Open Access Article
Open Access ArticleErasable and regenerated multicomponent patterned polymer brushes
Yuhong
Cui
a,
Baoluo
He
 a,
Qian
Ye
a,
Qian
Ye
 a,
Feng
Zhou
a,
Feng
Zhou
 b and
Bin
Li
b and
Bin
Li
 *bc
*bc
aState Key Laboratory of Solidification Processing, Center of Advanced Lubrication and Seal Materials, School of Materials Science and Engineering, Northwestern Polytechnical University, 710072 Xi'an, China
bState Key Laboratory of Solid Lubrication, Lanzhou Institute of Chemical Physics, Chinese Academy of Sciences, Tianshui Middle Road 18, 730000 Lanzhou, China. E-mail: binli@licp.cas.cn
cShandong Laboratory of Advanced Materials and Green Manufacturing at Yantai, Shandong 264006, China
First published on 7th January 2026
Abstract
Patterning polymer brushes represents a significantly controllable approach to surface modifications, capable of producing tailored interfacial properties. In particular, multi-component patterned polymer brushes consist of various polymer types, thereby offering enhanced versatility in surface functionalization and interface regulation. Here, we present a novel DNA hybridization-based microcontact printing technique (µCP) for the fabrication of patterned polymer brushes, which enhances the precision and controllability of the patterning process. Initially, µCP is employed to immobilize thiol end-functionalized single-stranded DNA (ssDNA) to a gold substrate. The immobilized ssDNA subsequently hybridizes with initiator-functionalized complementary ssDNA, facilitating surface-initiated atom transfer radical polymerization (SI-ATRP) within the delineated regions to fabricate patterned polymer brushes. This method enables precise control over the molecular weight, chemical composition, and functionality of polymer brushes, and also allows reversible grafting of polymer brushes by modulating the unwinding and rehybridization of double-stranded DNA (dsDNA). Furthermore, this surface grafting technique exhibits remarkable adaptability for constructing binary and ternary brush surfaces through the integration of diverse polymer types. Consequently, it provides a robust platform for developing multifunctional surfaces tailored to specific applications, such as biosensing and diagnostics.
Introduction
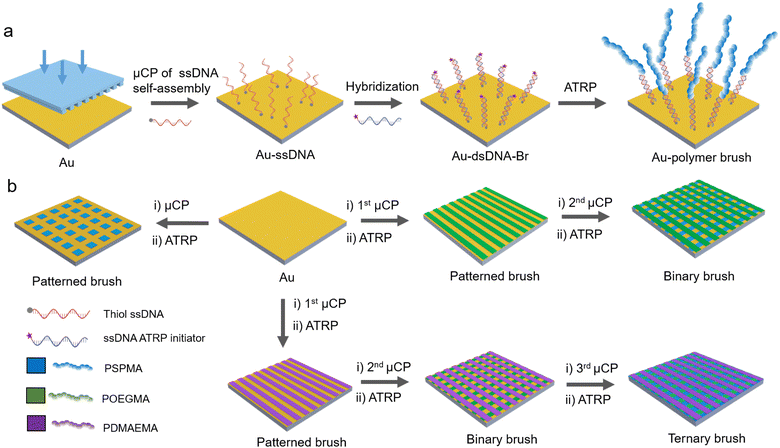
Polymer brushes are a type of brush-like interfacial material formed by chemically anchoring one end of polymer chains to a substrate surface, and can be used to regulate the physical and chemical properties of the material surface. The controlled fabrication of polymer brushes relies on surface-initiated polymerization techniques, such as atom transfer radical polymerization (ATRP), which enable precise control of the polymer chain length, grafting density, and chemical composition.1–3 Patterned polymer brushes integrate surface-initiated polymerizations (SIPs) with lithographic or other patterning methods. The preparation generally begins with the construction of a spatially resolved (patterned) initiator layer on the substrate, followed by controlled polymerization reactions to covalently grow polymer chains from the predetermined regions, thereby accurately transforming the two-dimensional initiator patterns into three-dimensional polymer brush arrays with high spatial resolution.4–12 Consequently, patterned polymer brushes have found widespread applications in many fields such as biosensing and microelectronics.13–16 Based on the complexity of the pattern, polymer brushes can be classified into unitary patterns and multi-component patterns. Unitary patterns refer to surfaces composed of a single type of polymer brush, characterized by uniform structures that are easy to control.17–21 Multi-component patterns consist of two or more different types of polymer brushes, which are typically arranged in alternating sequences or in a partitioned configuration. The construction of such multicomponent patterned systems from chemically or structurally different polymer brushes enables the realization of surface functionalities that cannot be achieved with simple, compositionally homogeneous coatings. This strategy enables the rational design of interfaces that integrate conflicting properties, such as combining antifouling with cell-adhesion domains to guide biological processes. Moreover, these systems can incorporate polymer brushes with disparate stimuli-responsiveness, yielding “smart” surfaces with programmable wettability, adhesion, and mass transfer characteristics for advanced bionic and diagnostic applications.22,23In order to advance the fabrication of micropatterned surfaces, a variety of surface modification techniques have been established for precise control over surface attributes, including nanoimprint lithography,24–26 femtosecond laser ablation,27,28 and µCP.29,30 Among these, µCP is characterized by its relative simplicity, high resolution and precision, cost-efficiency, and scalability. It patterns reactive molecules to a substrate surface using a soft template, typically polydimethylsiloxane (PDMS), thereby providing an accurate template for subsequent post-reactions.31–40 Moreover, µCP can be easily integrated with other nanofabrication techniques such as self-assembly, nanoparticle deposition, or optical lithography. In the early 1990s, Kumar and Whitesides pioneered the development of µCP and utilized it to prepare patterned arrays of self-assembled thiol monolayers on gold substrates.41 Despite the precision in pattern construction by µCP, the traditional µCP method faces certain challenges. For example, when printing multiple regions, maintaining clarity at the boundaries of different functional areas has been proven to be difficult, and diffusion-related cross-contamination between different inks is a great concern.42–44 Furthermore, when printing biomolecules, strict humidity control is essential to guarantee printing accuracy and biomolecule stability.45 To address these issues, the development of a more straightforward and efficient printing method is imperative. DNA is a double-helical biomacromolecule that is attractive due to its high specificity in hybridization and low nonspecific adsorption.46–53 These characteristics enable DNA molecules to precisely recognize and bind to target molecules in distinct regions, thereby enhancing precision and controllability, and fulfilling the rigorous requirements of biomolecular patterning.54,55 Moreover, He et al. demonstrated that surface-anchored DNA macroinitiators can significantly accelerate the growth rate of polymer brushes in the early stage of ATRP relative to conventional small-molecule initiators, owing to a synergistic effect of the charged phosphate backbone and the copper catalyst.56
The integration of µCP and highly controlled surface-initiated ATRP facilitates polymerization reactions to occur within precisely defined regions.57–67 Herein, we propose a novel surface patterning technique (Fig. 1). This approach involves the immobilization of the DNA initiator to the surface via µCP and integrating it with the ATRP reaction to facilitate the precise fabrication of multi-component patterned polymer brushes. This technique utilizes the unique molecular recognition and directional hybridization capabilities of DNA initiators to avoid the pattern blurring caused by initiator diffusion and non-specific adsorption in traditional processes, thus forming polymer brush patterns with clear boundaries. Through precise regulation of the ATRP process, the molecular weight and grafting density of polymer brushes can be controlled. Compared with traditional µCP, DNA hybridization exhibits a broader tolerance to ambient humidity and substantially enhances the robustness of the patterning process. Moreover, owing to the intrinsic reversibility of DNA hybridization, polymer brushes can be cleaved and regenerated.
Results and discussion
To synthesize the ATRP initiator, thiol-terminated ssDNA (ssDNA-SH) comprising 34 base pairs was initially attached to the surface of a gold substrate using a PDMS stamp to form a patterned monolayer. Subsequently, DBCO-terminated ssDNA (ssDNA-DBCO) with a complementary sequence was reacted with 2-azidoethyl 2-bromoisobutyrate through strain-promoted azide–alkyne “click” reaction (SPAAC), yielding Br-terminated ssDNA (ssDNA-Br) as the ATRP initiator. The molecular weight of DBCO-modified ssDNA was 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 028.7, following the reaction with 2-azidoethyl 2-bromoisobutyrate (Mw = 236.07); the molecular weight of ssDNA-Br increased to 11
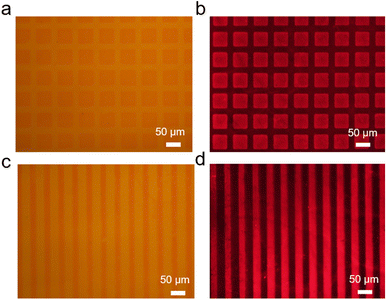
028.7, following the reaction with 2-azidoethyl 2-bromoisobutyrate (Mw = 236.07); the molecular weight of ssDNA-Br increased to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264.7, indicating the successful preparation of the ssDNA-Br initiator. ssDNA-Br was assembled with ssDNA-SH via complementary base pairing and was anchored on a gold surface (Fig. S4). ssDNA (5′-thiol, 3′-DBCO modified) reacted with azide-fluor-545 and was subsequently attached to the gold substrate using a 50 µm square-shaped PDMS stamp. The modified gold surface showed strong fluorescence, proving that ssDNA could self-assemble on the gold surface (Fig. S2 and S5). Two different PDMS stamp patterns, 50 µm square and 25 µm strip, were utilized to transfer ssDNA-SH, allowing for the creation of patterned ATRP initiators through hybridization with ssDNA-Br. With 3-sulfopropyl methacrylate potassium salt (SPMA) as the monomer, CuCl2·H2O as the catalyst, and ascorbic acid as the reducing agent, PSPMA brushes were obtained on the DNA initiator modified gold surface through surface-initiated atom transfer radical polymerization (SI-ATRP). Fig. 2 shows the optical and fluorescent microscopy images of PSPMA following acridine orange staining, with the intensely red fluorescent areas indicating the presence of PSPMA. To confirm the presence of PSPMA, the polymer on the surface was cleaved and collected for 1H-NMR and 13C-NMR measurements (Fig. S7). The peaks at 4.01, 2.86, and 1.99 ppm can be assigned to the methylene groups connecting the sulfonate group and the ester bond. The peak at 178.8 ppm in the 13C-NMR spectrum can be attributed to the carbonyl ester. These results suggest that PSPMA possesses a high degree of purity.
264.7, indicating the successful preparation of the ssDNA-Br initiator. ssDNA-Br was assembled with ssDNA-SH via complementary base pairing and was anchored on a gold surface (Fig. S4). ssDNA (5′-thiol, 3′-DBCO modified) reacted with azide-fluor-545 and was subsequently attached to the gold substrate using a 50 µm square-shaped PDMS stamp. The modified gold surface showed strong fluorescence, proving that ssDNA could self-assemble on the gold surface (Fig. S2 and S5). Two different PDMS stamp patterns, 50 µm square and 25 µm strip, were utilized to transfer ssDNA-SH, allowing for the creation of patterned ATRP initiators through hybridization with ssDNA-Br. With 3-sulfopropyl methacrylate potassium salt (SPMA) as the monomer, CuCl2·H2O as the catalyst, and ascorbic acid as the reducing agent, PSPMA brushes were obtained on the DNA initiator modified gold surface through surface-initiated atom transfer radical polymerization (SI-ATRP). Fig. 2 shows the optical and fluorescent microscopy images of PSPMA following acridine orange staining, with the intensely red fluorescent areas indicating the presence of PSPMA. To confirm the presence of PSPMA, the polymer on the surface was cleaved and collected for 1H-NMR and 13C-NMR measurements (Fig. S7). The peaks at 4.01, 2.86, and 1.99 ppm can be assigned to the methylene groups connecting the sulfonate group and the ester bond. The peak at 178.8 ppm in the 13C-NMR spectrum can be attributed to the carbonyl ester. These results suggest that PSPMA possesses a high degree of purity.
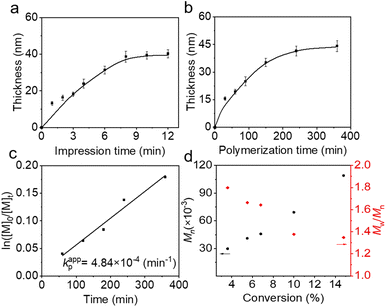
The growth dynamics of patterned polymer brushes were monitored using ellipsometry. As illustrated in Fig. 3a, the thickness of PSPMA increased with the extension of the printing time, reaching a maximum thickness at 8 minutes. Furthermore, the highest brush thickness was obtained when the ssDNA-SH content reached 70% (Fig. S8). By controlling the polymerization time, the thickness of polymer brushes can be adjusted. The logarithmic relationship between the initial and time-dependent monomer concentrations, ln([M]0/[M]t), demonstrated a linear growth with polymerization time and reached a plateau after 240 min (Fig. 3b and c). The evolution of molecular weight Mn with respect to monomer conversion was determined using gel permeation chromatography (GPC), and Mn increased linearly with monomer conversion. The monomer conversion rate reached 14.8% within 360 min, resulting in polymers with Mn ranging from 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 110
000 to 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and dispersity (Ɖ) ranging from 1.3 to 1.8, indicating a well-regulated living polymerization process enabled by the DNA-based ATRP initiator (Fig. 3d and S9).
000, and dispersity (Ɖ) ranging from 1.3 to 1.8, indicating a well-regulated living polymerization process enabled by the DNA-based ATRP initiator (Fig. 3d and S9).
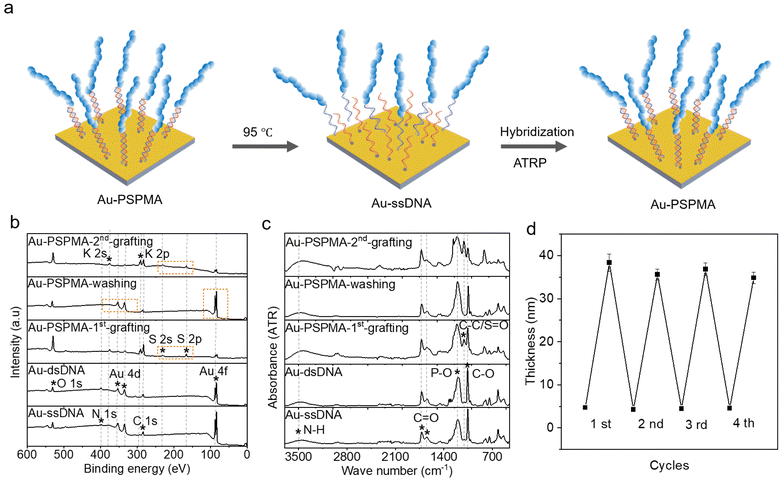
Polymer brushes can be detached from the gold surface under elevated temperature conditions (95 °C) by disrupting the hydrogen bonds between the complementary bases of dsDNA, wherein one strand of ssDNA remains anchored to the gold substrate through the gold–thiol bond, while the polymer brushes linked to another DNA strand become dissociated from the substrate. Consequently, upon subsequent rehybridization, the new ssDNA-Br initiator, possessing a complementary base sequence, can still reattach to the ssDNA-modified gold surface (Fig. 4a). The chemical composition of the gold surface after ssDNA assembly and hybridization, PSPMA growth, and detachment, was analyzed using X-ray photoelectron spectroscopy (XPS) and Fourier transform infrared spectroscopy (FTIR). The XPS spectrum revealed that after self-assembly of ssDNA on the gold surface, the N 1s signal was observed on the Au-ssDNA surface (Fig. 4b). The peaks at 398.3 and 399.8 eV correspond to the conjugated N and N–H/saturated N of the DNA base, respectively. Furthermore, the C 1s spectrum was characterized by three distinct peaks: C–C/C–H (284.8 eV), C–N/C–O (286.3 eV), and N–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–N (288.1 eV) (Fig. S10).
O)–N (288.1 eV) (Fig. S10).
Meanwhile, the signals corresponding to the N–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and P–O groups in DNA were also detected in the FTIR spectrum in Fig. 4c. The characteristic peaks of K and S elements of PSPMA were observed in the full XPS spectrum, while signals of Au 4d and Au 4f peaks were notably diminished. The peaks at 282.9, 284.7, 286.1, and 288.3 eV in the C 1s spectrum were attributed to –(C–C)n–, C–C/C–H, C–O, and O–C
O, and P–O groups in DNA were also detected in the FTIR spectrum in Fig. 4c. The characteristic peaks of K and S elements of PSPMA were observed in the full XPS spectrum, while signals of Au 4d and Au 4f peaks were notably diminished. The peaks at 282.9, 284.7, 286.1, and 288.3 eV in the C 1s spectrum were attributed to –(C–C)n–, C–C/C–H, C–O, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O moieties of PSPMA, respectively. The presence of the C–C/S
O moieties of PSPMA, respectively. The presence of the C–C/S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O signal was confirmed in the FTIR spectrum. These results showed that PSPMA was successfully immobilized to the surface of the gold. When the polymer brushes on the ssDNA were washed off, the signals of the S and K elements in the XPS spectrum disappeared, and the characteristic absorption peak of PSPMA disappeared in the FTIR spectrum. Following subsequent secondary hybridization and polymerization, these signals reappeared. The process was repeated four times, and the thickness of polymer brushes remained unaffected, demonstrating that polymer brushes prepared with the DNA initiator can be repeatedly erased and regenerated on the gold surface.
O signal was confirmed in the FTIR spectrum. These results showed that PSPMA was successfully immobilized to the surface of the gold. When the polymer brushes on the ssDNA were washed off, the signals of the S and K elements in the XPS spectrum disappeared, and the characteristic absorption peak of PSPMA disappeared in the FTIR spectrum. Following subsequent secondary hybridization and polymerization, these signals reappeared. The process was repeated four times, and the thickness of polymer brushes remained unaffected, demonstrating that polymer brushes prepared with the DNA initiator can be repeatedly erased and regenerated on the gold surface.
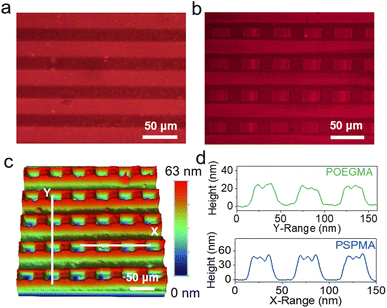
In addition, we have demonstrated the capability to precisely control the distribution of various polymer brush types on one single substrate surface, thereby facilitating the creation of complex binary and ternary pattern structures. The preparation of binary polymer brushes, which comprise two distinct polymer brushes, was initiated by employing a PDMS stamp with a 25 µm stripe pattern to print ssDNA-SH, followed by the growth of the first polymer brush, poly(ethylene glycol) methacrylate (POEGMA). For the fabrication of the second polymer brush (PSPMA), the same stamp of 25 µm stripe pattern was used. The gold substrate was subsequently rotated by 90°, and ssDNA-SH was stamped. ssDNA-SH was only printed onto regions where the PDMS stamp made contact with the gold substrate that had not been modified with POEGMA. Fig. 5a and b show the fluorescence images of the gold surface following each printing step and the growth of the polymer brush. During the initial printing stage, POEGMA was grown on the gold surface in horizontal stripes, forming a regular and uniform pattern. Subsequently, PSPMA was integrated based on the original POEGMA configuration to develop a POEGMA-PSPMA binary polymer brush surface featuring a cross arrangement. Nevertheless, at the periphery of the patterned polymer brushes, contact was compromised, preventing the transfer of ssDNA-SH and consequently leading to gaps between adjacent polymer brushes. The surface topography of the binary polymer brush surface was imaged using a 3D profilometer. As shown in Fig. 5c and d, the thickness of POEGMA was approximately 25 nm, whereas that of PSPMA was about 45 nm. The FTIR spectrum showed the characteristic absorption peaks corresponding to the C–C and S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional groups of PSPMA. Concurrently, the XPS spectrum revealed the presence of the S element of the sulfonic acid group in the PSPMA structure; these results confirmed the successful construction of the POEGMA-PSPMA binary polymer brush surface (Fig. S12).
O functional groups of PSPMA. Concurrently, the XPS spectrum revealed the presence of the S element of the sulfonic acid group in the PSPMA structure; these results confirmed the successful construction of the POEGMA-PSPMA binary polymer brush surface (Fig. S12).
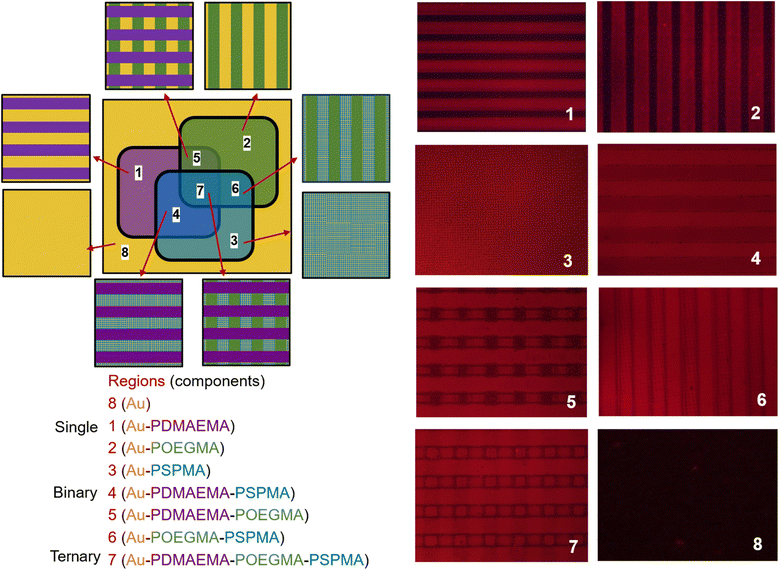
The versatility, efficiency, and adaptability of µCP technology render it an excellent tool for patterning complex structures utilizing various types of polymer brushes. To achieve a more intricate ternary brush pattern, the initial print employed a 25 µm stripe pattern. Subsequently, the second print involved rotating the gold wafer by 90° and utilizing the same 25 µm strip pattern to facilitate cross-printing. Finally, the third print adopted a 4 µm square pattern for backfilling the initiator. Fig. 6 schematically illustrates the patterned polymer brushes produced in different regions following the three printing steps. Starting with the initial printing of ssDNA-SH, and followed by ssDNA-Br hybridization and an ATRP reaction, polymer brushes exhibiting distinct patterns were successfully fabricated. These patterns comprised poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) (25 µm strips), POEGMA (25 µm strips), and PSPMA (4 µm squares). Through sequential cross-printing processes, ternary polymer brush surfaces can be fabricated. Subsequent to any two cross-printings, surfaces incorporating combinations of PDMAEMA-PSPMA, PDMAEMA-POEGMA, and POEGMA-PSPMA binary brushes can be achieved. Following three cross-printings, a surface of PDMAEMA-POEGMA-PSPMA ternary brushes was successfully fabricated. Fluorescence images showed the spatial distribution of various polymer brushes on the gold substrate. The analysis revealed that polymer brushes exhibited good surface coverage and demonstrated high uniformity. Despite differences in the physical and chemical properties of these polymer brushes, they retained well-defined boundary contours under fluorescence imaging. This observation suggests that cross-diffusion between adjacent regions was effectively suppressed, thereby ensuring that each polymer brush was confined to its designated area. Moreover, the precise spatial organization between distinct regions further proved the accuracy and controllability of the DNA initiator based printing technology (Fig. S13). Due to their unique structures and adjustable properties, these highly controllable polymer brushes hold significant potential for various applications, including biosensing, cell culture, and nanotechnology. Their versatility and customizability make them valuable tools for addressing complex challenges in surface science.
Conclusion
In summary, we have demonstrated a new DNA initiator based µCP method that is suitable for the preparation of multi-component patterned polymer brushes on gold substrates. This approach utilizes ssDNA and a custom-designed PDMS stamp, facilitating its attachment to the gold substrate through printing, which is restricted to areas where the two interfaces are in mutual contact. Subsequently, the ATRP initiator-terminated ssDNA is reversibly anchored to the complementary ssDNA-modified gold surface through dsDNA hybridization. Patterned polymer brushes are grown on the initiator-modified gold substrate through the ATRP reaction. These regions can undergo further patterning utilizing µCP to construct binary and ternary polymer brushes with complex and spatially defined structures. This innovative surface patterning technique enables the precise modulation of the molecular weight and grafting density of polymer brushes, and incorporates a unique capability for reversible surface modification. By leveraging the controlled unwinding of dsDNA and subsequent re-hybridization, polymer brushes can be repeatedly erased and regenerated from the gold substrate.Author contributions
B. Li, Q. Ye, and F. Zhou conceived and supervised the research; Y. Cui and B. He executed the experiments and analyzed the experimental data; Y. Cui and Q. Ye wrote the manuscript; B. Li and F. Zhou performed manuscript edits and revisions. All authors participated in discussions and approved the final manuscript.Conflicts of interest
The authors declare no conflicts of interest.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: experimental procedures, NMR spectra, SEM, and molecular weight. See DOI: https://doi.org/10.1039/d5sc08183a.Acknowledgements
This work was financially supported by the National Science Foundation of China (22472186), the Shandong Provincial Natural Science Foundation (ZR2023JQ023 and ZR2024MB117), and the Natural Science Foundation of Gansu Province (24JRRA048), China.Notes and references
- P. Chmielarz, M. Fantin, S. Park, A. A. Isse, A. Gennaro, A. J. D. Magenau, A. Sobkowiak and K. Matyjaszewski, Prog. Polym. Sci., 2017, 69, 47–78 Search PubMed.
- M. Abbasi, L. Faust and M. Wilhelm, Adv. Mater., 2019, 31, 1806484 Search PubMed.
- B. Li, B. Yu, W. T. S. Huck, F. Zhou and W. Liu, Angew. Chem., Int. Ed., 2012, 51, 5092–5095 Search PubMed.
- Z. Hassan, D. Varadharajan, C. Zippel, S. Begum, J. Lahann and S. Bräse, Adv. Mater., 2022, 34, 2201761 Search PubMed.
- C. Carbonell, D. Valles, A. M. Wong, A. S. Carlini, M. A. Touve, J. Korpanty, N. C. Gianneschi and A. B. Braunschweig, Nat. Commun., 2020, 11, 1244 CrossRef CAS PubMed.
- T. Chen, I. Amin and R. Jordan, Chem. Soc. Rev., 2012, 41, 3280–3296 Search PubMed.
- M. Flejszar, K. Ślusarczyk, P. Chmielarz, K. Wolski, A. A. Isse, A. Gennaro, M. Wytrwal-Sarna and M. Oszajca, Polymer, 2022, 255, 125098 CrossRef CAS.
- J. Ge, L.-H. Rong, X. Cheng, Y. Tang, D. J. Pochan, E. B. Caldona and R. C. Advincula, Macromolecules, 2025, 58, 3289–3297 Search PubMed.
- A. Johnson, J. Madsen, P. Chapman, A. Alswieleh, O. Al-Jaf, P. Bao, C. R. Hurley, M. L. Cartron, S. D. Evans, J. K. Hobbs, C. N. Hunter, S. P. Armes and G. J. Leggett, Chem. Sci., 2017, 8, 4517–4526 Search PubMed.
- C. G. Wang, C. Chen, K. Sakakibara, Y. Tsujii and A. Goto, Angew. Chem., Int. Ed., 2018, 57, 13504–13508 CrossRef CAS PubMed.
- C. G. Wang, H. W. Yong and A. Goto, ACS Appl. Mater. Interfaces, 2019, 11, 14478–14484 Search PubMed.
- Y.-H. Weng, H.-K. Tsao and Y.-J. Sheng, J. Phys. Chem. C, 2019, 123, 3560–3567 Search PubMed.
- Y. Hoshi, Y. Xu and C. K. Ober, Polymer, 2013, 54, 1762–1767 CrossRef CAS.
- K. L. Matos, Y. S. Zholdassov, M. A. Shlain, A. R. Cerullo, M. Barry, R. S. Deol, I. Kymissis and A. B. Braunschweig, Adv. Funct. Mater., 2025, 2502414 CrossRef CAS.
- B. Yu, B. S. Chang, W. S. Loo, S. Dhuey, P. O'Reilly, P. D. Ashby, M. D. Connolly, G. Tikhomirov, R. N. Zuckermann and R. Ruiz, ACS Nano, 2024, 18, 7411–7423 CrossRef CAS PubMed.
- A. Rajabi-Abhari, M. Soltani, K. Golovin and N. Yan, Nano Energy, 2023, 115, 108752 CrossRef CAS.
- Y. Liu, H. Hu, W. Ye, F. Zhou and J. Hao, J. Mater. Chem. C, 2013, 1, 902–907 RSC.
- P. Akarsu, R. Grobe, J. Nowaczyk, M. Hartlieb, S. Reinicke, A. Böker, M. Sperling and M. Reifarth, ACS Appl. Polym. Mater., 2021, 3, 2420–2431 CrossRef CAS PubMed.
- J. Ge, E. B. Caldona and R. C. Advincula, Polymer, 2025, 317, 127958 CrossRef CAS.
- S. Helfert, T. Zandrini, A. Rohatschek, M. Rufin, P. Machata, A. Zahoranová, O. G. Andriotis, P. J. Thurner, A. Ovsianikov, R. Liska and S. Baudis, Small Sci., 2024, 5, 2400263 CrossRef PubMed.
- S. Lamping, C. Buten and B. J. Ravoo, Acc. Chem. Res., 2019, 52, 1336–1346 CrossRef CAS PubMed.
- W. Sheng, B. Li, X. Wang, B. Dai, B. Yu, X. Jia and F. Zhou, Chem. Sci., 2015, 6, 2068–2073 RSC.
- F. Léonforte and M. Müller, Macromolecules, 2014, 48, 213–228 CrossRef.
- K. T. P. Lim, H. Liu, Y. Liu and J. K. W. Yang, Nat. Commun., 2019, 10, 25 CrossRef CAS PubMed.
- X. Nie, M. Xiong, J. Zeng, C. Li, Y. Chen, Z. Xu and W. Fan, Adv. Funct. Mater., 2024, 2416742 Search PubMed.
- L. Wang, H. Li, C. Zhao, L. Zhang, J. Li, S. U. Din, Z. Wang, J. Sun, S. A. G. Torres, Z. Fan and L. Wen, Nat. Mater., 2025, 24, 39–47 Search PubMed.
- S. Xuan, L. Zhuo, G. Li, Q. Zeng, J. Liu, J. Yu, L. Chen, Y. Yang, S. Liu, Y. Wang and K. Yin, Small, 2024, 20, 2404979 CrossRef CAS PubMed.
- D. Zhu, P. Zuo, F. Li, H. Tian, T. Liu, L. Hu, H. Huang, J. Liu and X. Qian, Colloid Interface Sci. Commun., 2024, 59, 100770 CrossRef CAS.
- T. B. Saw, A. Doostmohammadi, V. Nier, L. Kocgozlu, S. Thampi, Y. Toyama, P. Marcq, C. T. Lim, J. M. Yeomans and B. Ladoux, Nature, 2017, 544, 212–216 CrossRef CAS PubMed.
- B. Xu, M. Zhu, W. Zhang, X. Zhen, Z. Pei, Q. Xue, C. Zhi and P. Shi, Adv. Mater., 2016, 28, 3333–3339 CrossRef CAS PubMed.
- M. Reifarth, M. Sperling, R. Grobe, P. Akarsu, F. Schmitt, M. Schmette, S. Tank, K. M. Arndt, S. Chiantia, M. Hartlieb and A. Böker, Adv. Funct. Mater., 2025, 2423495 Search PubMed.
- N. Pallab, S. Reinicke, J. Gurke, R. Rihm, S. Kogikoski, M. Hartlieb and M. Reifarth, Polym. Chem., 2024, 15, 853–867 RSC.
- K. C. Pradel and N. Fukata, Nano Energy, 2021, 83, 105856 CrossRef CAS.
- B. Chang, D. Zhao and H. Sun, ACS Appl. Mater. Interfaces, 2022, 14, 23944–23950 CrossRef CAS PubMed.
- L. Fang, J. Zhang, Y. Chen, S. Liu, Q. Chen, A. Ke, L. Duan, S. Huang, X. Tian and Z. Xie, Adv. Funct. Mater., 2021, 31, 2100447 CrossRef CAS.
- L. Liu, Z. Cai, S. Xue, H. Huang, S. Chen, S. Gou, Z. Zhang, Y. Guo, Y. Yao, W. Bao and P. Zhou, Nat. Electron., 2025, 8, 135–146 Search PubMed.
- H. Wu, L. Wu, X. Zhou, B. Liu and B. Zheng, Small, 2018, 14, 1802128 CrossRef PubMed.
- E. P. Yalcintas, K. B. Ozutemiz, T. Cetinkaya, L. Dalloro, C. Majidi and O. B. Ozdoganlar, Adv. Funct. Mater., 2019, 29, 1906551 CrossRef CAS.
- X. Wang, M. Sperling, M. Reifarth and A. Böker, Small, 2020, 16, 1906721 CrossRef CAS PubMed.
- B. Li, B. Yu, W. T. S. Huck, W. Liu and F. Zhou, J. Am. Chem. Soc., 2013, 135, 1708–1710 CrossRef CAS PubMed.
- A. Kumar and G. M. Whitesides, Appl. Phys. Lett., 1993, 63, 2002–2004 CrossRef CAS.
- N. Pallab, E. Sperlich, M. Schenderlein, A. Krüger-Genge, J. Li, L. Zeininger, Z. Tošner, M. Uchman and M. Reifarth, Angew. Chem., Int. Ed., 2025, 64, e202501759 CrossRef CAS PubMed.
- I. Hirata, U. Zschieschang, T. Yokota, K. Kuribara, M. Kaltenbrunner, H. Klauk, T. Sekitani and T. Someya, Org. Electron., 2015, 26, 239–244 CrossRef CAS.
- O. Akbulut, A. A. Yu and F. Stellacci, Chem. Soc. Rev., 2010, 39, 30–37 RSC.
- S. Qiu, J. Ji, W. Sun, J. Pei, J. He, Y. Li, J. J. Li and G. Wang, Smart Mater. Med., 2021, 2, 65–73 Search PubMed.
- F. Huang, X. Zhou, D. Yao, S. Xiao and H. Liang, Small, 2015, 11, 5800–5806 CrossRef CAS PubMed.
- A. B. Marciel, D. J. Mai and C. M. Schroeder, Macromolecules, 2015, 48, 1296–1303 CrossRef CAS.
- N. Siegel, H. Hasebe, G. Chiarelli, D. Garoli, H. Sugimoto, M. Fujii, G. P. Acuna and K. Koł
![[a with combining cedilla]](https://www.rsc.org/images/entities/char_0061_0327.gif) taj, J. Am. Chem. Soc., 2024, 146, 17250–17260 CrossRef CAS PubMed.
taj, J. Am. Chem. Soc., 2024, 146, 17250–17260 CrossRef CAS PubMed. - Y. Tokura, Y. Jiang, A. Welle, M. H. Stenzel, K. M. Krzemien, J. Michaelis, R. Berger, C. Barner-Kowollik, Y. Wu and T. Weil, Angew. Chem., Int. Ed., 2016, 55, 5692–5697 CrossRef CAS PubMed.
- J. S. Wang and D. Y. Zhang, Nat. Chem., 2015, 7, 545–553 CrossRef CAS.
- F. Li, X. Ding, Z. Lv, J. Li and D. Yang, Nano Today, 2024, 54, 102061 CrossRef CAS.
- X. Lu, H. Fu, K. C. Shih, F. Jia, Y. Sun, D. Wang, Y. Wang, S. Ekatan, M. P. Nieh, Y. Lin and K. Zhang, J. Am. Chem. Soc., 2020, 142, 10297–10301 CrossRef CAS PubMed.
- A. R. Chandrasekaran, Nat. Rev. Chem., 2021, 5, 225–239 CrossRef CAS PubMed.
- X. Lou, M. S. Lewis, C. B. Gorman and L. He, Anal. Chem., 2005, 77, 4698–4705 CrossRef CAS PubMed.
- H. Qian and L. He, Anal. Chem., 2009, 81, 4536–4542 CrossRef CAS PubMed.
- X. Lou and L. He, Langmuir, 2006, 22, 2640–2646 CrossRef CAS PubMed.
- K. Matyjaszewski, Adv. Mater., 2018, 30, 1706441 CrossRef PubMed.
- J. O. Zoppe, N. C. Ataman, P. Mocny, J. Wang, J. Moraes and H.-A. Klok, Chem. Rev., 2017, 117, 1105–1318 CrossRef CAS PubMed.
- R. Wang, Q. Wei, W. Sheng, B. Yu, F. Zhou and B. Li, Angew. Chem., Int. Ed., 2023, 62, e202219312 CrossRef CAS PubMed.
- M.-N. Antonopoulou, R. Whitfield, N. P. Truong, D. Wyers, S. Harrisson, T. Junkers and A. Anastasaki, Nat. Chem., 2021, 14, 304–312 CrossRef PubMed.
- F. Lorandi, M. Fantin and K. Matyjaszewski, J. Am. Chem. Soc., 2022, 144, 15413–15430 CrossRef CAS PubMed.
- K. Parkatzidis, H. S. Wang, N. P. Truong and A. Anastasaki, Chem, 2020, 6, 1575–1588 CAS.
- G. Szczepaniak, L. Fu, H. Jafari, K. Kapil and K. Matyjaszewski, Acc. Chem. Res., 2021, 54, 1779–1790 CrossRef CAS PubMed.
- W. Yan, S. Dadashi-Silab, K. Matyjaszewski, N. D. Spencer and E. M. Benetti, Macromolecules, 2020, 53, 2801–2810 CrossRef CAS.
- Y. Liu, Y. Liu, Y. Wu and F. Zhou, ACS Nano, 2025, 19, 11576–11603 CrossRef CAS PubMed.
- D. A. Corbin, K. O. Puffer, K. A. Chism, J. P. Cole, J. C. Theriot, B. G. McCarthy, B. L. Buss, C. H. Lim, S. R. Lincoln, B. S. Newell and G. M. Miyake, Macromolecules, 2021, 54, 4507–4516 CrossRef CAS PubMed.
- B. Li, B. Yu, Q. Ye and F. Zhou, Acc. Chem. Res., 2014, 48, 229–237 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |