Open Access Article
Open Access ArticleConformational adaptability enabled higher-order self-sorting processes in coordination cages
Minaz
Parbin
ab,
Vellaiyadevan
Sivalingam
 ab,
Ramkumar
Venkatachalam
a and
Dillip Kumar
Chand
ab,
Ramkumar
Venkatachalam
a and
Dillip Kumar
Chand
 *ab
*ab
aDepartment of Chemistry, Indian Institute of Technology Madras, Chennai 600036, India. E-mail: dillip@zmail.iitm.ac.in
bIoE Center of Molecular Architecture, Department of Chemistry, Indian Institute of Technology Madras, Chennai 600036, India
First published on 11th December 2025
Abstract
Understanding the key role of conformational adaptability in biological processes is crucial to mimic the remarkable applications inherent to biological systems. Motivated by the efficacy of conformational adaptability, we equipped a conformationally-adaptive ligand in low-symmetry cis-Pd2La2Lx2-type coordination cages. A family of five cis-Pd2La2Lx2-type cages was assembled by complementary ligand pairing of a conformationally adaptable converging ligand (La-type) in combination with diverging rigid ligands (Lx-type) of different lengths. Integrative self-sorting of the individual cages showed that the converging ligand adapts to three distinct conformations in the Pd2La2Lx2-type architecture, to accommodate Lx-type ligands of varying sizes. Through a series of experiments, we found a higher order, i.e., 2-fold heteromeric completive self-sorting outcomes of two co-existing Pd2La2Lx2-type cages, where two chosen complementary rigid ligands could induce any two different conformations of the converging ligand in the co-existing cages. Then we further pushed the intricacy and demonstrated the unprecedented 3-fold heteromeric completive self-sorting in coordination cage systems, where the conformationally-adaptive ligand adapts three distinct conformations in three co-existing Pd2La2Lx2-type cages. This study paves the way for the utility of conformational adaptability to achieve switchable size, shape, and functionality in supramolecular systems toward bio-relevant applications.
Introduction
Coordination-driven self-assembly has been one of the most convenient strategies for constructing captivating nanostructured coordination architectures with applications such as molecular recognition, drug delivery, separation, catalysis, etc.1,2 Besides self-assembly, the controlled formation of well-defined coordination assemblies relies on the ability of the different components to associate together by mutual recognition, also known as self-sorting.3 Inspired by biological systems, mixed ligated coordination cages were achieved through self-sorting using various design strategies (e.g., geometric complementarity, coordination sphere engineering, endohedral-functionalization, guest-templation) that employ more than one type of ligand component.4 A rational combination of up to four different ligands was used to selectively achieve discrete Pd(II)-based binuclear assemblies (i.e., Pd2La3Lb, cis-/trans-Pd2La2Lb2, cis-/trans-Pd2La2LbLc, and Pd2LaLbLcLd-type) via integrative self-sorting.5 However, in biological self-assembly, the encoded subcomponents assemble into more than one co-existing ensemble via orthogonal self-sorting.6Recently, Clever group reported stoichiometrically controlled co-formation of two mixed ligated cages of Pd2La2Lb2-and Pd2La2Lc2-type, also known as 2-fold heteromeric completive self-sorting by both direct ligand assembly with Pd(II) or cage fusion reaction of the three high-symmetry homoleptic assemblies.7 Later, we demonstrated a 2-fold heteromeric completive self-sorting outcome using a combination of two low-symmetry homoleptic cages and a high-symmetry homoleptic cage.8
Further, exploring increasingly-complex, higher-order self-sorting processes using coordination cages could be a formidable challenge, as it will require comprehensive pre-programming of the individual components along with interference-free interplay of multiple metal–ligand interactions.6,9 However, we intend to develop a design strategy to demonstrate the co-formation of three distinct Pd2La2Lx2-type mixed ligated assemblies (i.e., Pd2La2Lb2, Pd2La2Lc2, and Pd2La2Ld2-type, where x = b, c, and d) having a common La-type ligand. Such a higher-order self-sorting outcome, termed as “3-fold heteromeric completive self-sorting” is illustrated in Fig. 1. Designing the mutually shared ligand (La-type) is crucial for enabling such complexity in self-sorting processes. We presume that conformationally adaptable units embedded in a mutually shared ligand could enable the possibility of different co-existing conformations suitable for other complementary ligands (Lx-type, x = b, c, d, etc.) to form mixed ligated Pd2La2Lx2-type cages using an adaptive shape-complementary approach.
Though conformational adaptability plays a key role in various biological processes (such as enzymatic catalysis, allosteric signalling, ligand binding in proteins, etc.), achieving conformational control in synthetic supramolecular hosts requires substantial effort.10,11 We envisaged that the conformational flexibility of amides stemming from the possible free rotation around CR1–Ccarbonyl and CR2–Namide bonds could potentially serve this purpose.12 A significant challenge in utilizing amide-incorporated ligands for constructing mixed ligated cages would be controlling the inherently flexible nature of the ligands. However, we believe, a delicate balance between flexibility and rigidity in the ligand designs may bring the required structural diversity in mixed ligated assemblies to achieve a 3-fold heteromeric completive self-sorting outcome.
In this report, we have designed a di-amide incorporated converging bidentate ligand (La-type) and explored the conformationally adaptive trait of the ligand with a series of pillar-type diverging bidentate ligands (Lx-type) of appropriate lengths (Fig. 2) to construct a family of cis-Pd2La2Lx2-type mixed ligated assemblies. The conformation of the La-type ligand (syn–syn, syn–anti, or anti–anti) in the formed coordination assembly is dictated by the length of the Lx-type diverging ligands employed for complexation. The conformational adaptability of the La-type ligand was further utilized to demonstrate the first example of a 3-fold heteromeric completive self-sorting in coordination cage systems. The three distinct coexisting binuclear assemblies (Pd2La2Lb2, Pd2La2Lc2, and Pd2La2Ld2-type) were achieved by cage fusion of four carefully chosen homoleptic assemblies (Pd2La4, Pd4Lb8, Pd4Lc8, and Pd4Ld8-type), where the mutually shared La-type ligand is locked in three different conformations in the three mixed ligated cages. Further, we have also studied the conformational adaptability of three converging ligands having di-, mono-, and non-amide moieties in their backbone for constructing cis-Pd2La2Lx2-type mixed ligated assemblies.
Results and discussion
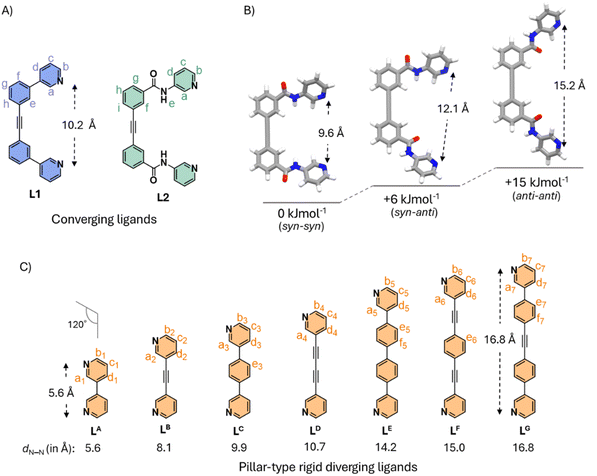
To study the utility of conformational adaptability in shape complementary cis-Pd2La2Lx2-type mixed ligated cages,5b,13 first, we designed a pair of converging bidentate ligands (La-type) L1 (rigid) and L2 (conformationally adaptable) (Fig. 2A). The converging ligands were designed to understand the relevance of rigidity vs. adaptability in the assembly of the desired mixed ligated cages, in combination with well-suited rigid diverging ligands (Lx-type) and Pd(II). Ligand L2 was designed by embedding di-amide functionality in the 1,2-diphenylacetylene spacered ligand L1. We presume that the bidentate ligand bearing di-amide moieties (L2) could possibly orient in three distinct conformational states (namely syn–syn, syn–anti, or anti–anti) in mixed ligated Pd2La2Lx2-type assembly, when paired alongside carefully designed diverging ligands (Lx-type, x = A–G) and Pd(II).Due to the partial delocalization across the amide bond of L2, the rotation around the CO–NH bond will be constrained; however, rotation around Cphenyl–Ccarbonyl as well as Cpyridyl–Namide bonds could result in several possible conformations. Out of all the possible conformations of L2, the three chosen conformations are shown in Fig. 2B. The three conformations were selected for their potential ability to form a shape complementary cis-Pd2La2Lx2-type assembly when paired with a suitable diverging ligand and Pd(II). Initial DFT (B3LYP/6-31g(d)) calculations revealed the approximate Npy⋯Npy distance of L2 in different conformations to be around 9.6 Å (syn–syn,L2SS), 12.1 Å (syn–anti, L2SA) and 15.2 Å (anti–anti, L2AA) (Fig. 2B). In a cis-Pd2La2Lx2-type assembly, L2 could adapt any of the three conformations dictated by the complementary diverging ligand. Hence, we have chosen a series of pillar-type rigid ligands (LA–LG) of varied lengths (Npy⋯Npy distance ranging from around 5.6 to 16.8 Å). These ligands possess an approximate bent angle of 120°, which could potentially dictate the conformation of L2 in the cis-Pd2La2Lx2-type cage (Fig. 2C).14–19 The combination of L2 and the rigid pillar-type ligands (LA–LG) was expected to result in a library of cis-Pd2La2Lx2-type mixed ligated cages. Also, L1 could form cis-Pd2La2Lx2-type assemblies when paired with a diverging ligand of appropriate length (from the series of LA–LG) and Pd(II), owing to the shape complementary nature. However, the number of cis-Pd2La2Lx2-type assemblies and extent of adaptability will be limited due to the rigid backbone of L1. To achieve our objective, we synthesized the ligands, and the detailed synthetic procedure is given in the SI, see Section S2.
Initially, we explored the self-assembly of individual ligand components (L1 and L2) with Pd(II). Complexation of Pd(NO3)2 with L1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio) in DMSO-d6 at room temperature was monitored by 1H NMR spectroscopy (Fig. S12). After 12 hours, a single species showing a characteristic complexation-induced downfield shift of the pyridine-αCH protons (ΔδComplex-Ligand (Ha) = 0.93 ppm & ΔδComplex-Ligand (Hb) = 0.76 ppm) was observed in the 1H NMR spectrum. Formation of the cage [Pd2(L1)4](NO3)4, 1·4NO3 was supported based on ESI-MS data. To attain a parallel coordination vector for a Pd2L4-type assembly, the converging ligand L1 needs to undergo twisting around the two Pd(II) centres. Hence, the formed cage was expected to be a helical Pd2L4-type assembly (Fig. S186). Likewise, the self-assembly of L2 with Pd(II) was anticipated to result in a helical Pd2L4-type architecture (Fig. S186).5a,13 Complexation of Pd(NO3)2 with L2 (1
2 ratio) in DMSO-d6 at room temperature was monitored by 1H NMR spectroscopy (Fig. S12). After 12 hours, a single species showing a characteristic complexation-induced downfield shift of the pyridine-αCH protons (ΔδComplex-Ligand (Ha) = 0.93 ppm & ΔδComplex-Ligand (Hb) = 0.76 ppm) was observed in the 1H NMR spectrum. Formation of the cage [Pd2(L1)4](NO3)4, 1·4NO3 was supported based on ESI-MS data. To attain a parallel coordination vector for a Pd2L4-type assembly, the converging ligand L1 needs to undergo twisting around the two Pd(II) centres. Hence, the formed cage was expected to be a helical Pd2L4-type assembly (Fig. S186). Likewise, the self-assembly of L2 with Pd(II) was anticipated to result in a helical Pd2L4-type architecture (Fig. S186).5a,13 Complexation of Pd(NO3)2 with L2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio) in DMSO-d6 at room temperature for 12 hours led to the formation of a discrete product, as confirmed by 1H NMR spectroscopy (Fig. S17). ESI-MS analysis confirms the composition [Pd2(L2)4](NO3)4, 2·4NO3 for the resultant cage assembly. Further, 1H–1H NOE spectrum analysis of the cage showed cross-peak correlations of amide protons (NHe) with outward-pointing protons Hd and Hg (Fig. S21), suggesting an all-(anti–anti) conformer of L2 in the cage structure.
2 ratio) in DMSO-d6 at room temperature for 12 hours led to the formation of a discrete product, as confirmed by 1H NMR spectroscopy (Fig. S17). ESI-MS analysis confirms the composition [Pd2(L2)4](NO3)4, 2·4NO3 for the resultant cage assembly. Further, 1H–1H NOE spectrum analysis of the cage showed cross-peak correlations of amide protons (NHe) with outward-pointing protons Hd and Hg (Fig. S21), suggesting an all-(anti–anti) conformer of L2 in the cage structure.
Pd(II)-based coordination complexes featuring LB, LC, LE, LF, and LG have been previously reported.16,19–21 In this work, we carried out the individual complexation of all the ligands with Pd(NO3)2 (reported cages were reproduced; see SI Section S2.4). Among the reported cases, LB and LC gave their respective Pd4L8 (tetrahedron)-type (i.e., B·8NO3 and C·8NO3, respectively) architecture upon complexation with Pd(NO3)2.16,20 When LA was treated with Pd(NO3)2 at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in DMSO-d6 at room temperature for 24 hours, the 1H NMR spectrum showed unassignable complicated signals due to the formation of a mixture of products. Complexation of Pd(NO3)2 with LD at a 1
1 ratio in DMSO-d6 at room temperature for 24 hours, the 1H NMR spectrum showed unassignable complicated signals due to the formation of a mixture of products. Complexation of Pd(NO3)2 with LD at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio in DMSO-d6 at room temperature for 3 hours resulted in a well-resolved 1H NMR spectrum consisting of two products. The resultant assemblies are proposed to be a mixture of Pd4L8 (tetrahedron) (major) and Pd3L6 (minor)-type cages (i.e., D·8NO3 and D′·6NO3. Similarly, LE and LF also produced a mixture of Pd4L8 (tetrahedron) (major) and Pd3L6 (minor)-type cages (i.e., E·8NO3 and E′·6NO3/F·8NO3 and F′·6NO3).16,21 However, when a longer ligand LG was treated with Pd(NO3)2 (2
2 ratio in DMSO-d6 at room temperature for 3 hours resulted in a well-resolved 1H NMR spectrum consisting of two products. The resultant assemblies are proposed to be a mixture of Pd4L8 (tetrahedron) (major) and Pd3L6 (minor)-type cages (i.e., D·8NO3 and D′·6NO3. Similarly, LE and LF also produced a mixture of Pd4L8 (tetrahedron) (major) and Pd3L6 (minor)-type cages (i.e., E·8NO3 and E′·6NO3/F·8NO3 and F′·6NO3).16,21 However, when a longer ligand LG was treated with Pd(NO3)2 (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio), a single set of peaks were observed in the 1H NMR spectrum, suggesting the formation of a single species, a Pd3L6-type (G·6NO3) architecture was further confirmed by ESI-MS data analysis (Fig. S96).19
1 ratio), a single set of peaks were observed in the 1H NMR spectrum, suggesting the formation of a single species, a Pd3L6-type (G·6NO3) architecture was further confirmed by ESI-MS data analysis (Fig. S96).19
Next, we explored the mixed ligand complexation behavior of L1 and L2 when paired with a rigid diverging ligand of suitable length and Pd(II). Based on the DFT calculated structure, we presume that L1 (Npy⋯Npy = 10.2 Å) and LB (Npy⋯Npy = 8.1 Å) could be the best fit for a cis-Pd2La2Lx2-type arrangement (Fig. S186).22 Self-assembly of Pd(NO3)2 with L1 and LB in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in DMSO-d6 at room temperature for 30 minutes resulted in the clean formation of a discrete cage, as demonstrated by 1H NMR spectroscopy. Further evidence for the formation of the mixed ligated cage [Pd2(L1)2(LB)2](NO3)4, 1B·4NO3 was provided by ESI-MS data, which shows isotopic peak patterns at m/z = 681.09 and 433.30 corresponding to the loss of two and three NO3− ions, respectively. Subsequently, we performed complexation of Pd(NO3)2 with L1 and LA/LC (shorter/longer than LB) (Fig. 3). Mixing Pd(NO3)2, L1 and LA in a 1
1 ratio in DMSO-d6 at room temperature for 30 minutes resulted in the clean formation of a discrete cage, as demonstrated by 1H NMR spectroscopy. Further evidence for the formation of the mixed ligated cage [Pd2(L1)2(LB)2](NO3)4, 1B·4NO3 was provided by ESI-MS data, which shows isotopic peak patterns at m/z = 681.09 and 433.30 corresponding to the loss of two and three NO3− ions, respectively. Subsequently, we performed complexation of Pd(NO3)2 with L1 and LA/LC (shorter/longer than LB) (Fig. 3). Mixing Pd(NO3)2, L1 and LA in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in DMSO-d6 showed formation of a mixture of assemblies with one dominant species in the 1H NMR spectrum (Fig. S31). ESI-MS data showing characteristic peak patterns for the sequential loss of NO3− ions supported the composition of [Pd2(L1)2(LA)2](NO3)4, 1A·4NO3. However, pairing the rigid ligand L1 with a relatively longer ligand LC and Pd(NO3)2 resulted in a mixture of homoleptic assemblies (narcissistic self-sorting), as evident from the 1H NMR spectrum (Fig. S44). Further employing longer ligands LD–LG for complexation with Pd(NO3)2 and L1 also resulted in narcissistic self-sorting (Fig. S45–S48). Integratively self-sorted cis-Pd2La2Lx2-type assembly was only successful for the self-assembly L1 with LB and Pd(II), whereas due to the rigid backbone of L1 and incompatible lengths of the ligand pairs, the mixed ligand assembly of L1, and other ligands from the series, i.e., LA, LC–LG was unsuccessful.
1 ratio in DMSO-d6 showed formation of a mixture of assemblies with one dominant species in the 1H NMR spectrum (Fig. S31). ESI-MS data showing characteristic peak patterns for the sequential loss of NO3− ions supported the composition of [Pd2(L1)2(LA)2](NO3)4, 1A·4NO3. However, pairing the rigid ligand L1 with a relatively longer ligand LC and Pd(NO3)2 resulted in a mixture of homoleptic assemblies (narcissistic self-sorting), as evident from the 1H NMR spectrum (Fig. S44). Further employing longer ligands LD–LG for complexation with Pd(NO3)2 and L1 also resulted in narcissistic self-sorting (Fig. S45–S48). Integratively self-sorted cis-Pd2La2Lx2-type assembly was only successful for the self-assembly L1 with LB and Pd(II), whereas due to the rigid backbone of L1 and incompatible lengths of the ligand pairs, the mixed ligand assembly of L1, and other ligands from the series, i.e., LA, LC–LG was unsuccessful.
Then, we studied the adaptive self-assembly behavior of the ligand L2 in the presence of a range of diverging ligands towards the assembly of cis-Pd2La2Lx2-type cages. We thought LB (Npy⋯Npy = 8.1 Å) would be suitable for the syn–syn conformation (L2SS, Npy⋯Npy = 9.6 Å). Upon treating L2 and LB with Pd(NO3)2 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in DMSO-d6 at room temperature for 30 minutes, the 1H NMR spectrum showed a single set of signals for the formation of a discrete species. The spectrum showed the characteristic complexation-induced downfield shift in the position of the inwards pointed pyridine-αCHIN of both the ligands (ΔδComplex-Ligand (Ha) = 0.51 ppm & ΔδComplex-Ligand (Ha2) = 1.41 ppm for L2 and LB respectively) (Fig. 4B(iii)). The formation of complex [Pd2(L2)2(LB)2](NO3)4, 2B·4NO3 was further investigated by ESI-MS data analysis, where isotopic peak patterns at m/z = 490.73 and 352.55 was observed for the loss of three and four NO3− ions, respectively. Further, a single band in the 1H DOSY spectrum confirmed that all the protons correspond to a single species in the solution with a diffusion coefficient of 1.21 × 10−10 m2 s−1 (Fig. S118).
1 ratio in DMSO-d6 at room temperature for 30 minutes, the 1H NMR spectrum showed a single set of signals for the formation of a discrete species. The spectrum showed the characteristic complexation-induced downfield shift in the position of the inwards pointed pyridine-αCHIN of both the ligands (ΔδComplex-Ligand (Ha) = 0.51 ppm & ΔδComplex-Ligand (Ha2) = 1.41 ppm for L2 and LB respectively) (Fig. 4B(iii)). The formation of complex [Pd2(L2)2(LB)2](NO3)4, 2B·4NO3 was further investigated by ESI-MS data analysis, where isotopic peak patterns at m/z = 490.73 and 352.55 was observed for the loss of three and four NO3− ions, respectively. Further, a single band in the 1H DOSY spectrum confirmed that all the protons correspond to a single species in the solution with a diffusion coefficient of 1.21 × 10−10 m2 s−1 (Fig. S118).
To investigate the conformationally adaptive nature of ligand L2, we performed its complexation with the next ligand in the series LC (Npy⋯Npy = 9.9 Å) and Pd(NO3)2 at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in DMSO-d6. After 30 minutes, the 1H NMR spectrum indicated the formation of a discrete product corresponding to cage [Pd2(L2)2(LC)2](NO3)4, 2C·4NO3 where the pyridine−αCHIN showed complexation-induced downfield shift (ΔδComplex-Ligand (Ha) = 0.98 ppm & ΔδComplex-Ligand (Ha3) = 1.46 ppm for L2 and LC, respectively) for both the ligands (Fig. 4B(iv)). We presume LC might be slightly longer to fit the syn–syn conformation of L2 in the cis-Pd2La2Lx2-type cage structure. Thus, L2 might have adapted syn–anti conformation (L2SA) during the self-assembly process to form cage 2C·4NO3 selectively. To further test our hypothesis for the conformational adaptability of L2, we performed complexation of Pd(NO3)2 with L2 and LD (Npy⋯Npy = 10.7 Å) (1
1 ratio in DMSO-d6. After 30 minutes, the 1H NMR spectrum indicated the formation of a discrete product corresponding to cage [Pd2(L2)2(LC)2](NO3)4, 2C·4NO3 where the pyridine−αCHIN showed complexation-induced downfield shift (ΔδComplex-Ligand (Ha) = 0.98 ppm & ΔδComplex-Ligand (Ha3) = 1.46 ppm for L2 and LC, respectively) for both the ligands (Fig. 4B(iv)). We presume LC might be slightly longer to fit the syn–syn conformation of L2 in the cis-Pd2La2Lx2-type cage structure. Thus, L2 might have adapted syn–anti conformation (L2SA) during the self-assembly process to form cage 2C·4NO3 selectively. To further test our hypothesis for the conformational adaptability of L2, we performed complexation of Pd(NO3)2 with L2 and LD (Npy⋯Npy = 10.7 Å) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) in DMSO-d6. After stirring the reaction at room temperature for 30 minutes, a discrete assembly was observed in the 1H NMR spectrum (Fig. 4B(v)). The composition of the formed product was found to be [Pd2(L2)2(LD)2](NO3)4, 2D·4NO3 from ESI-MS data analysis. The facile formation of complex 2D·4NO3 confirms the ability of L2 to adapt to a different conformation to form an entropically more favourable mixed ligated cage.
1 ratio) in DMSO-d6. After stirring the reaction at room temperature for 30 minutes, a discrete assembly was observed in the 1H NMR spectrum (Fig. 4B(v)). The composition of the formed product was found to be [Pd2(L2)2(LD)2](NO3)4, 2D·4NO3 from ESI-MS data analysis. The facile formation of complex 2D·4NO3 confirms the ability of L2 to adapt to a different conformation to form an entropically more favourable mixed ligated cage.
Having found an adaptive ligand capable of switching its conformation to adopt a length suitable for fitting a complementary ligand in a Pd2La2Lx2-type cage, we next decided to attempt the complexation of Pd(NO3)2 with L2 and LE (Npy⋯Npy = 14.2 Å) or LF (Npy⋯Npy = 15.0 Å). A clean formation of the cage [Pd2(L2)2(LE)2](NO3)4, 2E·4NO3 and [Pd2(L2)2(LF)2](NO3)4, 2F·4NO3 was obtained via self-assembly of L2 with LE/LF and Pd(NO3)2 respectively (Fig. 4B(vi and vii)). The formed mixed ligated assemblies were characterized by 1D and 2D NMR spectroscopy techniques and ESI-MS data. Based on the Npy⋯Npy distances of LE and LF, we could infer that the L2 is likely to be present in the anti–anti conformation (L2AA). To examine the extent of conformational adaptability of L2 for the formation of Pd2La2Lx2-type assembly, we tried the complexation with the shortest/longest ligand in the chosen series, i.e., LA (Npy⋯Npy = 5.6 Å) and LG (Npy⋯Npy = 16.8 Å). Expectedly, both ligands were found to yield either a narcissistic self-sorting outcome or a mixture of unidentified assemblies.
The comparative 1H NMR spectral analysis of all five cis-Pd2La2Lx2-type cages, 2B·4NO3−2F·4NO3, showed distinctive complexation-induced chemical shift changes (ΔδComplex-Ligand) for Ha and Hd (Fig. 4B). While the 1H NMR spectrum of 2B·4NO3 exhibited ΔδComplex-Ligand for Ha about 0.51 ppm (downfield shift), the cages 2C·4NO3 and 2D·4NO3 displayed even higher ΔδComplex-Ligand for Ha (0.98 ppm and 1.00 ppm). However, for the cages 2E·4NO3 and 2F·4NO3, we found more pronounced ΔδComplex-Ligand for Ha (1.52 ppm and 1.58 ppm). Likewise, a similar observation was found for the proton Hd, which showed gradual upfield shifts from 2B·4NO3 to 2F·4NO3. This distinctive chemical shift change may be due to the intramolecular hydrogen bonding between Ha/Hd and carbonyl oxygen. As the conformational switching progressively changes from syn–syn to anti–anti via syn–anti conformation, Ha/Hd could potentially move closer/away from C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively.
O, respectively.
After successfully achieving a series of cages 2B·4NO3–2F·4NO3 through integrative self-sorting of individual ligand components and Pd(II) using an adaptive shape-complementary approach, we were curious to inspect whether the self-assembled products are pathway-dependent or at the thermodynamic minimum. Hence, we performed the cage-to-cage transformations from the corresponding homoleptic assemblies. Mixing 2 equiv. of 2·4NO3 with 1 equiv. of corresponding homoleptic assemblies of Lx (x = B–F), i.e., B·8NO3–F·8NO3 (Pd4L8-type cages D·8NO3, E·8NO3 and F·8NO3 exist along with the Pd3L6-type cages D′·6NO3, E′·6NO3 and F′·6NO3), individually in DMSO-d6, resulted in the mixed ligated assembly 2X·4NO3 under thermodynamic control (Fig. 4A). The robustness of the mixed ligated cage 2B·4NO3–2F·4NO3 was further demonstrated by concentration variation studies (SI Section S8 and Fig. S151–S155).
Thereafter, we were curious to inspect the conformational preference of ligand L2 in the Pd2La2Lx2-type cage assemblies. For this purpose, we performed ligand exchange reactions, in which to a chosen mixed ligated complex, a calculated amount of another ligand (from the series LB–LF) was added. In some cases, the bound ligand has been replaced partially or entirely by the incoming ligand (SI Section S9, Scheme S32 and Fig. S156–S175). Based on these experiments, the relative ligand selectivity of L2 to form mixed ligated cages was found to be LE > LC > LF > LB ≃ LD. Based on the electrostatic potential map analysis of the ligands LB–LF, we presume this outcome could possibly be due to the basicity of the rigid complementary ligands rather than the conformation of the L2 (Fig. S185).
The striking difference in the adaptability of L1 and L2 to self-assemble into cis-Pd2La2Lx2-type cages stems from the embedded amide moieties in L2. When L1 was paired with a set of diverging ligands (Lx, Npy⋯Npy = 5.6–16.8 Å), an absolute integrative self-sorted product was obtained only for the case of LB. Whereas integrative self-sorting of L2 was successful for a series of five ligands, i.e., LB–LF (Npy⋯Npy = 8.1–15.0 Å), resulting in a family of five cis-Pd2La2Lx2-type assemblies (2B·4NO3–2F·4NO3). Thus, we were curious to study the adaptability of a converging ligand with a single amide moiety embedded. Hence, we chose a mono-amide functionalized ligand L3, for the complexation with LA–LG.
In our previous report,8 we utilized L3 (a structurally-constrained ligand) along with suitable symmetrical/unsymmetrical diverging ligands and Pd(II) to develop two highly anisotropic binuclear mixed ligated cages. However, the conformational adaptability of L3 was unexplored. To investigate the full extent of adaptability of L3 in the presence of suitable diverging ligands, we attempted the complexation of Pd(NO3)2 with L3 and LA–LG. We previously obtained the mixed ligated cage, i.e., [Pd2(L3)2(LB)2](NO3)4, 3B·4NO3 as the minor product upon complexation of L3 and LB with Pd(II). Whereas in the case of LC, an exclusive formation of [Pd2(L3)2(LC)2](NO3)4, 3C·4NO3 was achieved. Among the newly attempted cases (i.e., LA, LD–LG), we successfully obtained discrete mixed ligated assembly [Pd2(L3)2(LD)2]](NO3)4, 3D·4NO3 from the combination of L3, LD and Pd(NO3)2, as indicated by 1H NMR spectroscopy and ESI-MS data (Fig. S82 and S112). Since L3 is unsymmetrical in nature, the mixed ligated cage was obtained as an isomeric mixture of two possible isomers of cage 3D·4NO3 (Fig. S186). For all the other cases, the mixed ligated assembly of L3 and the diverging ligands (LA, LE−LG) were unsuccessful. So, the mono-amide ligand L3 can adapt to two different diverging ligands (LC and LD), producing the desired cis-Pd2La2Lx2-type cages (where L3 is in anti-conformation in the cage structures, Fig. S85); while the rigid L1 yielded only one cis-Pd2La2Lx2-type cage through integrative self-sorting. Hence, we infer that the conformational adaptability of L3 falls somewhere between L1 and L2. This showcases that the extent of conformational adaptability is proportional to the number of embedded amide units in the ligand backbone (Fig. 5A).
In the Pd2La2Lx2-type assembly, the identical ligands coordinate to the Pd(II) centres in a cis fashion, where L2 can adapt to any of the three conformations such as syn–syn (L2SS), syn–anti (L2SA) and anti–anti (L2AA) in the cage structure. Earlier, we postulated the existence of three conformations based on the complexation-induced chemical shift changes in protons Ha and Hd, presumably due to possible intramolecular hydrogen bonding interactions with carbonyl oxygen (Fig. 4B). Next, we used 1H–1H NOE spectroscopy to verify the existing conformations of L2 in all five mixed-ligated cages. In the NOE spectrum of 2B·4NO3, the amide proton (N–He) shows strong cross-peak correlations with Ha and Hf, indicating that the L2 is present in a syn–syn conformation (L2SS) (Fig. 5B(i)). Similarly, we found NOE correlations for the amide protons (N–He) of 2E·4NO3 and 2F·4NO3 with Hd and Hg, no correlations were found with Ha/Hf, indicating the ligand conformation to be anti–anti (L2AA) in the cage (Fig. 5B(iii) and S73). Understandably, the cages 2C·4NO3 and 2D·4NO3 showed multiple NOE correlations for the amide N–He with Ha, Hd, Hf, and Hg suggesting a syn–anti (L2SA) conformation of L2 in the cage (Fig. 5B(ii) and S60). In addition to the intra-ligand contacts, the NOE spectrum of all the cages also showed several cross-peaks for the inter-ligand through-space contacts between L2 and Lx (x = B–F).
The syn–syn (L2SS) and anti–anti (L2AA) conformation of L2 in 2B·4NO3 and 2E·4NO3/2F·4NO3 contributes to the overall higher symmetry of the cages. On the other hand, though the syn–anti (L2SA) conformation of L2 in 2C·4NO3 and 2D·4NO3 could lower the symmetry of the cage, we obtained a single set of signals in the 1H NMR spectrum, presumably due to the fast exchange between the conformations at room temperature. To restrict the ligand conformation in the cage assembly, we attempted a variable temperature NMR study (lowering the temperature up to −30 °C) using 2C·4BF4 and 2D·4BF4 in CD3CN. However, we could not observe any significant splitting of the 1H NMR signals, suggesting the existence of fast exchange process even at −30 °C (Fig. S176 and S180). Further, the two units of L2 in cages 2C·4NO3 and 2D·4NO3 can have either a parallel/antiparallel arrangement in the Pd2La2Lx2-type structure (Fig. S187 and S188). DFT calculation of the two possible isomers at the B3LYP/LanL2DZ, 6-31G(d) level in gas and implicit solvent (DMSO) phase suggested a lower energy for isomer-I, i.e., parallel arrangement of L2 in both the cages (SI Section S11, Table S1 and S2).
To further corroborate the conformational control in five cis-Pd2La2Lx2-type cages, [Pd2(L2)2(Lx)2]4+ by single crystal X-ray diffraction (SC-XRD) analysis, concerted efforts were made to crystallize the cages. We used different solvents and solvent mixtures as well as varied the counteranions. After numerous attempts, we could get crystal data suitable for SC-XRD analysis of [2C]4+ and [2D]4+ with NO3− as counteranion by vapour diffusion of benzene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile and toluene
acetonitrile and toluene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) t-butanol in DMSO solution of 2C·4NO3/2D·4NO3, respectively. SC-XRD analysis confirmed syn–anti conformation of L2 in both the cage structures (Fig. 6). Subsequently, we collected the crystal data for [2F]4+ with BF4− as counteranion. The single crystals were obtained by vapour diffusion of THF in a mixture of DMSO
t-butanol in DMSO solution of 2C·4NO3/2D·4NO3, respectively. SC-XRD analysis confirmed syn–anti conformation of L2 in both the cage structures (Fig. 6). Subsequently, we collected the crystal data for [2F]4+ with BF4− as counteranion. The single crystals were obtained by vapour diffusion of THF in a mixture of DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH3CN (2
CH3CN (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution of 2F·4BF4. Structural analysis reiterated the existence of L2 in anti–anti conformation in [2F]4+ (Fig. 6C). In cages [2B]4+–[2F]4+, the angle of intersection of the Pd(Npy)4 planes (usually parallel for typical Pd2La4-type cages) gradually decreased when we progressively increased the length of rigid complementary ligands.
1) solution of 2F·4BF4. Structural analysis reiterated the existence of L2 in anti–anti conformation in [2F]4+ (Fig. 6C). In cages [2B]4+–[2F]4+, the angle of intersection of the Pd(Npy)4 planes (usually parallel for typical Pd2La4-type cages) gradually decreased when we progressively increased the length of rigid complementary ligands.
We successfully introduced conformational adaptability in cis-Pd2La2Lx2-type systems with embedded amide moiety in La-type ligand and suitable rigid diverging ligands (Lx-type), which could dictate the requisite conformation of La in the cage. The control over the choice of conformations based on the length of rigid diverging ligands opens the possibility of exploring the higher-order self-sorting processes using Pd2La2Lx2-type cages. We presume that three distinct conformations of L2 (where L2 can be used as a common ligand) could enable up to an unprecedented 3-fold heteromeric completive self-sorting (i.e., three co-existing mixed ligated assemblies) in coordination cages.
We set out to explore the conformationally adaptive nature of ligand L2 to demonstrate the self-assembly of three different ligands and Pd(II), targeting the assembly of two co-existing Pd2La2Lx2-type cages, also known as 2-fold heteromeric completive self-sorting.6,7 In the co-existing mixed ligated assemblies, we take the conformationally adaptive L2 as a common ligand, while we use the combination of any two rigid ligands from a set of five ligands, LB−LF. For all the experiments, we treated Pd(NO3)2 with L2 and two chosen ligands amongst LB−LF in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in DMSO-d6 and stirred at room temperature. In the cases where 2 consecutive ligands (i.e., LB
1 ratio in DMSO-d6 and stirred at room temperature. In the cases where 2 consecutive ligands (i.e., LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC, LC
LC, LC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LD, LD
LD, LD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LE and LE
LE and LE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LF) were selected alongside L2 and Pd(NO3)2 (Fig. 7A), we observed two different scenarios: (i) 2-fold heteromeric completive self-sorting outcome for the cases I and III, where a mixture of co-existing mixed-ligated cages (2B·4NO3 + 2C·4NO3 and 2D·4NO3 + 2E·4NO3, respectively) was observed; (ii) mixed self-sorting (both integrative and heteromeirc) outcome for the cases II and IV, where we observed an additional trileptic mixed-ligated assembly5c,d ([Pd2(L2)2LCLD](NO3)4 and [Pd2(L2)2LELF](NO3)4, respectively), along with heteromeric products (2C·4NO3 + 2D·4NO3 and 2E·4NO3 + 2F·4NO3) (Fig. S125, S129, S132 and S134). Also, in case II the trileptic cage [Pd2(L2)2LCLD](NO3)4 was observed as the dominating species in the mixture of assemblies. ESI-MS analysis for case II confirmed the existence of three co-existing assemblies, 2C·4NO3, 2D·4NO3 and [Pd2(L2)2LCLD](NO3)4. Likewise, a combination of 2E·4NO3, 2F·4NO3 and [Pd2(L2)2LELF](NO3)4 was supported by ESI-MS data for case IV (Fig. 7B). However, all the other diverging ligand combinations (i.e., LB
LF) were selected alongside L2 and Pd(NO3)2 (Fig. 7A), we observed two different scenarios: (i) 2-fold heteromeric completive self-sorting outcome for the cases I and III, where a mixture of co-existing mixed-ligated cages (2B·4NO3 + 2C·4NO3 and 2D·4NO3 + 2E·4NO3, respectively) was observed; (ii) mixed self-sorting (both integrative and heteromeirc) outcome for the cases II and IV, where we observed an additional trileptic mixed-ligated assembly5c,d ([Pd2(L2)2LCLD](NO3)4 and [Pd2(L2)2LELF](NO3)4, respectively), along with heteromeric products (2C·4NO3 + 2D·4NO3 and 2E·4NO3 + 2F·4NO3) (Fig. S125, S129, S132 and S134). Also, in case II the trileptic cage [Pd2(L2)2LCLD](NO3)4 was observed as the dominating species in the mixture of assemblies. ESI-MS analysis for case II confirmed the existence of three co-existing assemblies, 2C·4NO3, 2D·4NO3 and [Pd2(L2)2LCLD](NO3)4. Likewise, a combination of 2E·4NO3, 2F·4NO3 and [Pd2(L2)2LELF](NO3)4 was supported by ESI-MS data for case IV (Fig. 7B). However, all the other diverging ligand combinations (i.e., LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LD, LB
LD, LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LE, LB
LE, LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LF, LC
LF, LC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LE, LC
LE, LC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LF and LD
LF and LD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LF) apart from the cases I−IV, displayed 2-fold heteromeric completive self-sorting outcome. So, it is understandable that the heteromeric assembly using two different diverging ligands together with L2 and Pd(NO3)2 yielded a 2-fold heteromeric completive self-sorting outcome, where the two diverging ligands could induce two different conformations of L2. On the other hand, when two different diverging ligands that can induce the same conformations produce mixed self-sorting (a combination of integrative and heteromeric) outcomes. A similar self-sorting outcome was observed for cage fusion reaction (case I–IV) of the three homoleptic assemblies, i.e., 2·4NO3 and the two chosen X·8NO3 (X = B–F). The 1H NMR spectra of all the cage mixtures obtained through 2-fold heteromeric completive self-sorting are comparable to that of the respective mixed-ligated Pd2La2Lx2-type cages (Fig. S125–S134).
LF) apart from the cases I−IV, displayed 2-fold heteromeric completive self-sorting outcome. So, it is understandable that the heteromeric assembly using two different diverging ligands together with L2 and Pd(NO3)2 yielded a 2-fold heteromeric completive self-sorting outcome, where the two diverging ligands could induce two different conformations of L2. On the other hand, when two different diverging ligands that can induce the same conformations produce mixed self-sorting (a combination of integrative and heteromeric) outcomes. A similar self-sorting outcome was observed for cage fusion reaction (case I–IV) of the three homoleptic assemblies, i.e., 2·4NO3 and the two chosen X·8NO3 (X = B–F). The 1H NMR spectra of all the cage mixtures obtained through 2-fold heteromeric completive self-sorting are comparable to that of the respective mixed-ligated Pd2La2Lx2-type cages (Fig. S125–S134).
Next, we intend to extend the self-sorting processes to a more intricate level, i.e., 3-fold heteromeric completive self-sorting. Achieving three discrete co-existing mixed-ligated assemblies via the self-assembly of four distinct ligands could be a fascinating study. We deduced that using the diverging ligand pairs inducing two different conformations in L2 is essential for attaining orthogonality in the coordination cage systems to achieve 2-fold heteromeric completive self-sorting. Hence, we decided to pick a mix of three diverging ligands that induce three distinct conformations of L2 (i.e., L2SS, L2SA and L2AA) to achieve 3-fold heteromeric completive self-sorting. Firstly, we have chosen diverging ligands LB, LD and LF, such that L2 would adapt to L2SS, L2SA and L2AA conformations in their respective mixed ligated cages. A mixture of L2, LB, LD, LF and Pd(NO3)2 (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) in DMSO-d6 was stirred at room temperature for 12 hours (path A). Very interestingly, the 1H NMR spectrum showed clean formation of three different Pd2La2Lx2-type assemblies, 2B·4NO3, 2D·4NO3 and 2F·4NO3 co-existing in the solution (Fig. S142). Here, the three different mixed-ligated assemblies share a common ligand (L2). However, they differ in the relative orientation of the amide moiety in the cage structures, where the ligand L2 is locked in syn–syn, syn–anti and anti–anti conformations. Alternatively, the cage fusion of the four high-symmetry homoleptic assemblies 2·4NO3, B·8NO3, D·8NO3 and F·8NO3 (both D·8NO3 and F·8NO3 exist alongside a minor product (Pd3L6)) also resulted in the simultaneous formation of three low-symmetry mixed ligated assemblies (path B) representing the 3-fold heteromeric completive self-sorting as shown in Fig. 8. Though the system involves simultaneous utilization of multiple metal–ligand interactions, full orthogonality within the Pd2La2Lx2-type cages was achieved by geometric complementarity and conformational adaptability of L2 in the presence of other rigid diverging ligands (Lx). A similar 3-fold heteromeric self-sorting process was also observed for the other combinations of the diverging ligands (LB
3) in DMSO-d6 was stirred at room temperature for 12 hours (path A). Very interestingly, the 1H NMR spectrum showed clean formation of three different Pd2La2Lx2-type assemblies, 2B·4NO3, 2D·4NO3 and 2F·4NO3 co-existing in the solution (Fig. S142). Here, the three different mixed-ligated assemblies share a common ligand (L2). However, they differ in the relative orientation of the amide moiety in the cage structures, where the ligand L2 is locked in syn–syn, syn–anti and anti–anti conformations. Alternatively, the cage fusion of the four high-symmetry homoleptic assemblies 2·4NO3, B·8NO3, D·8NO3 and F·8NO3 (both D·8NO3 and F·8NO3 exist alongside a minor product (Pd3L6)) also resulted in the simultaneous formation of three low-symmetry mixed ligated assemblies (path B) representing the 3-fold heteromeric completive self-sorting as shown in Fig. 8. Though the system involves simultaneous utilization of multiple metal–ligand interactions, full orthogonality within the Pd2La2Lx2-type cages was achieved by geometric complementarity and conformational adaptability of L2 in the presence of other rigid diverging ligands (Lx). A similar 3-fold heteromeric self-sorting process was also observed for the other combinations of the diverging ligands (LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC
LC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LF, LB
LF, LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC
LC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LE and LB
LE and LB![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LD
LD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LE) with L2 and Pd(NO3)2. In all the cases, the 1H NMR spectrum showed clean formation of the desired co-existing mixed ligated assemblies (Fig. S139–S141).
LE) with L2 and Pd(NO3)2. In all the cases, the 1H NMR spectrum showed clean formation of the desired co-existing mixed ligated assemblies (Fig. S139–S141).
Conclusions
In summary, we have demonstrated a substantial change in the mixed ligand complexation behavior of a converging ligand by incorporating di-amide functionality on its backbone. While L1 undergoes absolute integrative self-sorting with only one ligand from a series of diverging ligands (LA–LG) to form a cis-Pd2La2Lx2-type cage. On the other hand, the integration of amide moieties on both sides, i.e., L2, enables a family of five cis-Pd2La2Lx2-type architectures via self-assembly of the conformationally adaptive ligand and complementary ligands of varied lengths ranging from 8.1 to 15 Å. Fascinatingly, adaptability of L2 with three distinct switchable conformational states enabled us to explore the higher order 2-/3-fold heteromeric completive self-sorting. An unprecedented, cage fusion of four different homoleptic assemblies resulted in a 3-fold heteromeric completive self-sorting, where three distinct cis-Pd2La2Lx2-type assemblies were found to co-exist in solution. The error-free orthogonality in the coordination cage system involving multiple metal–ligand interactions was a result of the ability of the conformationally adaptive ligand L2 to attain three different conformations (L2SS, L2SA and L2AA) in the cis-Pd2La2Lx2-type cages. This work illustrates that the introduction of switchable conformational states in the ligand backbone paves the way for constructing new architectures with modulable cavity size, shape and functions.Author contributions
M. P., V. S., and D. K. C. designed the project. M. P. and V. S. carried out the research and analyzed the data. M. P., V. S., and D. K. C. wrote the manuscript. V. R. refined the SC-XRD data and contributed to the preparation of the manuscript. D. K. C. is the principal investigator and managed the project. All the authors reviewed the manuscript and have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
CCDC 2466372 (L2) and 2466383–2466385 (2C·4NO3, 2D·4NO3, and 2F·4BF4) contain the supplementary crystallographic data for this paper.23a–dThe data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc05568g.
Acknowledgements
D. K. C. thanks the Anusandhan National Research Foundation (ANRF) (formerly Science and Engineering Research Board (SERB)), Government of India (Project no. CRG/2022/004413), for financial support. D. K. C. thanks IIT Madras for financial support through a Mid-Career Institute Research and Development Award (Project no. IRDA-2019) and through a Center under Institute of Eminence program (IoE Center of Molecular Architecture) (Project no. IoE Phase II). M. P. thanks UGC for a fellowship. We also thank the CoE on Molecular Materials and Functions and IoE Center of Molecular Architecture for IoE funded single crystal X-ray diffraction facility. We thank the Department of Chemistry, IIT Madras, for NMR facility and DST-FIST funded ESI-MS facility. We thank Ayan Ghosh, Dr. D. Elias Jesu Packiam, for rendering help in single-crystal X-ray data collection, and Dr. R. Baskar for DOSY NMR measurements. We thank the P. G. Senapathy Centre for Computing Resources, IIT Madras, for providing access to the Gaussian16 package.References
- (a) M. Ruben, J. Rojo, F. J. Romero-Salguero, L. H. Uppadine and J.-M. Lehn, Angew. Chem., Int. Ed., 2004, 43, 3644–3662 Search PubMed; (b) M. Fujita, M. Tominaga, A. Hori and B. Therrien, Acc. Chem. Res., 2005, 38, 371–380 Search PubMed; (c) T. R. Cook and P. J. Stang, Coord. Chem. Rev., 2015, 115, 7001–7045 CrossRef CAS PubMed; (d) S. D. P. Fielden, D. A. Leigh and S. L. Woltering, Angew. Chem., Int. Ed., 2017, 56, 11166–11194 CrossRef CAS PubMed; (e) Q. Chen and K. Zhu, Chem. Soc. Rev., 2024, 53, 5677–5703 Search PubMed; (f) P. J. Stang and B. Olenyuk, Acc. Chem. Res., 1997, 30, 502–518 CrossRef CAS; (g) J. Dong, Y. Liu and Y. Cui, Acc. Chem. Res., 2021, 54, 194–206 Search PubMed; (h) K. Acharyya and P. S. Mukherjee, Angew. Chem., Int. Ed., 2019, 58, 8640–8653 Search PubMed; (i) J. R. Nitschke, Acc. Chem. Res., 2007, 40, 103–112 CrossRef CAS PubMed; (j) Y.-F. Han and G.-X. Jin, Acc. Chem. Res., 2014, 47, 3571–3579 CrossRef CAS PubMed.
- (a) D. Zhang, T. K. Ronson, Y.-Q. Zou and J. R. Nitschke, Nat. Rev. Chem., 2021, 5, 168–182 CrossRef CAS PubMed; (b) A. Galan and P. Ballester, Chem. Soc. Rev., 2016, 45, 1720–1737 RSC; (c) S. He, L. Wu, X. Li, H. Sun, T. Xiong, J. Liu, C. Huang, H. Xu, H. Sun, W. Chen, R. Gref and J. Zhang, Acta Pharm. Sin. B, 2021, 11, 2362–2395 CrossRef CAS PubMed; (d) Y. Xue, X. Hang, J. Ding, B. Li, R. Zhu, H. Pang and Q. Xu, Coord. Chem. Rev., 2021, 430, 213656 CrossRef CAS; (e) V. Sivalingam, S. Krishnaswamy and D. K. Chand, Chem.-Eur.J., 2023, 29, e202300891 CrossRef CAS PubMed.
- A. Wu and L. Isaacs, J. Am. Chem. Soc., 2003, 125, 4831–4835 CrossRef CAS PubMed.
- (a) T. K. Ronson, D. A. Roberts, S. P. Black and J. R. Nitschke, J. Am. Chem. Soc., 2015, 137, 14502–14512 CrossRef CAS PubMed; (b) Y.-R. Zheng, Z. Zhao, M. Wang, K. Ghosh, J. B. Pollock, T. R. Cook and P. J. Stang, J. Am. Chem. Soc., 2010, 132, 16873–16882 CrossRef CAS PubMed; (c) S. Mukherjee and P. S. Mukherjee, Chem. Commun., 2014, 50, 2239–2248 RSC; (d) C.-B. Tian and Q.-F. Sun, Chem.-Eur.J., 2023, 29, e202300195 CrossRef CAS PubMed; (e) J. A. Davies, T. K. Ronson and J. R. Nitschke, J. Am. Chem. Soc., 2024, 146, 5215–5223 CrossRef CAS PubMed; (f) D. Preston and J. D. Evans, Angew. Chem., Int. Ed., 2023, 62, e202314378 CrossRef CAS PubMed; (g) S. Pullen, J. Tessarolo and G. H. Clever, Chem. Sci., 2021, 12, 7269–7293 RSC; (h) Q.-F. Sun, S. Sato and M. Fujita, Angew. Chem., Int. Ed., 2014, 53, 13510–13513 CrossRef CAS PubMed; (i) S. Sudan, R.-J. Li, S. M. Jansze, A. Platzek, R. Rudolf, G. H. Clever, F. Fadaei-Tirani, R. Scopelliti and K. Severin, J. Am. Chem. Soc., 2021, 143, 1773–1778 CrossRef CAS PubMed; (j) S. Prusty, K. Yazaki, M. Yoshizawa and D. K. Chand, Chem.-Eur.J., 2017, 23, 12456–12461 Search PubMed; (k) S. Samantray, S. Krishnaswamy and D. K. Chand, Nat. Commun., 2020, 11, 880 CrossRef CAS PubMed.
- (a) D. A. McMorran and P. J. Steel, Angew. Chem., Int. Ed., 1998, 37, 3295–3297 CrossRef CAS; (b) W. M. Bloch, J. J. Holstein, W. Hiller and G. H. Clever, Angew. Chem., Int. Ed., 2017, 56, 8285–8289 CrossRef CAS PubMed; (c) Y. Liu, S.-H. Liao, W.-T. Dai, Q. Bai, S. Lu, H. Wang, X. Li, Z. Zhang, P. Wang, W. Lu and Q. Zhang, Angew. Chem., Int. Ed., 2023, 62, e202217215 CrossRef CAS PubMed; (d) T. Abe, N. Sanada, K. Takeuchi, A. Okazawa and S. Hiraoka, J. Am. Chem. Soc., 2023, 145, 28061–28074 CrossRef CAS PubMed; (e) K. Wu, E. Benchimol, A. Baksi and G. H. Clever, Nat. Chem., 2024, 16, 584–591 CrossRef CAS PubMed.
- M. L. Saha, S. De, S. Pramanik and M. Schimttel, Chem. Soc. Rev., 2013, 42, 6860–6909 RSC.
- E. Benchimol, I. Regeni, B. Zhong, M. Kabiri, J. J. Holstein and G. H. Clever, J. Am. Chem. Soc., 2024, 146, 6905–6911 CrossRef CAS PubMed.
- M. Parbin, V. Sivalingam and D. K. Chand, Angew. Chem., Int. Ed., 2024, 63, e202410219 CrossRef CAS PubMed.
- M. L. Saha and M. Schmittel, Org. Biomol. Chem., 2012, 10, 4651–4684 RSC.
- (a) T. Haliloglu and I. Bahar, Curr. Opin. Struct. Biol., 2015, 35, 17–23 CrossRef CAS PubMed; (b) S. Corless and N. Gilbert, Biophys. Rev., 2016, 8, 245–258 CrossRef CAS PubMed; (c) S. Raman, Biochemistry, 2018, 57, 376–382 CrossRef CAS PubMed.
- (a) Y.-H. Huang, Y.-L. Lu, J. Ruan, S.-P. Zheng, X.-D. Zhang, C.-H. Liu, Y.-H. Qin, Z.-M. Cao, Z. Jiao, H.-S. Xu and C.-Y. Su, J. Am. Chem. Soc., 2023, 145, 23361–23371 CrossRef CAS PubMed; (b) D. Zhang, Q. Gan, A. J. Plajer, R. Lavendomme, T. K. Ronson, Z. Lu, J. D. Jensen, B. W. Laursen and J. R. Nitschke, J. Am. Chem. Soc., 2022, 144, 1106–1112 CrossRef CAS PubMed; (c) J. Mendez-Arroyo, J. Barroso-Flores, A. M. Lifschitz, A. A. Sarjeant, C. L. Stern and C. A. Mirkin, J. Am. Chem. Soc., 2014, 136, 10340–10348 CrossRef CAS PubMed; (d) A. P. Gia, A. de. Juan, D. Aranda, F. G. Guijarro, J. Aragó, E. Ortí, M. García-Iglesias and D. González-Rodríguez, J. Am. Chem. Soc., 2025, 147, 918–931 CrossRef CAS; (e) H. Xu, T. K. Ronson, A. W. Heard, P. C. P. Teeuwen, L. Schneider, P. Pracht, J. D. Thoburn, D. J. Wales and J. R. Nitschke, Nat. Chem., 2025, 17, 289–296 CrossRef CAS PubMed; (f) W. L. Jiang, B. Huang, X. L. Zhao, X. Shi and H. B. Yang, Chem, 2023, 9, 2655–2668 CrossRef CAS; (g) J. Mosquera, T. K. Ronson and J. R. Nitschke, J. Am. Chem. Soc., 2016, 138, 1812–1815 CrossRef CAS PubMed.
- (a) R. Yamasaki, A. Tanatani, I. Azumaya, S. Saito, K. Yamaguchi and H. Kagechika, Org. Lett., 2003, 5, 1265–1267 CrossRef CAS; (b) J. K. R. Deka, B. Sahariah, K. Baruah, A. K. Bar and B. K. Sarma, Chem. Commun., 2020, 56, 4874–4877 RSC; (c) P. Xing, Y. Liu, B. Li, Z.-Y. Dong, H.-J. Qian and L. Wang, Inorg. Chim. Acta., 2022, 536, 120854 CrossRef CAS.
- (a) W. M. Bloch, Y. Abe, J. J. Holstein, C. M. Wandtke, B. Dittrich and G. H. Clever, J. Am. Chem. Soc., 2016, 138, 13750–13755 CrossRef CAS PubMed; (b) A. Platzek, S. Juber, C. Yurtseven, S. Hasegawa, L. Schneider, C. Drechsler, K. E. Ebbert, R. Rudolf, Q.-Q. Yan, J. J. Holstein, L. V. Schafer and G. H. Clever, Angew. Chem., Int. Ed., 2022, 61, e202209305 CrossRef CAS PubMed.
- R. S. Rarig, R. Lam, P. Y. Zavalij, J. K. Ngala, R. L. LaDuca, J. E. Greedan and J. Zubieta, Inorg. Chem., 2002, 41, 2124–2133 CrossRef CAS PubMed.
- E. Bosch and C. L. Barnes, New J. Chem., 2001, 25, 1376–1378 RSC.
- D. K. Chand, K. Biradha, M. Kawano, S. Sakamoto, K. Yamaguchi and M. Fujita, Chem.–Asian J., 2006, 1, 82–90 CrossRef CAS PubMed.
- E. Merkul, D. Urselmann and T. J. J. Müller, Eur. J. Org Chem., 2011, 2011, 238–242 CrossRef.
- K. J. Kilpin, M. L. Gower, S. G. Telfer, G. B. Jameson and J. D. Crowley, Inorg. Chem., 2011, 50, 1123–1134 CrossRef CAS PubMed.
- H. Yu, Z. Guo, J. Tang, N. Han, J. Shi, M. Li, H. Zhang and M. Wang, Chem. Sci., 2025, 16, 6114–6120 RSC.
- R.-J. Li, J. de Montmollin, F. Fadaei-Tirani, R. Scopelliti and K. Severin, Dalton Trans., 2023, 52, 6451–6456 RSC.
- J. A. Findlay, K. M. Patil, M. G. Gardiner, H. I. MacDermott-Opeskin, M. L. O'Mara, P. E. Kruger and D. Preston, Chem.–Asian J., 2022, 17, e202200093 CrossRef CAS PubMed.
- A. Tarzia, W. Shan, V. Posligua, C. J. T. Cox, L. Male, B. D. Egleston, R. L. Greenaway, K. E. Jelfs and J. E. M. Lewis, Chem.-Eur.J., 2025, 31, e202403336 CrossRef CAS PubMed.
- (a) CCDC 2466372: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2nsgdb; (b) CCDC 2466383: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2nsgrp; (c) CCDC 2466384: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2nsgsq; (d) CCDC 2466385: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2nsgtr.
| This journal is © The Royal Society of Chemistry 2026 |