationship between the absorbance and concentration of Ir1, Ir3, Ir4 and Ir1a–Ir4a up to 100 µM in PBS buffer (with 1% DMSO, Fig. S28), demonstrating their sufficient aqueous solubility in aqueous media under the experimental conditions. The aqueous solubility of Ir2 is within 40 µM, which can be attributed to its superior lipophilic character limiting its water solubility.
Po/w of Ir1a–Ir4a range between −0.76 and −0.02 (Table S2), which indicates that Ir1–Ir4 possess superior lipophilicity and biocompatibility to Ir1a–Ir4a. The UV/Vis absorption spectra exhibit a linear relationship between the absorbance and concentration of Ir1, Ir3, Ir4 and Ir1a–Ir4a up to 100 µM in PBS buffer (with 1% DMSO, Fig. S28), demonstrating their sufficient aqueous solubility in aqueous media under the experimental conditions. The aqueous solubility of Ir2 is within 40 µM, which can be attributed to its superior lipophilic character limiting its water solubility.
Job's plots of emission spectra confirm that Ir1–Ir4 have 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 binding stoichiometry with Zn2+ (Fig. S29). Moreover, 1H NMR spectra showed that H atoms of two DPA moieties display a low-field shift and the signal gradually weakens upon titration with Zn2+, suggesting that Ir1–Ir4 bind with Zn2+ through six N atoms of ditopic DPA moieties (Fig. 1C and S30). A hypochromic effect is observed for UV/Vis titration of Ir1–Ir4 with Zn2+, and the binding constants of Ir1, Ir2, Ir3 and Ir4 calculated by using the Benesi–Hildebrand equation are 2.34 × 1010, 9.89 × 109, 8.26 × 109 and 1.21 × 1010 M−2, respectively (Fig. 1D, S31, S32 and Table S3).67 Red shifts are observed upon the phosphorescence titration of Ir1–Ir4 with Zn2+, and the emission intensity of Ir3 increases, whereas the other complexes show phosphorescence quenching. The binding constants of Ir1, Ir2, Ir3 and Ir4 obtained by emission titration are 2.01 × 1010, 1.08 × 1010, 1.02 × 1010 and 3.81 × 1010 M−2, respectively (Fig. 1E, S31 and S32). These binding constants demonstrate that they have high Zn2+ binding affinity compared with the Zn2+ sensors reported previously.68–70 Upon titration with Zn2+, the phosphorescence decay curves show that Zn2+ can decrease the lifetimes of Ir1–Ir4 (Fig. 1E, S33 and Tables S3–S6). For Ir1, the short lifetime component (τ1) varies slightly in the range of about 70.74–198.04 ns. The long lifetime component (τ2) quenches as the ratio of Zn2+/Ir1 increases, which can be utilized for intracellular two-photon phosphorescence lifetime imaging microscopy (TPPLIM) to analyze the cellular labile Zn2+ level (Fig. 1F). The control complexes Ir1a–Ir4a showed no response towards Zn2+, further verifying that Zn2+ binds with Ir1–Ir4 through DPA moieties (Fig. S34).
2 binding stoichiometry with Zn2+ (Fig. S29). Moreover, 1H NMR spectra showed that H atoms of two DPA moieties display a low-field shift and the signal gradually weakens upon titration with Zn2+, suggesting that Ir1–Ir4 bind with Zn2+ through six N atoms of ditopic DPA moieties (Fig. 1C and S30). A hypochromic effect is observed for UV/Vis titration of Ir1–Ir4 with Zn2+, and the binding constants of Ir1, Ir2, Ir3 and Ir4 calculated by using the Benesi–Hildebrand equation are 2.34 × 1010, 9.89 × 109, 8.26 × 109 and 1.21 × 1010 M−2, respectively (Fig. 1D, S31, S32 and Table S3).67 Red shifts are observed upon the phosphorescence titration of Ir1–Ir4 with Zn2+, and the emission intensity of Ir3 increases, whereas the other complexes show phosphorescence quenching. The binding constants of Ir1, Ir2, Ir3 and Ir4 obtained by emission titration are 2.01 × 1010, 1.08 × 1010, 1.02 × 1010 and 3.81 × 1010 M−2, respectively (Fig. 1E, S31 and S32). These binding constants demonstrate that they have high Zn2+ binding affinity compared with the Zn2+ sensors reported previously.68–70 Upon titration with Zn2+, the phosphorescence decay curves show that Zn2+ can decrease the lifetimes of Ir1–Ir4 (Fig. 1E, S33 and Tables S3–S6). For Ir1, the short lifetime component (τ1) varies slightly in the range of about 70.74–198.04 ns. The long lifetime component (τ2) quenches as the ratio of Zn2+/Ir1 increases, which can be utilized for intracellular two-photon phosphorescence lifetime imaging microscopy (TPPLIM) to analyze the cellular labile Zn2+ level (Fig. 1F). The control complexes Ir1a–Ir4a showed no response towards Zn2+, further verifying that Zn2+ binds with Ir1–Ir4 through DPA moieties (Fig. S34).
We screened the selectivity of the phosphorescence response of the complexes for various essential metal ions (K+, Na+, Ca2+, Mg2+, Mn2+, Fe3+, Fe2+, Ni2+, Co2+, Cr3+, Cu+ and Cu2+) and common biomolecules (GSH, BSA, RNA and DNA; Fig. 1H and S35A–C). Fe2+, Fe3+ and Mn2+ can diminish the emission intensity of Ir1–Ir4 to various degrees, and the biomolecules also have certain effects on them. After adding 2.0 equiv. of Zn2+, Ir1–Ir4 can reverse the phosphorescence changes and retain their response to Zn2+, demonstrating their preferential binding selectivity for Zn2+. Paramagnetic Ni2+, Co2+ and Cu2+ can partially quench the emission of Ir1–Ir4 and affect their responses to Zn2+, and Cu+ will completely quench the emission of Ir1–Ir4, which is similar to the Zn2+ sensors previously reported.53,55,60,61 Most metal ions and biomolecules cannot affect the phosphorescence lifetime response of Ir1–Ir4 to Zn2+, except for paramagnetic Ni2+, Co2+ and Cu2+ inhibiting the lifetime quenching effect of Zn2+ towards Ir1–Ir4 to a certain degree (Fig. 1I and S33D–F). However, the significant disparity between the cellular concentrations of free copper (≈10−3 fM)71,72 and cellular labile zinc (≈0.1–104 pM)46 implies that Cu2+ and Cu+ will show little effect on the phosphorescence responses of Ir1 to intracellular Zn2+.
The photophysical–chemical process of Ir1–Ir4 titrated with Zn2+ can be explained by the photoinduced electron transfer (PET) mechanism.73 Generally, Zn2+ will inhibit the PET process and enhance the emission intensity of “turn-on” sensors, such as Ir3.55,60,61 For Ir1, Ir2 and Ir4, the coordination of DPA moieties with Zn2+ will strengthen their PET process and quench the phosphorescence emission (Fig. 1J).53,73,74 However, considering that there are two coordination centers in one complex molecule, the intramolecular interactions among different ligands and metal centers are too intricate to partition the complexes into phosphorescence structures and ionophore structures.75 Thus, the PET mechanism can only crudely explain the phosphorescence response of the compounds towards Zn2+.
Therefore, density functional theory (DFT) and time-dependent density functional theory (TD-DFT) were further applied to examine the energy change of Ir1 titrated with Zn2+. Ir1 has a group of triplet excited states composed of complicated intramolecular electron transitions (Table S7). The first triplet state T1 is constructed from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) transition, assignable as triplet metal-to-ligand charge transfer (3MLCT) and triplet ligand-to-ligand charge transfer (3LLCT, Fig. S36). The second triplet state T2 arises from HOMO−8 (27.2%), HOMO−2 (20.7%), HOMO−4 (14.6%) to the LUMO, which can be attributed to 3MLCT, 3LC^NLN^NCT and triplet intra-ligand charge transfer (3ILN^NCT, Fig. S36).60 DFT calculations of the adduct of Ir1 with 2 equiv. of Zn2+ (Ir1-Zn2) show that the HOMO energy level of Ir1-Zn2 is close to that of Ir1, while the LUMO energy level of Ir1-Zn2 is lower than that of Ir1 (Fig. S37). The narrowed energy gap explains the red-shift emission of Ir1 upon titration with Zn2+.74 In addition, HOMO−8, HOMO−2, and HOMO−4 orbitals of Ir1-Zn2 are stabilized, which is significantly different from those of complex Ir1, implying that the binding of Zn2+ causes a significant change in the intramolecular charge distribution of Ir1 (Fig. S37). The composition of higher-energy 3LC^NLN^NCT decreases, while the composition of lower-energy 3ILN^NCT increases, resulting in a decrease in the energy of T2, shortening the phosphorescence lifetime and reducing the emission intensity of T2 (Fig. 1K). Additionally, a significant reduction in the T2 excited state energy to a similar energy of T1 interferes with the photon release from T1 to S0. The increase of the energy gap between S2 and T2 is unfavorable for the intersystem crossing (ISC) of electrons and the accumulation of triplet excitons, which further leads to the weakening of the phosphorescence emission (Fig. 1K).
Ir1–Ir4 show potent antiproliferative activity in vitro
It has been reported that the zinc content in breast cancer cells is abnormally higher than that in normal breast tissue,51 which encourages us to explore the antitumor activities of Ir1–Ir4 on breast cancer cell lines. The antiproliferative activity of Ir1–Ir4 on human triple negative breast cancer (MDA-MB-231), human breast cancer (MCF-7), mouse triple negative breast cancer (4T1), human normal breast epithelial cells (MCF-10A) and cervical cancer cells (HeLa) was assessed by MTT assay (Table 1). The growth inhibitory activities of Ir1–Ir4 in MDA-MB-231 cells are about 12.3–51.7 times that of cisplatin. The order of growth inhibitory activities of Ir1–Ir4 towards MDA-MB-231 cells is as follows: Ir2 ≈ Ir4 > Ir1 > Ir3. However, the IC50 of complexes in MCF-10A cells is ordered as follows: Ir2 > Ir3 > Ir1 ≈ Ir4. The IC50 of Ir1a–Ir4a on MDA-MB-231 cells falls within the range of 3.3–6.3 µM, in which Ir1a is 7.4-fold less active than Ir1 (Table S8). Additionally, studies indicate that cervical cancer cells exhibit low zinc levels,76 which is validated by inductively coupled plasma-mass spectrometry (ICP-MS, Fig. S38). MTT assays demonstrate that Ir1–Ir4 and Ir1a–Ir4a exhibit comparable toxicity in HeLa cells (Table 1 and S8), suggesting that the zinc-targeting design of Ir1–Ir4 enhances their cytotoxic effects in breast cancer cells with elevated zinc content. These results prove that Ir1–Ir4 possess strong anti-proliferative activities against breast cancer cell lines.| Complex | MDA-MB-231 | MCF-7 | 4T1 | MCF-10A | HeLa |
|---|---|---|---|---|---|
| a Cells were incubated with the compounds for 72 h. Data are presented as the means ± standard deviations (SD). | |||||
| Ir1 | 0.85 ± 0.05 | 3.72 ± 0.16 | 8.70 ± 0.49 | 2.85 ± 0.28 | 1.88 ± 0.35 |
| Ir2 | 0.39 ± 0.02 | 1.60 ± 0.05 | 3.02 ± 0.48 | 1.29 ± 0.09 | 1.12 ± 0.02 |
| Ir3 | 1.64 ± 0.10 | 2.30 ± 0.16 | 4.83 ± 0.90 | 2.41 ± 0.25 | 1.17 ± 0.05 |
| Ir4 | 0.42 ± 0.04 | 3.53 ± 0.71 | 4.60 ± 0.74 | 2.88 ± 0.11 | 1.27 ± 0.44 |
| Cisplatin | 20.16 ± 5.60 | 18.30 ± 5.74 | 25.50 ± 2.52 | 15.50 ± 4.33 | 8.35 ± 0.86 |
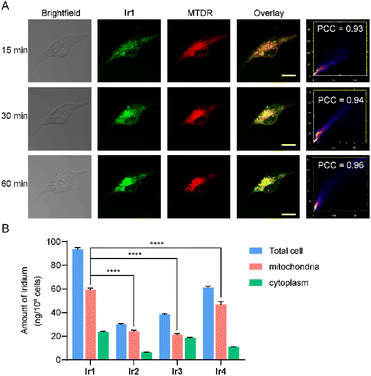
Ir1 colocalizes with Mito-Tracker Deep Red (MTDR) with a high Pearson's correlation coefficient (PCC = 0.93–0.96, Fig. 2A), while it localizes weakly with ER-Tracker Red (ETR, PCC = 0.78) or Lyso-Tracker Red (LTR, PCC = 0.52, Fig. S39). Significantly, Ir1 is already localized to mitochondria after 15-minute incubation in MDA-MB-231 cells, and MTDR exhibits mitochondrial filamentous structures. Mitochondrial damage occurs in MDA-MB-231 cells treated with Ir1 for 30 minutes, revealing some punctate structures. At 1 hour, MTDR displays a greater number of punctate structures, confirming that Ir1 caused severe mitochondrial damage. Meanwhile, colocalization experiments show that Ir2–Ir4 and Ir1a–Ir4a are mainly localized in mitochondria after 1-h incubation (Fig. S40).
The cellular uptake capabilities of Ir1–Ir4 in MDA-MB-231 cells were measured by ICP-MS (Fig. 2B). Although all the complexes are accumulated in the mitochondria of MDA-MB-231 cells, Ir1 shows the highest cellular uptake levels. The total cellular uptake of Ir1 is 1.5–3.0 times that of Ir2–Ir4, while its mitochondrial uptake is 1.3–2.8 times that of Ir2–Ir4. We further investigated the cellular uptake levels of Ir1a–Ir4a, which were lower than those of Ir1–Ir4 (Fig. S41). Considering the antitumor activities and mitochondrial accumulation levels of Ir1–Ir4, Ir1 was selected for further study.
Ir1 can image chelatable Zn2+ by TPPLIM and affect intracellular zinc homeostasis
As the long-lifetime component (τ2) value of Ir1 shows a good linear response to the change of the proportion of bound Zn2+, we chose Ir1 for further studies. Two-photon phosphorescence lifetime imaging microscopy (TPPLIM) shows that the fitted value of τ2 is approximately 900 ns at 15 min, which corresponds to the τ2 lifetime of Ir1 alone. After 1 h, τ2 of Ir1 decreases to approximately 600 ns, and the TPPLM image at 6 h is similar to that at 1 h (Fig. 3A). By correspondence analysis with the previous phosphorescence lifetime measurements (Fig. 3B), these results show that the concentration ratio of Zn2+ to Ir1 was about 0.6 (Fig. 3C), indicating that about 30% of Ir1 binds to labile Zn2+ in mitochondria at this point. ICP-MS measurement shows that both cellular zinc content and mitochondrial zinc content are increased in Ir1-treated cells, confirming that Ir1 induces the accumulation of zinc in mitochondria (Fig. 3D). Furthermore, the treatment with Ir1 elevated zinc levels in the nucleus, but did not significantly alter ER zinc content (Fig. S42). This compartment-specific effect indicates dysregulation of cellular zinc homeostasis. In contrast, although Ir1a also tends to accumulate in mitochondria, it shows minimal impact on zinc contents in whole cells and mitochondria (Fig. 3D).To further confirm the effects of Ir1 on Zn2+ cellular homeostasis, we analyzed the proteins related to zinc homeostasis regulation from RNA-sequencing (RNA-seq) data. A series of Zn2+-regulated proteins is dysregulated in Ir1-treated cells, confirming the dysregulation of zinc homeostasis due to intervention in the expression of zinc-related proteins within cells (Table S11). Gene set enrichment analysis (GSEA) shows that the response to zinc ions is upregulated in the Ir1-treated group (Fig. 3E). The alterations in the expression of zinc-related proteins caused by Ir1 treatment are also detected by the real-time quantitative polymerase chain reaction (RT-qPCR, Fig. 3F). After treatment with Ir1, the ZIP family protein SLC39A5 is up-regulated by 2.04-fold, whereas the expression of ZnT family protein SLC30A10 is significantly down-regulated by about 1.80-fold. Most of the MT proteins for Zn2+ buffering and storage are also up-regulated to various degrees. Western blot detects that the expression levels of SLC39A5 and MT protein MT1X are up-regulated, conforming with the results of the RT-qPCR (Fig. 3G). Given that MT1X is mainly expressed in the cell nucleus,77 its upregulation can explain the increase in zinc content in the cell nucleus induced by Ir1. From these data, we propose that Ir1-treatment increases the intracellular level of Zn2+ by dysregulating zinc transporter proteins and metallothioneins, and further sequesters mitochondrial labile zinc by targeting mitochondria.
Ir1 acts as a mtDNA-targeted nuclease mimic upon Zn2+ binding
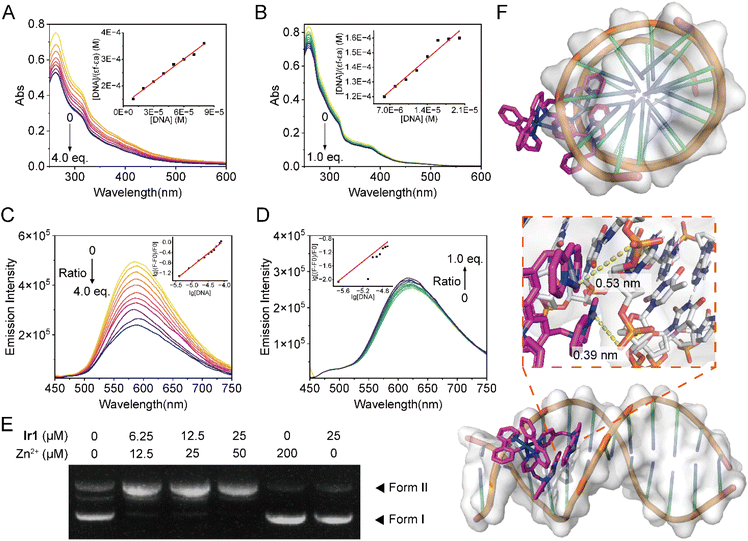
Due to the fact that zinc is the active center of nucleases, and it has been reported that simple zinc complexes can mimic the activity of nucleases,78 we then studied the DNA binding and cleavage activities of Ir1 and its Zn2+-binding form Ir1-Zn2. The binding constants of Ir1 and Ir1-Zn2 towards calf thymus DNA (ct-DNA) measured by UV/Vis titration are (2.18 ± 0.17) × 105 M−1 and (3.11 ± 0.37) × 105 M−1, respectively, which indicates that they possess moderate DNA binding affinities (Fig. 4A and B). The binding constants of Ir1 and Ir1-Zn2 obtained by emission titration are (1.24 ± 0.46) × 105 M−1 and (3.15 ± 1.23) × 105 M−1, respectively (Fig. 4C and D), which also indicates that binding of Zn2+ can increase the DNA binding affinity of Ir1. Upon DNA binding, the emission intensity of Ir1 and Ir1-Zn2 is decreased and increased, respectively, which implies their different DNA binding modes. Agarose gel electrophoresis shows that Ir1 or Zn2+ alone can't cleave plasmid DNA, while Ir1-Zn2 can cause DNA degradation (Fig. 4E), indicating the nuclease mimetic activity of Ir1-Zn2.Molecular docking studies show that the binding energies of Ir1 and Ir1-Zn2 with double-stranded DNA (ds-DNA) are (−8.32 ± 0.49) and (−9.77 ± 0.68) kcal mol−1, respectively, which further proves that Zn2+ coordination increases the DNA-binding capabilities of Ir1 (Tables S9 and S10). The most stabilized configuration from docking shows that Ir1 and Ir1-Zn2 are located within the minor groove (Fig. 4F and S41). The viscosity of ct-DNA decreases slightly in the presence of Ir1 or Ir1-Zn2, which verifies that compounds are more likely to bind within the DNA grooves, causing the DNA helix to bend and twist, thereby reducing its effective length and consequently lowering the viscosity (Fig. S42).79 Hydrophobic interactions between ds-DNA and Ir1-Zn2 stabilized the docking form, prompting the DPA moieties of Ir1-Zn2 to intercalate into ds-DNA more thoroughly than Ir1. Generally, zinc complexes cleave DNA through hydrolysis of phosphodiester bonds and nucleophilic attack.80 The molecular docking calculation shows that the distances between Zn ions and oxygen atoms of phosphodiester bonds are 0.39 and 0.53 nm (Fig. 4F), which implies the existence of the weak interactions and the potential trend for DNA hydrolysis.
Ir1 activates the cGAS-STING pathway by damaging mtDNA
Transmission electron microscopy (TEM) observation reveals that Ir1 triggers necrosis, with notable mitochondrial damage and numerous cytoplasmic vacuoles (Fig. 5A). The type of cell death of Ir1 was investigated by measuring the cell viabilities of MDA-MB-231 cells with pretreatment of different cell death inhibitors for 1 h and further treatment of Ir1 (10 µM, 24 h). Only the necrostatin-1 (Nec-1, necroptosis inhibitor) and necrosulfonamide (NSA, necroptosis inhibitor) can improve the cell viabilities of MDA-MB-231 cells with treatment of Ir1, suggesting that Ir1 can induce necrosis (Fig. 5B). Otherwise, Z-VAD-FMK (pan-caspase inhibitor), 3-methyladenine (3-MA, autophagy inhibitor), ferrostatin-1 (Fer-1, ferroptosis inhibitor) and liproxstatin-1 (Lip-1, ferroptosis inhibitor) show limited effects on inhibition of antiproliferative activities of Ir1.The reactive oxygen species (ROS) levels in MDA-MB-231 cells increase in a dose-dependent manner after Ir1 treatment (Fig. 5C). As measured by 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) staining and flow cytometry, the cellular ROS values increase 7-fold in MDA-MB-231 cells incubated with Ir1 at a concentration of 16 µM for 6 h. In addition, 96% of MDA-MB-231 cells show depolarized mitochondria with lost mitochondrial membrane potential (MMP) after 4-µM Ir1 treatment for 6 h, as measured by 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl-carbocyanine iodide (JC-1) staining and flow cytometry (Fig. 5D). The human mitochondrial genome encodes 37 genes, including 13 critical genes that encode essential components of the oxidative phosphorylation (OXPHOS) system.81 RNA-seq reveals that the expression of the 13 genes encoded by the mitochondrial genome is attenuated (|fold change| ≥ 2; false discovery rate ≤ 0.05, Table S12). The real-time quantitative polymerase chain reaction (RT-qPCR) shows that the expression of all 13 genes in Ir1-treated MDA-MB-231 cells is down-regulated by 1.50–2.51 times (fold change >2, Fig. 5E), further demonstrating that Ir1 can effectively damage mitochondrial function through generation of ROS and disruption of mitochondrial gene expression.
In addition, the morphology of mitochondria gradually disperses from a clear filamentous structure in Ir1-treated cells (Fig. 5F). After 15–30 min treatment, the fluorescence of PicoGreen gradually separates from that of MTDR, suggesting that mtDNA is released in the cytoplasm. In contrast, MTDR and PicoGreen show a good co-localization pattern in control cells. Additionally, Ir1a can't induce the dispersion of fluorescence of MTDR and PicoGreen, indicating its disability to damage mtDNA (Fig. S43). After Ir1 treatment (2.0 µM), the copy number of mtDNA is reduced by about 58% compared with that of nuclear DNA (nucDNA; Fig. 5G). Gel electrophoresis reveals that Ir1 causes significant mtDNA fragmentation in MDA-MB-231 cells (Fig. 5H). Meanwhile, the expression of γH2A.X, a marker of nuclear DNA damage and repair, is dose-dependently up-regulated after Ir1 treatment (Fig. 5I). Incubation of N-acetylcysteine (NAC, an ROS scavenger) can partially reduce the expression of γH2A.X, which indicates that Ir1 also induces nucDNA damage through generation of ROS (Fig. 5K). Western blot detects that the expression levels of cGAS and p-STING are elevated (Fig. 5I), while NAC treatment partially abrogates cGAS and p-STING expression (Fig. 5K), demonstrating that Ir1 activates the cGAS-STING pathway through nucDNA damage as an ROS producer and mtDNA damage as nuclease mimics. It has been reported that PD-L1 expression is related to ROS accumulation.82 Western blot confirms that Ir1 can increase the expression of PD-L1 in MDA-MB-231 cells (Fig. 5J), and NAC can substantially reduce Ir1-induced PD-L1 upregulation (Fig. 5K), which inspires us to investigate whether Ir1 can synergize with immune checkpoint inhibition in vivo.
Impact of Ir1 on the transcriptome
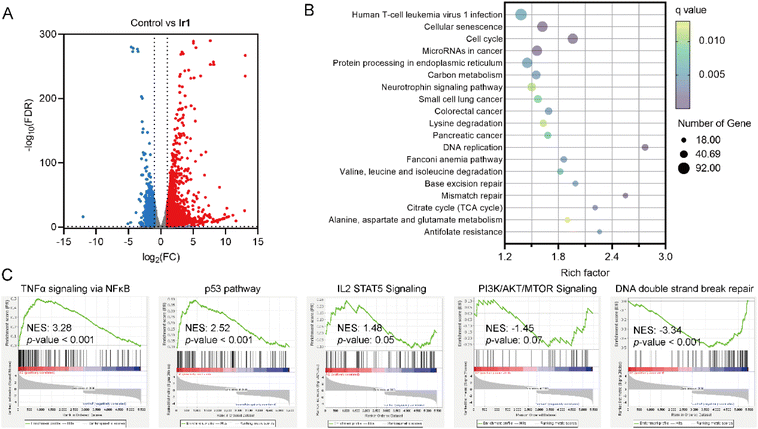
RNA-sequencing (RNA-seq) was performed to further investigate the impact of Ir1 on the gene expression pattern. The correlation coefficients between every two individual samples from the same group are greater than 0.95 (Fig. S44), indicating that the data are repeatable. Cluster analysis and the heatmap show that the differentially expressed genes (DEGs) induced in the Ir1-treated cells present a significant difference in expression patterns compared to the control group, suggesting the reproducibility of the data (Fig. S45). 4756 significantly differentially expressed genes (|fold change| ≥ 2; false discovery rate ≤0.05; upregulated: 3149 genes; downregulated: 1607 genes) are detected in Ir1-treated cells compared with the control cells (Fig. 6A).Gene Ontology (GO) enrichment analysis reveals that Ir1 mainly influences the DNA replication, cell cycle, nucleus, mitochondrion, DNA binding, ATP binding and ion channel binding (Fig. S46–S48). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis shows that Ir1 mainly influences DNA replication, cell cycle and mismatch repair (Fig. 6B). Gene set enrichment analysis (GSEA) shows that TNF-α signaling via the NF-κB, p53 pathway and IL2 STAT5 signaling are upregulated in the Ir1-treated group, while DNA double strand break repair and PI3K/AKT/mTOR signaling are dysregulated in the Ir1-treated group (Fig. 6C). These results suggest that Ir1 mainly exerts its anticancer effects through the induction of DNA damage and immune responses.
Ir1 synergistically acts with ICIs to effectively improve the immune TMEs
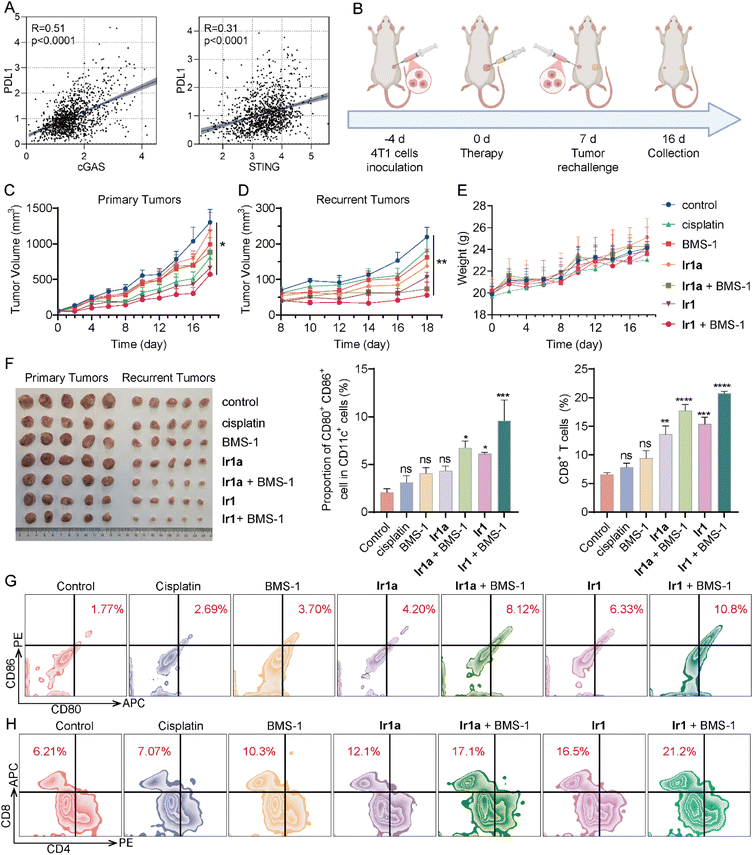
Recent studies highlight that the depletion of PD-L1 is associated with STING activation, which can inhibit cancer growth.83 The Cancer Genomic Atlas (TCGA) database analysis shows that the correlation R-values of PD-L1 with cGAS and STING reach 0.51 and 0.31 (p-values <0.001, Fig. 7A), respectively, indicating that they are closely associated. As one of the small-molecule PD-L1 inhibitors, BMS-1 has been extensively utilized in various antitumor studies,84–86 and it is chosen as the synergistic ICI drug with Ir1.To assess the biodistribution of Ir1, we administered Ir1 (5 mg kg−1) via intratumoral injection to tumor-bearing mice and measured drug concentrations in tissues, tumors, and blood at 24 hours, 48 hours, and 7 days post-administration (Fig. S49). ICP-MS analysis revealed iridium accumulation within tumors at 24 h, decreasing by approximately 27.2% at 48 hours, with residual intertumoral levels of 2.9% at 7 days. Following administration, the drug accumulated in the liver and kidneys, with both organs showing significant reduction by day 7. Concentrations in blood decreased by approximately 93.2% at day 7 compared to 24 h post-administration. These results confirm that the drug undergoes metabolism via the liver and kidneys and there is almost no drug residue in the mice after 7 days of administration. Blood routine tests further demonstrate that hematological parameters are all in the normal range, indicating that Ir1 has no obvious toxicity in mice (Fig. S50).
We established a tumor rechallenge model to evaluate immunization vaccination, thereby evaluating the effectiveness of tumor immunotherapy (Fig. 7B). 4T1 cells (1× 106 cells/100 µL PBS) were inoculated subcutaneously into BALB/C female mice as primary tumors. 4 days later, mice were randomized into 7 groups (control, cisplatin, BMS-1, Ir1, Ir1a, Ir1 + BMS-1, and Ir1a + BMS-1), followed by a single intertumoral treatment with corresponding drugs (5 mg kg−1). After 7 days, 4T1 cells (2× 106 cells/100 µL PBS) were implanted contralaterally as recurrent tumors. Tumor volume and body weight were monitored for 18 days post-administration.
At the end of treatment, the suppression effects on primary tumors are ordered as follows: Ir1 + BMS-1 > Ir1 > cisplatin > BMS-1 ≈ Ir1a + BMS-1 > Ir1a (Fig. 7C and F), suggesting that Ir1 alone or in combination with BMS-1 has an optimal antitumor effect compared to cisplatin and Ir1a alone or in combination with BMS-1. Simultaneously, the inhibition rates of Ir1 + BMS-1 and Ir1a + BMS-1 on recurrent tumors are 74.5% and 66.4%, showing better performance compared with the inhibition rates of Ir1, Ir1a (37.2%), cisplatin (16.9%), and BMS-1 (25.9%, Fig. 7D and F). These results indicate that the combination of complexes with ICIs can significantly improve the antitumor immune response. All the groups maintained stable body weights without significant differences (Fig. 7E), and HE staining shows that no serious structural or pathological alterations are observed in the major organs of mice in all treatment groups (Fig. S51), indicating that Ir1 alone or in combination with BMS-1 has good biocompatibility.
Flow cytometry reveals an elevation in CD86+ CD80+ cells in the Ir1 + BMS-1 treatment group (from about 1.77% to 10.8%; Fig. 7G), suggesting that the combination therapy promotes the maturation of DC cells. The impact of either cisplatin or BMS-1 on DC cells is not obvious. Furthermore, the treatment of Ir1 + BMS-1 induces an increase in the percentage of CD8+ T cells, from 6.21% to 21.2% (Fig. 7H). Both Ir1a + BMS-1 and Ir1 treatment groups show certain effects on DC cell maturation and CD8+ T cell infiltration.
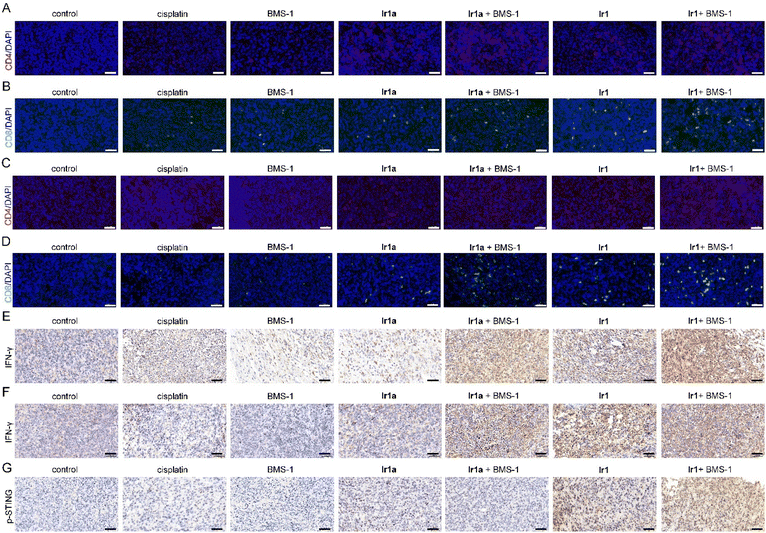
Immunofluorescence (IF) staining shows that Ir1, Ir1 + BMS-1, and Ir1a + BMS-1 treatment stimulates CD4/CD8 T cell infiltration into tumor tissues in primary and recurrent tumors, which is consistent with the results of flow cytometry (Fig. 8A–D). Immunohistochemistry (IHC) demonstrates that the combined treatment of metal complexes and BMS-1 increases the expression of interferon-γ (IFN-γ), one of the cytokines for tumor immune regulation (Fig. 8E and F). In addition, Ir1 effectively elevates the expression of p-STING on primary tumors, whereas Ir1a or cisplatin can't activate the STING pathway in vivo (Fig. 8G). These results indicate that Ir1 can effectively inhibit tumor growth and activate immune responses, and it further can synergistically act with immune checkpoint inhibitors to effectively improve the immune TMEs.
Conclusions
We have designed a series of iridium complexes that can sequester zinc in mitochondria. By utilizing the specific response phosphorescence lifetimes of Ir1 to zinc and TPPLIM, we prove that Ir1 can chelate zinc in mitochondria. Interestingly, the in situ formation of the heteronuclear complex Ir1-Zn2 exhibits nuclease-mimetic activity, enabling the degradation of mtDNA. This process facilitates the cytoplasmic release of mtDNA fragments to activate the cGAS-STING signaling pathway. As a consequence, Ir1 can induce tumor cell necrosis by increasing mitochondrial ROS and breaking the zinc homeostasis. RNA-seq also proves that Ir1 modulates intracellular zinc ion homeostasis and regulates cancer immune signaling pathways. In vivo experiments reveal that Ir1 synergizes with ICIs to inhibit primary tumors and exert vaccine effects, thus improving the immune TMEs and inhibiting the growth of dismal tumors. By combining the mitochondria-targeting and imaging capabilities of Ir(III) complexes, we constructed the first phosphorescent metal complex capable of disrupting intracellular zinc homeostasis and generating a nuclease in situ, providing an efficient strategy to activate the cGAS-STING immune pathway.Ethical statement
The animal experiment procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University and performed in accordance with the ethical guidelines of Sun Yat-sen University. The accreditation number is SYSU-IACUC-2025-000460.Author contributions
Zhi-Yuan Li: conceptualization, data curation, formal analysis, visualization, writing – original draft. Long-Bo Yu: data curation, formal analysis, visualization. Qing-Hua Shen: data curation, formal analysis, visualization. Liang Hao: formal analysis, funding acquisition, visualization. Peng Wang: data curation. Xiao-Xiao Chen: visualization. Yu-Yi Ling: conceptualization, data curation, formal analysis, visualization, writing – review & editing. Cai-Ping Tan: conceptualization, funding acquisition, project administration, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
The data generated in this study are available within the article, supplementary information (SI), source data, and from corresponding authors upon request. Source data are provided in this paper. Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc07181j.Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 22177142 and 22207136), the Guangdong Basic and Applied Basic Research Foundation (No. 2024B1515040028) and the Fundamental Research Funds for the Central Universities.References
- H. Ishikawa and G. N. Barber, Nature, 2008, 455, 674 CrossRef CAS PubMed.
- A. Ablasser, M. Goldeck, T. Cavlar, T. Deimling, G. Witte, I. Röhl, K.-P. Hopfner, J. Ludwig and V. Hornung, Nature, 2013, 498, 380 CrossRef CAS PubMed.
- G. N. Barber, Nat. Rev. Immunol., 2015, 15, 760 CrossRef CAS.
- S.-R. Woo, L. Corrales and T. F. Gajewski, Trends Immunol., 2015, 36, 250 CrossRef CAS.
- N. Samson and A. Ablasser, Nat. Cancer, 2022, 3, 1452 CrossRef CAS PubMed.
- K. R. B. Lanng, E. L. Lauridsen and M. R. Jakobsen, Nat. Immunol., 2024, 25, 1144 CrossRef CAS PubMed.
- C. Y. Ding, Z. L. Song, A. C. Shen, T. T. Chen and A. Zhang, Acta Pharm. Sin. B, 2020, 10, 2272 CrossRef CAS.
- L. Corrales, L. H. Glickman, S. M. McWhirter, D. B. Kanne, K. E. Sivick, G. E. Katibah, S.-R. Woo, E. Lemmens, T. Banda, J. J. Leong, K. Metchette, T. W. Dubensky and T. F. Gajewski, Cell Rep., 2015, 11, 1018 CrossRef CAS.
- T. Benkovics, F. Peng, E. M. Phillips, C. An, R. S. Bade, C. K. Chung, Z. E. X. Dance, P. S. Fier, J. H. Forstater, Z. Liu, Z. Liu, P. E. Maligres, N. M. Marshall, N. Salehi Marzijarani, J. A. McIntosh, S. P. Miller, J. C. Moore, A. J. Neel, J. V. Obligacion, W. Pan, M. T. Pirnot, M. Poirier, M. Reibarkh, B. D. Sherry, Z. J. Song, L. Tan, B. W. H. Turnbull, D. Verma, J. H. Waldman, L. Wang, T. Wang, M. S. Winston and F. Xu, J. Am. Chem. Soc., 2022, 144, 5855 CrossRef CAS.
- W. Chang, M. D. Altman, C. A. Lesburg, S. A. Perera, J. A. Piesvaux, G. K. Schroeder, D. F. Wyss, S. Cemerski, Y. Chen, E. DiNunzio, A. M. Haidle, T. Ho, I. Kariv, I. Knemeyer, J. E. Kopinja, B. M. Lacey, J. Laskey, J. Lim, B. J. Long, Y. Ma, M. L. Maddess, B.-S. Pan, J. P. Presland, E. Spooner, D. Steinhuebel, Q. Truong, Z. Zhang, J. Fu, G. H. Addona, A. B. Northrup, E. Parmee, J. R. Tata, D. J. Bennett, J. N. Cumming, T. Siu and B. W. Trotter, J. Med. Chem., 2022, 65, 5675 CrossRef CAS.
- J. M. Ramanjulu, G. S. Pesiridis, J. Yang, N. Concha, R. Singhaus, S.-Y. Zhang, J.-L. Tran, P. Moore, S. Lehmann, H. C. Eberl, M. Muelbaier, J. L. Schneck, J. Clemens, M. Adam, J. Mehlmann, J. Romano, A. Morales, J. Kang, L. Leister, T. L. Graybill, A. K. Charnley, G. Ye, N. Nevins, K. Behnia, A. I. Wolf, V. Kasparcova, K. Nurse, L. Wang, A. C. Puhl, Y. Li, M. Klein, C. B. Hopson, J. Guss, M. Bantscheff, G. Bergamini, M. A. Reilly, Y. Lian, K. J. Duffy, J. Adams, K. P. Foley, P. J. Gough, R. W. Marquis, J. Smothers, A. Hoos and J. Bertin, Nature, 2018, 564, 439 CrossRef CAS PubMed.
- Z. L. Song, X. Y. Wang, Y. Zhang, W. T. Gu, A. C. Shen, C. Y. Ding, H. Li, R. X. Xiao, M. Y. Geng, Z. Q. Xie and A. Zhang, J. Med. Chem., 2021, 64, 1649 CrossRef CAS.
- B.-S. Pan, S. A. Perera, J. A. Piesvaux, J. P. Presland, G. K. Schroeder, J. N. Cumming, B. W. Trotter, M. D. Altman, A. V. Buevich, B. Cash, S. Cemerski, W. Chang, Y. Chen, P. J. Dandliker, G. Feng, A. Haidle, T. Henderson, J. Jewell, I. Kariv, I. Knemeyer, J. Kopinja, B. M. Lacey, J. Laskey, C. A. Lesburg, R. Liang, B. J. Long, M. Lu, Y. Ma, E. C. Minnihan, G. O’Donnell, R. Otte, L. Price, L. Rakhilina, B. Sauvagnat, S. Sharma, S. Tyagarajan, H. Woo, D. F. Wyss, S. Xu, D. J. Bennett and G. H. Addona, Science, 2020, 369, eaba6098 CrossRef CAS PubMed.
- E. N. Chin, C. Yu, V. F. Vartabedian, Y. Jia, M. Kumar, A. M. Gamo, W. Vernier, S. H. Ali, M. Kissai, D. C. Lazar, N. Nguyen, L. E. Pereira, B. Benish, A. K. Woods, S. B. Joseph, A. Chu, K. A. Johnson, P. N. Sander, F. Martínez-Peña, E. N. Hampton, T. S. Young, D. W. Wolan, A. K. Chatterjee, P. G. Schultz, H. M. Petrassi, J. R. Teijaro and L. L. Lairson, Science, 2020, 369, 993 CrossRef CAS.
- J. Li, S. M. Canham, H. Wu, M. Henault, L. Chen, G. Liu, Y. Chen, G. Yu, H. R. Miller, V. Hornak, S. M. Brittain, G. A. Michaud, A. Tutter, W. Broom, M. E. Digan, S. M. McWhirter, K. E. Sivick, H. T. Pham, C. H. Chen, G. S. Tria, J. M. McKenna, M. Schirle, X. Mao, T. B. Nicholson, Y. Wang, J. L. Jenkins, R. K. Jain, J. A. Tallarico, S. J. Patel, L. Zheng, N. T. Ross, C. Y. Cho, X. Zhang, X.-C. Bai and Y. Feng, Nat. Chem. Biol., 2023, 20, 365 CrossRef PubMed.
- X. J. Zhao, R. Zheng, B. B. Zhang, Y. Zhao, W. L. Xue, Y. Fang, Y. W. Huang and M. Z. Yin, Angew. Chem., Int. Ed., 2024, 63, e202318799 CrossRef CAS.
- S. X. Li, M. Luo, Z. H. Wang, Q. Feng, J. Wilhelm, X. Wang, W. Li, J. Wang, A. Cholka, Y.-X. Fu, B. D. Sumer, H. T. Yu and J. M. Gao, Nat. Biomed. Eng., 2021, 5, 455 CrossRef CAS.
- M. Luo, H. Wang, Z. H. Wang, H. C. Cai, Z. G. Lu, Y. Li, M. J. Du, G. Huang, C. S. Wang, X. Chen, M. R. Porembka, J. Lea, A. E. Frankel, Y.-X. Fu, Z. J. Chen and J. M. Gao, Nat. Nanotechnol., 2017, 12, 648 CrossRef CAS.
- Y. Jia, W. Jia, Z. Tang, Y. Wu, W. Yang, W. Ye, H. Ren, Y. Xie, Y. Chen and M. Yu, Angew. Chem., Int. Ed., 2025, 202514516 Search PubMed.
- K. Shang, D. Tang, M. Cui, H. Hou, B. Xiao, J. Liu, R. Zhang, R. Kuai, J. Liu, H. Xiao, F. Huang and J. Wang, Adv. Mater., 2025, 202506011 Search PubMed.
- P. Dosta, A. M. Cryer, M. Prado and N. Artzi, Nat. Rev. Bioeng., 2025, 3, 660 CrossRef CAS.
- Q. Yu, S. Sun, N. Yang, Z. Pei, Y. Chen, J. Nie, H. Lei, L. Wang, F. Gong and L. Cheng, J. Am. Chem. Soc., 2025, 147, 3161 CrossRef CAS PubMed.
- Q.-R. Li, X. Zhang, C. Zhang, Y. Zhang, M.-T. Niu, Z. Chen, S.-M. Zhang, J. He, W.-H. Chen and X.-Z. Zhang, J. Am. Chem. Soc., 2025, 147, 24555 CrossRef CAS PubMed.
- C. G. Wang, Y. K. Guan, M. Z. Lv, R. Zhang, Z. Y. Guo, X. M. Wei, X. X. Du, J. Yang, T. Li, Y. Wan, X. D. Su, X. J. Huang and Z. F. Jiang, Immunity, 2018, 48, 675 CrossRef CAS PubMed.
- Y. Yang, H. H. Fan, X. Y. Xu, S. K. Yao, W. H. Yu and Z. J. Guo, CCS Chem., 2024, 6, 2210 CrossRef CAS.
- M. Du and Z. J. Chen, Science, 2018, 361, 704 CrossRef CAS.
- Y.-Y. Ling, X.-Y. Xia, L. Hao, W.-J. Wang, H. Zhang, L.-Y. Liu, W. Liu, Z.-Y. Li, C.-P. Tan and Z.-W. Mao, Angew. Chem., Int. Ed., 2022, 61, e202210988 CrossRef CAS.
- S. R. Zhang, D. F. Song, W. H. Yu, J. Li, X. Y. Wang, Y. C. Li, Z. H. Zhao, Q. Xue, J. Zhao, J. P. Li and Z. J. Guo, Natl. Sci. Rev., 2024, 11, nwae020 CrossRef CAS.
- Y. Chen, S. Feng, L. Li, G. Liu, Q. Ma, Z. Liu, Y. Zhao and Q. Wang, J. Med. Chem., 2025, 68, 11928 CrossRef CAS.
- J.-Q. Wang, X.-M. Liu, Z.-S. Zhu, Z. Li, C.-Z. Xie, X. Qiao, Y.-K. Feng and J.-Y. Xu, J. Med. Chem., 2025, 68, 9661 CrossRef CAS.
- T.-Z. Ma, L.-Y. Liu, Y.-L. Zeng, K. Ding, H. Zhang, W. Liu, Q. Cao, W. Xia, X. Xiong, C. Wu and Z.-W. Mao, Chem. Sci., 2024, 15, 9756 RSC.
- Y. Zheng, X.-X. Chen, D.-Y. Zhang, W.-J. Wang, K. Peng, Z.-Y. Li, Z.-W. Mao and C.-P. Tan, Chem. Sci., 2023, 14, 6890 RSC.
- Y. L. Zeng, L. Y. Liu, T. Z. Ma, Y. Liu, B. Liu, W. Liu, Q. H. Shen, C. Wu and Z. W. Mao, Angew. Chem., Int. Ed., 2024, 63, e202410803 CrossRef CAS PubMed.
- Y. Li, B. Liu, Y. Zheng, M. Hu, L.-Y. Liu, C.-R. Li, W. Zhang, Y.-X. Lai and Z.-W. Mao, J. Med. Chem., 2024, 67, 16235 CrossRef CAS.
- Y.-Y. Ling, Z.-Y. Li, X. Mu, Y.-J. Kong, L. Hao, W.-J. Wang, Q.-H. Shen, Y.-B. Zhang and C.-P. Tan, Eur. J. Med. Chem., 2024, 275, 116638 CrossRef CAS.
- G. Xu, Q. Liang, L. Gao, S. Xu, W. Luo, Q. Wu, J. Li, Z. Zhang, H. Liang and F. Yang, J. Med. Chem., 2024, 67, 19573 CrossRef CAS.
- L. X. Cai, Y. Wang, Y. Y. Chen, H. H. Chen, T. Yang, S. R. Zhang, Z. J. Guo and X. Y. Wang, Chem. Sci., 2023, 14, 4375 RSC.
- F. Li, Z. Wen, C. Wu, Z. Yang, Z. Wang, W. Diao, D. Chen, Z. Xu, Y. Lu and W. Liu, J. Med. Chem., 2024, 67, 1982 CrossRef CAS PubMed.
- X.-X. Chen, Z.-Y. Li, Q.-H. Shen, K. Peng, P. Wang, L.-B. Yu, Y.-Y. Han and C.-P. Tan, ACS Nano, 2025, 19, 26953 CrossRef CAS.
- W. Qiao, J. Q. Chen, H. Y. Zhou, C. Hu, S. Dalangood, H. J. Li, D. D. Yang, Y. Yang and J. Gui, Adv. Sci., 2024, 11, 202305979 Search PubMed.
- Y. M. Xing, A. Peng, J. H. Yang, Z. F. Cheng, Y. Yue, F. L. Liu, F. H. Li, Y. Liu and Q. Liu, Adv. Sci., 2024, 11, 202309583 Search PubMed.
- L. Yi, X. Jiang, Z. G. Zhou, W. Xiong, F. Xue, Y. Liu, H. Z. Xu, B. Fan, Y. Li and J. L. Shen, Adv. Mater., 2024, 36, 202304328 Search PubMed.
- B. R. Kimmel, K. Arora, N. C. Chada, V. Bharti, A. J. Kwiatkowski, J. E. Finkelstein, A. Hanna, E. N. Arner, T. L. Sheehy, L. E. Pastora, J. Yang, H. M. Pagendarm, P. T. Stone, E. Hargrove-Wiley, B. C. Taylor, L. A. Hubert, B. M. Fingleton, K. N. Gibson-Corley, J. C. May, J. A. McLean, J. C. Rathmell, A. Richmond, W. K. Rathmell, J. M. Balko and J. T. Wilson, Nat. Biomed. Eng., 2025, 9, 1719 CrossRef CAS.
- W. Maret, Adv. Nutr., 2013, 4, 82 CrossRef CAS.
- T. Kambe, T. Tsuji, A. Hashimoto and N. Itsumura, Physiol. Rev., 2015, 95, 749 CrossRef CAS.
- H. B. Fang, Y. H. Li, X. Z. Yang, Y. C. Chen, Z. J. Guo and W. J. He, Curr. Opin. Chem. Biol., 2023, 76, 102378 CrossRef CAS PubMed.
- E. Bafaro, Y. Liu, Y. Xu and R. E. Dempski, Signal Transduction Targeted Ther., 2017, 2, 17029 CrossRef PubMed.
- A. K. Baltaci, K. Yuce and R. Mogulkoc, Biol. Trace Elem. Res., 2017, 183, 22 CrossRef PubMed.
- B. Chen, P. Yu, W. N. Chan, F. Xie, Y. Zhang, L. Liang, K. T. Leung, K. W. Lo, J. Yu, G. M. K. Tse, W. Kang and K. F. To, Signal Transduction Targeted Ther., 2024, 9, 6 CrossRef CAS PubMed.
- J. Wang, H. H. Zhao, Z. L. Xu and X. X. Cheng, Cancer Biol. Med., 2020, 17, 612 CAS.
- S. Alam and S. L. Kelleher, Nutrients, 2012, 4, 875 CrossRef CAS.
- J. M. Goldberg and S. J. Lippard, FEBS Lett., 2022, 597, 151 CrossRef.
- H. Woo, S. Cho, Y. Han, W.-S. Chae, D.-R. Ahn, Y. You and W. Nam, J. Am. Chem. Soc., 2013, 135, 4771 CrossRef CAS PubMed.
- Y. You, S. Cho and W. Nam, Inorg. Chem., 2014, 53, 1804 CrossRef CAS.
- S. Y. Ryu, M. Huh, Y. You and W. Nam, Inorg. Chem., 2015, 54, 9704 CrossRef CAS PubMed.
- K. Kikuchi, K. Komatsu and T. Nagano, Curr. Opin. Chem. Biol., 2004, 8, 182 CrossRef CAS PubMed.
- J. C. Shen, T. W. Rees, L. N. Ji and H. Chao, Coord. Chem. Rev., 2021, 443, 214016 CrossRef CAS.
- S. Pete, N. Roy, B. Kar and P. Paira, Coord. Chem. Rev., 2022, 460, 214462 CrossRef CAS.
- B. Kar, U. Das, N. Roy and P. Paira, Coord. Chem. Rev., 2023, 474, 214860 CrossRef CAS.
- Y. You, S. Lee, T. Kim, K. Ohkubo, W.-S. Chae, S. Fukuzumi, G.-J. Jhon, W. Nam and S. J. Lippard, J. Am. Chem. Soc., 2011, 133, 18328 CrossRef CAS.
- C. L. Zhang, M. S. Liu, S. X. Liu, H. Yang, Q. Zhao, Z. P. Liu and W. J. He, Inorg. Chem., 2018, 57, 10625 CrossRef CAS.
- R.-R. Ye, C.-P. Tan, L. He, M.-H. Chen, L.-N. Ji and Z.-W. Mao, Chem. Commun., 2014, 50, 10945 RSC.
- F. Lafolet, S. Welter, Z. Popović and L. D. Cola, J. Mater. Chem., 2005, 15, 2820 RSC.
- M. Bandini, M. Bianchi, G. Valenti, F. Piccinelli, F. Paolucci, M. Monari, A. Umani-Ronchi and M. Marcaccio, Inorg. Chem., 2010, 49, 1439 CrossRef CAS PubMed.
- X. Y. Yi, C. S. Zhang, S. Guo, J. Ma and J. Z. Zhao, Dalton Trans., 2014, 43, 1672 RSC.
- P.-K. Lee, W. H.-T. Law, H.-W. Liu and K. K.-W. Lo, Inorg. Chem., 2011, 50, 8570 CrossRef CAS PubMed.
- H. A. Benesi and J. H. Hildebrand, J. Am. Chem. Soc., 1949, 71, 2703 CrossRef CAS.
- E. Tomat and S. J. Lippard, Inorg. Chem., 2010, 49, 9113 CrossRef CAS.
- D. Buccella, J. A. Horowitz and S. J. Lippard, J. Am. Chem. Soc., 2011, 133, 4101 CrossRef CAS.
- R. J. Radford, W. Chyan and S. J. Lippard, Chem. Sci., 2014, 5, 4512 RSC.
- T. D. Rae, P. J. Schmidt, R. A. Pufahl, V. C. Culotta and T. V. O'Halloran, Science, 1999, 284, 805 CrossRef CAS.
- F. Bulcke, R. Dringen and I. F. Scheiber, in Neurotoxicity of Metals, ed. M. Aschner and L. G. Costa, Springer International Publishing, Cham, 2017, pp. 313, DOI:10.1007/978-3-319-60189-2_16.
- W. Sun, M. Li, J.-L. Fan and X.-J. Peng, Acc. Chem. Res., 2019, 52, 2818 CrossRef CAS PubMed.
- Z. Liu, W. He and Z. Guo, Chem. Soc. Rev., 2013, 42, 1568 RSC.
- Q. Zhao, F. Y. Li and C. H. Huang, Chem. Soc. Rev., 2010, 39, 3007 RSC.
- C. Han, J. Jing, X. Zhao, J. Guo, S. Zheng and L. Du, Biol. Trace Elem. Res., 2003, 94, 113 CrossRef CAS.
- S. Mididoddi, J. P. McGuirt, M. A. Sens, J. H. Todd and D. A. Sens, Toxicol. Lett., 1996, 85, 17 CrossRef CAS.
- D. Desbouis, I. P. Troitsky, M. J. Belousoff, L. Spiccia and B. Graham, Coord. Chem. Rev., 2012, 256, 897 CrossRef CAS.
- T. Sarwar, S. U. Rehman, M. A. Husain, H. M. Ishqi and M. Tabish, Int. J. Biol. Macromol., 2015, 73, 9 CrossRef CAS PubMed.
- P. Kumar, S. Tomar, K. Kumar and S. Kumar, Dalton Trans., 2023, 52, 6961 RSC.
- E. A. Schon, S. DiMauro and M. Hirano, Nat. Rev. Genet., 2012, 13, 878 CrossRef CAS PubMed.
- C. Bailly, Life Sci., 2020, 246, 117403 CrossRef CAS.
- J.-J. Lee, S. Y. Kim, S. H. Kim, S. Choi, B. Lee and J.-S. Shin, Cell Death Dis., 2022, 13, 791 CrossRef CAS.
- T. Yu, W. Nie, Z. Hong, Y. He, J. Chen, X. Mi, S. Yang, X. Li, B. Wang, Y. Lin and X. Gao, Adv. Funct. Mater., 2021, 31, 202100715 Search PubMed.
- J.-Y. Zhou, Q.-H. Shen, X.-J. Hong, W.-Y. Zhang, Q. Su, W.-G. Li, B. Cheng, C.-P. Tan and T. Wu, Chem. Eng. J., 2023, 474, 145516 CrossRef CAS.
- Q.-H. Shen, L.-B. Yu, S.-M. Yao, Z.-Y. Li, Q.-Q. Zhang, P. Wang and C.-P. Tan, J. Med. Chem., 2025, 68, 16284 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2026 |