 Open Access Article
Open Access ArticleDevelopment of novel salicylic acid derivatives with dual anti-inflammatory and anti-arthritic potentials: synthesis, in vitro bio-evaluation, and in silico toxicity prediction with molecular modeling simulations
Kholoud Heshama,
Wael M. Aboulthanab and
Ahmed Ragab *cd
*cd
aNanoscience and Technology Program, Faculty of Science, Galala University, Galala City, Suez 43511, Egypt
bBiochemistry Department, Biotechnology Research Institute, National Research Centre, Dokki, Cairo 12622, Egypt
cChemistry Department, Faculty of Science (boys), Al-Azhar University, Nasr City, Cairo 11884, Egypt. E-mail: ahmed_ragab@azhar.edu.eg;; ahmed_ragab7@ymail.com; Tel: +201009341359
dChemistry Department, Faculty of Science, Galala University, Galala City, Suez 43511, Egypt. E-mail: ahmed.abdelwahab@gu.edu.eg
First published on 15th January 2026
Abstract
Development of anti-inflammatory agents targeting COX and 5-LOX, along with anti-arthritic agents, is a crucial approach in drug discovery. In this study, we designed and synthesized novel azomethine salicylic acid derivatives 2–9 by condensing 4-aminosalicylic acid with various formyl or ketone groups attached to benzylidene or heterocyclic cores. The reaction was carried out under reflux conditions utilizing acetic acid as the solvent. Initially, target prediction was performed, and the results indicated that these derivatives have potential as inhibitors of enzymes, proteases, and kinases. Furthermore, the designed derivatives underwent evaluation to assess their anti-inflammatory activity through COX-1, COX-2, and 5-LOX, as well as their anti-arthritic properties. Three derivatives 2, 4, and 9 demonstrated the most significant activity, with IC50 values of 10.16 ± 0.18, 9.68 ± 0.17, and 10.13 ± 0.18 µg mL−1 for COX-1, and 7.68 ± 0.05, 7.32 ± 0.04, and 7.66 ± 0.05 µg mL−1 for COX-2, respectively. These results exhibited superior activity compared to Aspirin, which had IC50 values of 11.21 ± 0.12 and 8.45 ± 0.05 µg mL−1, while demonstrating competitive activity relative to Naproxen (IC50 = 8.13 ± 0.14 and 6.18 ± 0.04 µg mL−1) and Indomethacin (IC50 = 7.16 ± 0.05 and 5.47 ± 0.04 µg mL−1) for COX-1 and COX-2, respectively. For 5-LOX, compound 4 demonstrated the most potent activity, with an IC50 value of 11.64 ± 0.20 µg mL−1. This value is comparable to that of naproxen (IC50 = 9.65 ± 0.17 µg mL−1) and zileuton (IC50 = 8.43 ± 0.05 µg mL−1), while demonstrating greater efficacy than aspirin (IC50 = 13.68 ± 0.13 µg mL−1). These findings suggest that compound 4 may serve as a potent inflammatory mediator with multiple targets. In terms of arthritic activity, the synthesized derivatives demonstrated the ability to inhibit protein denaturation and proteinase activity, exhibiting moderate inhibitory effects. Finally, in silico toxicity predictions were conducted, demonstrating a safer profile compared to the utilized drugs. Additionally, docking simulations were performed for the most active derivatives, revealing higher binding affinities, supported by hydrogen bonding, arene-cation interactions, and hydrophobic interactions.
1 Introduction
Inflammation is a physiological response triggered by infection or injury, primarily aimed at eliminating harmful agents and promoting the repair of damaged tissues.1 During this process, various mediators are released sequentially, including eicosanoids, bradykinin, vasoactive amines, interleukins, cytokines, and growth factors.2 The inflammatory response is initiated by enzymes such as cyclooxygenases, lipoxygenases, and nitric oxide synthases (NOS), which are crucial for the biosynthesis of prostaglandins, thromboxanes, and prostacyclins, molecules essential to inflammatory processes.3,4 Prostaglandins (PGs) are critical precursors for numerous inflammatory mediators that initiate inflammatory responses in the body when their synthesis is inhibited by NSAIDs. Common adverse effects associated with NSAIDs include renal failure, gastrointestinal hemorrhage, and ulceration. These potential complications underscore the need for developing novel anti-inflammatory agents with enhanced therapeutic profiles.5 Five-lipoxygenase (5-LOX) is an enzyme that catalyzes the oxidation of polyunsaturated fatty acids to produce leukotrienes (LTs), which are key mediators of inflammation. These leukotrienes influence inflammation, being mediators of bronchoconstriction and hypersensitivity.6,7 Lipoxygenases are non-heme iron-containing dioxygenases that generate hydroperoxy metabolites (HPETEs) by converting substrates into hydroperoxy derivatives.8,9 These enzymes are found in various biological systems, including animals, plants, and fungi.10 5-Lipoxygenase (5-LOX) is expressed in various cell types, including basophils, neutrophils, eosinophils, leukocytes, and monocytes/macrophages. In quiescent cells, 5-LOX is localized either in the cytosol, as observed in neutrophils, or within a nuclear compartment associated with chromatin. Upon stimulation, 5-LOX translocates from the cytosol to the nuclear envelope and the perinuclear endoplasmic reticulum in neutrophils.11 There are several drugs that inhibit 5-LOX that can be taken orally, including zileuton, meclofenamate sodium (meclomen), and licofelone12 (Fig. 1). | ||
| Fig. 1 Rational study illustrates the structure of azomethine salicylic acid derivatives 2–9 and some common, as well as commercially available anti-inflammatory drugs. | ||
There are two cyclooxygenase enzymes (COXs): the constitutive COX-1 isozyme and the inducible COX-2 isozyme.13 The COX-1 isozyme is synthesized endogenously by various tissues and plays a crucial role in several physiological processes, including gastroprotection and vascular homeostasis.14 In contrast, COX-2 is upregulated in response to various mutagenic and pro-inflammatory stimuli and is involved in the biosynthesis of prostacyclin, which acts as an anti-platelet aggregation agent and vasodilator.15 Therefore, achieving dual inhibition of COX-1, COX-2, and 5-LOX is crucial for developing anti-inflammatory agents with fewer side effects.16 A moderately selective COX-2 inhibitor, such as celecoxib, is preferable to highly selective inhibitors like rofecoxib and valdecoxib, which were withdrawn from the market due to concerns regarding potential cardiovascular toxicity.17 Moreover, Aspirin or Indomethacin are non-steroidal anti-inflammatory drugs (NSAID), works by suppressing the biosynthesis of prostaglandins (PGs) through non-selective inhibition of the COX-1 and COX-2 enzymes18,19 (Fig. 1).
3Salicylates have garnered considerable attention for over a century due to their complex biological actions.20 Numerous diseases can be effectively treated with salicylic acid derivatives. One widely utilized nonsteroidal anti-inflammatory drug (NSAID) is acetylsalicylic acid, commonly known as aspirin.21 Acetylsalicylic acid (Aspirin) serves as a classic example of a selective COX-1 inhibitor, whereas naproxen and indomethacin function as non-selective COX inhibitors. Aspirin is derived from optimizing substituents around the salicylic acid nucleus and is used as an analgesic, antipyretic, and anti-inflammatory agent.22 The hybridization of salicylic acid derivatives with heterocyclic cores has demonstrated efficacy in targeting cancer cells as anticancer agents23,24 and has also shown potential as antibacterial agents.25,26 Furthermore, the incorporation of anilides into salicylic acid has demonstrated significant antifungal activity.27 Additionally, 4-aminosalicylic acid serves as an effective tuberculosis agent,28 while diflunisal, a derivative of 4-aminosalicylic acid, possesses potent analgesic and antipyretic properties.29 In the search for a superior alternative to aspirin, diflunisal, an unacetylated derivative of salicylic acid, was developed and exhibited prolonged anti-inflammatory and analgesic effects compared to aspirin and has reduced ulcerogenic properties.30 Schiff bases are characterized by the presence of an azomethine group, represented by the formula (R–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–R′), where R and R′ may consist of aryl, cycloalkyl, alkyl, or heterocyclic groups.31,32 These compounds are also referred to as anils and imines33 and are widely recognized for their versatility, displaying diverse chemical reactivity, coordination capabilities, and biological properties.34 Their structural flexibility and ease of synthesis have enabled their broad application in medicinal chemistry, catalysis, and materials science.35 Additionally, the significance of these compounds is mainly attributed to the nitrogen atom, which possesses a lone pair of electrons and is sp2 hybridized, thereby influencing their chemical and biological properties.36 The hydrolyzability of Schiff bases is influenced by several factors, including pH, the electronic properties of substituents, and the degree of conjugation with aromatic systems.37,38 Generally, Schiff bases derived from aromatic aldehydes with electron-withdrawing substituents exhibit enhanced stability and resistance to hydrolysis, especially at neutral or slightly basic pH.39,40 Additionally, intramolecular hydrogen bonding in ortho-substituted salicylaldimines significantly stabilizes the imine bond, reducing its susceptibility to nucleophilic attack.41,42 These compounds exhibited a wide range of biological activities, including anti-inflammatory, antioxidant, anticancer effects as VEGFR-2 inhibitors, antidiabetic properties, antibacterial effects, anti-arthritic activity, anti-Alzheimer effects, and more.43–48
N–R′), where R and R′ may consist of aryl, cycloalkyl, alkyl, or heterocyclic groups.31,32 These compounds are also referred to as anils and imines33 and are widely recognized for their versatility, displaying diverse chemical reactivity, coordination capabilities, and biological properties.34 Their structural flexibility and ease of synthesis have enabled their broad application in medicinal chemistry, catalysis, and materials science.35 Additionally, the significance of these compounds is mainly attributed to the nitrogen atom, which possesses a lone pair of electrons and is sp2 hybridized, thereby influencing their chemical and biological properties.36 The hydrolyzability of Schiff bases is influenced by several factors, including pH, the electronic properties of substituents, and the degree of conjugation with aromatic systems.37,38 Generally, Schiff bases derived from aromatic aldehydes with electron-withdrawing substituents exhibit enhanced stability and resistance to hydrolysis, especially at neutral or slightly basic pH.39,40 Additionally, intramolecular hydrogen bonding in ortho-substituted salicylaldimines significantly stabilizes the imine bond, reducing its susceptibility to nucleophilic attack.41,42 These compounds exhibited a wide range of biological activities, including anti-inflammatory, antioxidant, anticancer effects as VEGFR-2 inhibitors, antidiabetic properties, antibacterial effects, anti-arthritic activity, anti-Alzheimer effects, and more.43–48
Building upon previously established findings and the ongoing efforts of our research group to design and develop novel heterocyclic compounds utilizing hybridization approaches,49–53 this study focuses on the synthesis of a new salicylic acid pharmacophore that integrates aromatic and heterocyclic aldehydes and ketones to create a novel hybrid structure featuring Schiff base modifications. The designed salicylic acid derivatives encompass functional fragments such as pyrazole, quinoline, sulfonamide, salicylaldehyde, indole, furan, and indene, which are interconnected through azomethine groups and are anticipated to demonstrate enhanced biological properties. Additionally, all synthesized derivatives were evaluated for their inhibitory activity against cyclooxygenases (COX-1 and COX-2) and lipoxygenase, followed by an assessment of their anti-arthritic properties, including inhibition of protein denaturation and protease activity, to explore their potential as multi-target agents with both anti-inflammatory and anti-arthritic effects. Furthermore, in silico toxicity predictions were conducted, and molecular docking simulations were performed for the most promising derivatives against 5-lipoxygenase (5-LOX) to elucidate the binding modes and investigate the conformational dynamics of the tested derivatives within the active site.
2 Results and discussion
2.1 Chemistry
In this study, new series of Schiff bases derived from amino-salicylic acid were synthesized, and the structures of the resulting derivatives were thoroughly characterized using FT-IR, 1H and 13C NMR, and elemental analysis as described in Schemes 1 and 2. The formation of the azomethine group was classified according to the type of formyl or ketone group utilized in the condensation reaction. The first scheme involved the synthesis of simple benzylidene or inden-2-ylidene derivatives, whereas the second scheme resulted in the formation of Schiff bases that included heterocyclic moieties such as pyrazole, 2-oxoindoline, and benzo[h]quinoline derivatives.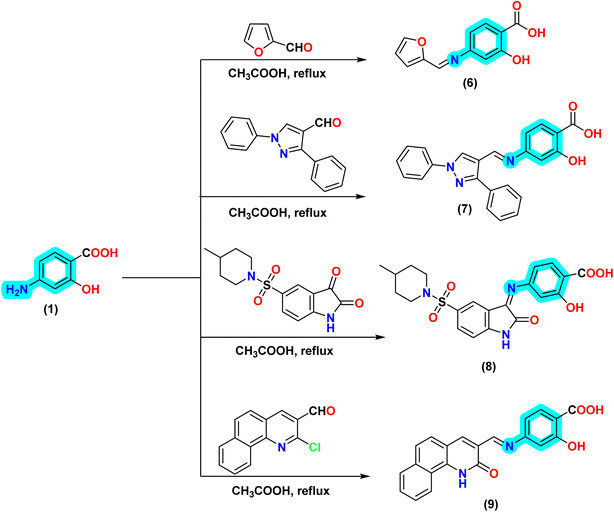 | ||
| Scheme 2 Synthesis of new salicylic acid Schiff base derivatives incorporating heterocyclic fragments, including furan, pyrazole, 2-oxoindoline, and benzo[h]quinoline derivatives. | ||
Firstly, an equimolar mixture of 4-aminosalicylic acid (4-ASA) (1) and 4-methoxybenzaldehyde and 4-isopropylbenzaldehyde (cuminaldehyde) in 10 mL of acetic acid was stirred under reflux conditions to afford compounds 2 and 3. The resulting solid product was obtained on hot. This solid product was then filtered while hot, recrystallized from methanol, and subsequently dried at room temperature to yield the final product as a solid powder. The IR spectrum of compound 2-hydroxy-4-((4-isopropylbenzylidene) amino)benzoic acid (3) displayed frequencies at 3359, 2976, 2923, 1688, and 1605 cm−1 related to hydroxyl groups, aliphatic-H (CH–sp3), carbonyl of carboxylic group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and azomethine group (C
O), and azomethine group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) that confirm the formation of Schiff base. The 1H NMR spectrum of compound 3 represented three significant signals at δ 9.97, 9.76, and 9.12 ppm related to aromatic hydroxyl group, carboxylic–OH proton, and azomethine proton (CH = N–), respectively. Additionally, the two methyl groups of isopropyl group were observed as two doublets with coupling constant J = 6.8 Hz at δ 1.22, and 1.15 ppm with integration of three for each signal, while the methine-CH appears as singlet signal at δ 2.96 ppm. The seven aromatic protons of compound 3 were recorded in the expected region between δ 7.78 and 6.67 ppm. The 13C NMR spectrum of compound 3 indicated three significant deshielded signals at δ 172.50, 167.77, 154.08 ppm related to the carbonyl of carboxylic group, carbon attached to phenolic OH, and carbon of azomethine group (–CH
N) that confirm the formation of Schiff base. The 1H NMR spectrum of compound 3 represented three significant signals at δ 9.97, 9.76, and 9.12 ppm related to aromatic hydroxyl group, carboxylic–OH proton, and azomethine proton (CH = N–), respectively. Additionally, the two methyl groups of isopropyl group were observed as two doublets with coupling constant J = 6.8 Hz at δ 1.22, and 1.15 ppm with integration of three for each signal, while the methine-CH appears as singlet signal at δ 2.96 ppm. The seven aromatic protons of compound 3 were recorded in the expected region between δ 7.78 and 6.67 ppm. The 13C NMR spectrum of compound 3 indicated three significant deshielded signals at δ 172.50, 167.77, 154.08 ppm related to the carbonyl of carboxylic group, carbon attached to phenolic OH, and carbon of azomethine group (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–). In addition, the carbons of two phenyl rings appear between δ 129.93–115.42 ppm, while the isopropyl fragment displayed as three signals at δ 33.93, 24.02, and 21.52 ppm (Scheme 1).
N–). In addition, the carbons of two phenyl rings appear between δ 129.93–115.42 ppm, while the isopropyl fragment displayed as three signals at δ 33.93, 24.02, and 21.52 ppm (Scheme 1).
Furthermore, the synthesized 2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzylidene derivative 4 was obtained by the reaction of 4-aminosalicylic acid (4-ASA) (1) with 5-sulfonyl salicylaldehyde under reflux conditions in an acidic medium. Conversely, the treatment of 4-aminosalicylic acid (4-ASA) (1) with ninhydrin yields the corresponding Schiff base featuring indene nucleus, as appeared in compound 5. The IR spectrum of compound 4 exhibited characteristic absorption bands at 3371, 3077, 1630, 1601, 1328, and 1149 cm−1, corresponding to the phenolic –OH group, sp2 carbon of the aromatic rings, carbonyl of the carboxylic group, azomethine group, and sulfonyl group, respectively. The 1H NMR spectrum exhibited three singlet signals at δ 11.39, 10.32, and 9.72 ppm, which were attributed to the two aromatic hydroxyl protons and the carboxylic –OH proton, respectively. In addition to a characteristic signal at δ 8.19 ppm for the imine proton. The pyrrolidine ring was represented by two singlet signals in the shielded region at δ 3.10 and 1.65 ppm. The aromatic protons appeared as two singlet signals at 7.99 and 5.97 ppm, along with four doublet signals at 7.91, 7.41, 7.20, and 6.08 ppm. In the 13C NMR spectrum of compound 4, signals were observed at δ 172.55, 163.89, 156.29, and 150.62 ppm, corresponding to the carbonyl, two phenolic hydroxyl groups, and the azomethine group, respectively. The presence of the pyrrolidine-functionalized ring was confirmed by two signals at δ 48.30 and 25.13 ppm, while the aromatic carbons exhibited chemical shifts ranging from 135.07 to 98.98 ppm (Scheme 1).
Furthermore, to incorporate some heterocyclic nucleus as furan and pyrazole moiety into the salicylic acid derivative, our research was extended to encompass the condensation of 4-aminosalicylic acid (4-ASA) (1) with furan and 4-formyl pyrazole derivative, leading to the formation of the corresponding Schiff base derivative 6 and 7. The 1H NMR spectrum of compound 7 exhibited two signals at δ 10.03 and 9.34 ppm, which are attributed to aromatic hydroxyl group and carboxylic –OH proton, as well as a singlet signal at δ 7.59 ppm corresponding to the imino proton (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–), and a signal at δ 7.53 ppm associated with pyrazole-H5. Additionally, thirteen aromatic protons were identified within the range of δ 8.01 to 7.10 ppm. Conversely, the 13C NMR spectrum indicated the presence of four signals at δ 172.50, 166.11, 157.18, and 153.57 ppm, corresponding to the carbonyl carbon, the carbon attached to the phenolic hydroxyl group, and the carbon adjacent to the imino carbon, respectively. Furthermore, all aromatic carbons appeared within the range of δ 145.71 to 113.71 ppm (Scheme 2).
N–), and a signal at δ 7.53 ppm associated with pyrazole-H5. Additionally, thirteen aromatic protons were identified within the range of δ 8.01 to 7.10 ppm. Conversely, the 13C NMR spectrum indicated the presence of four signals at δ 172.50, 166.11, 157.18, and 153.57 ppm, corresponding to the carbonyl carbon, the carbon attached to the phenolic hydroxyl group, and the carbon adjacent to the imino carbon, respectively. Furthermore, all aromatic carbons appeared within the range of δ 145.71 to 113.71 ppm (Scheme 2).
Subsequently, the reaction of 4-aminosalicylic acid (4-ASA) (1) with 5-sulfonyl isatin derivatives yields the corresponding Schiff base derivative 8, with the resultant structure being confirmed through spectroscopic techniques. Conversely, the treatment of 2-chloro-3-formyl-benzo[h]quinoline with the amino group of 4-aminosalicylic acid (4-ASA) (1) produces the corresponding Schiff base derivative 9. The reaction, conducted in acetic acid, results in the hydrolysis of the chloro atom at the second position of benzo[h]quinoline, converting it into a hydroxyl group that tautomerizes to give the 2-oxo-1,2-dihydrobenzo[h]quinoline derivative 7. The IR spectrum of compound 8 represented bands at 3445, 3363, 3157, 3070, 2950, 2927, and 2847 cm−1 corresponding to hydroxyl, NH, and CH-sp2 stretching modes associated with the aromatic structure, while the latter three bands are indicative of CH-sp3 vibrations from the aliphatic ring of 4-methylpiperidine. Furthermore, notable bands were observed at 1730, 1618, 1355, and 1148 cm−1, corresponding to the carbonyl group, azomethine moiety, and the sulfonyl group, respectively. Additionally, the 1H NMR spectrum of compound 8 demonstrated three signals in the downfield region at δ 10.71, 10.39, and 9.46 ppm, which are attributed to aromatic hydroxyl group, carboxylic –OH proton, and one NH group from the amide functionality of the 2-oxoindole nucleus. Additionally, the 1H NMR spectrum further confirmed the incorporation of 4-methylpiperidine, presenting two signals at δ 3.00 and 2.04–2.06 ppm corresponding to the two methylene groups adjacent to the nitrogen atom. The remaining two other methylene groups appear as two protons as a singlet signal at δ 1.64 and two singlet signals at δ 1.25 and 1.07 ppm with integration one for each proton. The methyl group of 4-methylpiperidine was observed at δ 0.83 ppm, while the proton of the tertiary carbon to which it is attached was detected at δ 1.54 ppm. The 13C NMR spectrum of compound 8 confirmed the existence of 4-methylpiperidine, displaying five signals in the upfield region at δ 46.45, 44.07, 33.30, 29.91, and 22.72 ppm. Additionally, the carbonyl of the carboxylic group was noted at δ 173.54 ppm, the carbonyl of the 2-oxoindole ring at δ 164.14 ppm, and two signals at δ 161.78 and 153.53 ppm were associated with the carbonyl linked to the phenolic hydroxyl group and the imino carbon, respectively (Scheme 2).
2.2 Target prediction studies
The in silico target prediction of the designed Schiff base 2–9 were carried out using SwissTargetPrediction web-based tool that developed by the Swiss Institute of Bioinformatics (SIB) that predicts the protein targets of small molecule compounds as described previously.54,55 By using SwissTargetPrediction, the target classes for each compound can be identified, as illustrated in Fig. 2. Firstly. introduction of 4-methoxybenzylidene to salicylic acid to synthesize compound 2 suggests its potential as an enzyme inhibitor, with a probability of 33.3%. The primary enzyme targets identified include the insulin-degrading enzyme and Methionyl-tRNA synthetase. Furthermore, compound 2 is anticipated to exhibit lyase activity with a probability value of 26.7%, specifically targeting carbonic anhydrase I and II. Additionally, it is categorized as a target for kinases, oxidoreductases, electrochemical transporters, phosphodiesterases, and family A G protein-coupled receptors, each with a probability of 6.7%. For derivative 3, kinases represent the largest proportion of the remaining categories at 33.3%, indicating their prevalence. Other targets, including lyases, oxidoreductases, phosphatases, kinases, and nuclear receptors, together account for nearly 13.3%. In the case of derivative 4, a similar distribution pattern is observed, albeit with varied percentages. The highest percentage corresponds to family A G protein-coupled receptors, accounting for 20.0%. This category specifically targets the Endothelin receptor ET-A and the Prostanoid EP2 receptor. Other classes, including oxidoreductases, kinases, erasers, and nuclear receptors, each make up 13.3% and target Egl nine homolog 1, Casein kinase II alpha, Lysine-specific demethylase 4C, and Thyroid hormone receptor alpha, respectively. Additionally, voltage-gated ion channels and phosphodiesterases target the sodium channel protein type X alpha subunit (by homology) and phosphodiesterase 5A (Fig. 2). Notably, both the enzyme and phosphodiesterase targeting rates are 20.0%, specifically focusing on ATP-citrate synthase and phosphodiesterase 5A.Furthermore, compound 5 is predicted to have the highest target prediction for nuclear receptors and erasers, with a probability value of 20%, followed by oxidoreductases and kinases at 13.3%. Compound 6 revealed multiple targets, with enzyme inhibitors predicted at 33.3%, followed by protease inhibitors at approximately 26.7% and membrane receptors at approximately 13.3%. For compound 7, the highest percentage is for phosphatase at 40.0%, specifically targeting protein-tyrosine phosphatase 1C. The next highest target is an enzyme, with a targeting percentage of 20.0%, specifically targeting anandamide amidohydrolase. In the chart for derivative 8, notable percentages include Family A G protein-coupled receptors at 33.3% and lyases at 26.7%. Moreover, the introduction of benzo[h]quinoline-2-one, as represented in compound 9, is predicted to increase the percentage of kinase inhibitors to 40%, followed by Family A G protein-coupled receptors targeting adenosine A1 and A3 receptors, along with additional targets such as phosphatases, oxidoreductases, and kinases, each with probability values of 13.3%. Additionally, target prediction was conducted for native amino salicylic acid to assess the effect of substituents on target prediction probability. The 4-amino salicylic acid was found to target lyases with a probability value of 46.7%, protease inhibitors at 26.7%, enzyme inhibitors at 13.3%, and oxidoreductases and unclassified proteins at 6.7%.
Finally, we can conclude that the comparative analysis of the predicted behaviors of the synthesized compounds relative to native amino salicylic acid (compound 1) reveals that the reaction of the amino group to form an azomethine linkage with benzylidene or heterocyclic fragments significantly enhances the targeting efficacy toward enzymes, kinases, and proteases to varying extents.
2.3 Biological evaluation
The biological activity of the synthesized salicylic acid derivatives was evaluated by assessing their inhibitory effects on cyclooxygenases (COX-1 and COX-2) and lipoxygenase. We then investigated their anti-arthritic properties, focusing on their ability to inhibit protein denaturation and protease activity. This study aimed to explore their potential as multi-target agents with both anti-inflammatory and anti-arthritic activities.2.3.1.1 Cyclooxygenase inhibitory activity and structural–activity relationship (SAR). The potency of the newly designed azomethine salicylic acid derivatives 2–9 were evaluated against cyclooxygenase enzymes (COX-1 and COX-2) in terms of inhibitory percentage at 100 µg mL−1 and subsequently measured their IC50 (concentration that inhibits 50% of enzymes) expressed as µg mL−1, as represented in Table 1.
| Sample (µg mL−1) | Anti-inflammatory activitya | |||
|---|---|---|---|---|
| COX-1 | COX-2 | |||
| Inhib. (%) | IC50 (µg mL−1) | Inhib. (%) | IC50 (µg mL−1) | |
| a All previous values were calculated as mean + SE from n = 3 samples. | ||||
| 2 | 67.02 ± 0.67 | 10.16 ± 0.18 | 69.19 ± 0.67 | 7.68 ± 0.05 |
| 3 | 36.97 ± 0.57 | 18.44 ± 0.66 | 38.26 ± 0.66 | 13.58 ± 0.35 |
| 4 | 70.37 ± 0.70 | 9.68 ± 0.17 | 72.54 ± 0.70 | 7.32 ± 0.04 |
| 5 | 22.43 ± 0.73 | 30.45 ± 1.46 | 24.60 ± 0.73 | 21.63 ± 0.66 |
| 6 | 25.11 ± 0.90 | 27.20 ± 1.24 | 27.28 ± 0.90 | 19.53 ± 0.91 |
| 7 | 22.48 ± 0.70 | 30.37 ± 1.42 | 24.65 ± 0.70 | 21.58 ± 0.65 |
| 8 | 24.92 ± 0.79 | 27.39 ± 1.20 | 27.10 ± 0.79 | 19.63 ± 0.53 |
| 9 | 67.22 ± 0.67 | 10.13 ± 0.18 | 69.39 ± 0.67 | 7.66 ± 0.05 |
| Naproxen | 83.77 ± 0.83 | 8.13 ± 0.14 | 85.95 ± 0.83 | 6.18 ± 0.04 |
| Aspirin | 60.72 ± 0.76 | 11.21 ± 0.12 | 62.90 ± 0.76 | 8.45 ± 0.05 |
| Indomethacin | 95.02 ± 1.49 | 7.16 ± 0.05 | 97.20 ± 1.49 | 5.47 ± 0.04 |
The results concerning the inhibitory potential of the azomethine salicylic acid derivatives 2–9, as represented by IC50 values, indicated a comparatively enhanced inhibition of COX-2, suggesting higher selectivity for COX-2 alongside notable inhibitory potential against cyclooxygenase enzymes. Furthermore, the evaluated azomethine salicylic acid derivatives 2–9 exhibited moderate to significant inhibitory activity, with IC50 values ranging from 9.68 ± 0.17 to 30.37 ± 1.42 µg mL−1, in contrast to the nonsteroidal anti-inflammatory drug (NSAID) positive control agents Naproxen (IC50 = 8.13 ± 0.14 µg mL−1), Aspirin (IC50 = 11.21 ± 0.12 µg mL−1), and Indomethacin (IC50 = 7.16 ± 0.05 µg mL−1) against COX-1. Moreover, regarding COX-2, the azomethine salicylic acid derivatives 2–9 demonstrated IC50 values ranging from 7.32 ± 0.04 to 21.63 ± 0.66 µg mL−1, compared to Naproxen (IC50 = 6.18 ± 0.04 µg mL−1), Aspirin (IC50 = 8.45 ± 0.05 µg mL−1), and Indomethacin (IC50 = 5.47 ± 0.04 µg mL−1). Notably, three of the salicylic acid derivatives, specifically 2, 4, and 9, exhibited the highest activity against both COX-1 and COX-2, in the order of 4 > 9 > 2. Of particular interest, the derivative 2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzylidene 4 demonstrated the most potent activity, with IC50 values of 9.68 ± 0.17 µg mL−1 and 7.32 ± 0.04 µg mL−1 against COX-1 and COX-2, respectively indicated strong inhibition to both enzymes. This enhanced activity may be attributed to the presence of two phenolic hydroxyl groups and a sulfonyl group, which could facilitate the formation of hydrogen bonds within the active sites of COX-1 and COX-2.
The structure–activity relationship analysis revealed that the introduction of the 4-methoxybenzylidene as an electron-donating group to salicylic acid (compound 2) resulted in significant activity, with IC50 values of 10.16 ± 0.18 µg mL−1 and 7.68 ± 0.05 µg mL−1 against COX-1 and COX-2, respectively. These IC50 values are lower than those of Aspirin, yet slightly higher than those of Naproxen and Indomethacin. Furthermore, the substitution of the methoxy group with an isopropyl group in the 4-isopropylbenzylidene derivative (compound 3) led to a dramatic decrease in activity against both enzymes, displaying IC50 values of 18.44 ± 0.66 µg mL−1 for COX-1 and 13.58 ± 0.35 µg mL−1 for COX-2. Additionally, the introduction of more hydrophilic groups, specifically dioxo groups, in compound 5, which features a 1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene moiety, resulted in decreased activity, with IC50 values of 30.45 ± 1.46 µg mL−1 for COX-1 and 21.63 ± 0.66 µg mL−1 for COX-2. This observation indicates that the increase in hydrophobic character does not enhance cyclooxygenase activity.
Furthermore, the second series 6–9, which contains a heterocyclic moiety via the azomethine group, demonstrates that the incorporation of the benzo[h]quinoline nucleus into salicylic acid through the azomethine group enhances as displayed in compound 9 exhibited significant inhibitory activity against COX-1 and COX-2, with IC50 values of 10.13 ± 0.18 and 7.66 ± 0.05 µg mL−1, respectively, approaching that of the 4-methoxybenzylidene derivative 2. Moderate COX-1 and COX-2 inhibitory activities were observed in the compounds 4-((furan-2-ylmethylene)amino)-2-hydroxybenzoic acid (compound 6), 1,3-diphenyl-1H-pyrazole derivative (compound 7), and 4-((5-((4-methylpiperidin-1-yl)sulfonyl)-2-oxoindolin-3-ylidene)amino)benzoic acid (compound 8), with IC50 values of 27.20 ± 1.24, 30.37 ± 1.42, and 27.39 ± 1.20 µg mL−1 for COX-1, and IC50 values of 19.53 ± 0.91, 21.58 ± 0.65, and 19.63 ± 0.53 µg mL−1 for COX-2, respectively. These results indicate that the effect of pyrazole or 5-(sulfonyl)isatin, when incorporated into salicylic acid derivatives, exhibited similar activity, while the incorporation of furan core results in a slight reduction in activity compared to compounds 7 and 8.
In conclusion, the most promising derivatives 2, 4, and 9, exhibited enhanced activity relative to aspirin, evidenced by lower IC50 values. Furthermore, their activity was comparable to that of naproxen and indomethacin, differing by only 2 µg mL−1. These findings indicate that the most promising derivatives demonstrate competitive efficacy with standard nonsteroidal anti-inflammatory drugs (NSAIDs) in terms of COX-1 inhibition and present potential as anti-inflammatory candidates targeting COX-2. Moreover, the hybridization of 2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzylidene with salicylic acid via a spacer derived from the azomethine group demonstrated enhanced inhibitory activity, both in the presence of benzylidene fragments and heterocyclic cores. Furthermore, the potency of these compounds against cyclooxygenase can be ranked in the following order: 4 > 9 > 2 > 3 > 6 > 8 > 7 > 5. Additionally, the synthesized derivatives exhibited comparable inhibitory effects against COX-1 and COX-2 indicated that these derivatives exhibited dual targets.
2.3.1.2 In vitro 5-lipoxygenase inhibitory activity. 5-Lipoxygenase (5-LOX) is an enzyme that plays a crucial role in the reduction of leukotriene biosynthesis. Its function can be succinctly described as catalyzing the oxygenation of arachidonic acid, resulting in the production of 5-hydroperoxyeicosatetraenoic acid (5-HPETE). This compound is subsequently converted into leukotrienes (LTs), including LTB4, LTC4, LTD4, and LTE4, which are recognized as potent inflammatory mediators.56,57 The newly synthesized azomethine salicylic acid derivatives 2–9 were assessed for their inhibitory activity against 5-lipoxygenase (5-LOX) at a concentration of 100 µg mL−1. The IC50 values were subsequently calculated using a serial dilution method and the results presented in Table 2. Among the synthetized derivatives, it was found that 2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzylidene derivative 4 demonstrated the most potent activity with IC50 values of 11.64 ± 0.20 µg mL−1 closely to naproxen (IC50 = 9.65 ± 0.17 µg mL−1) and Zileuton (IC50 = 8.43 ± 0.05 µg mL−1), but more promising to Aspirin (IC50 = 13.68 ± 0.13 µg mL−1) indicating that the presence of pyrrolidin-1-ylsulfonyl at C5 and hydroxyl group at C2 could enhancing the bonding and electronic interaction with active site of 5-LOX.
| Sample (µg mL−1) | 5-LOXa | |
|---|---|---|
| Inhib. (%) | IC50 (µg mL−1) | |
| a All previous values were calculated as mean + SE from n = 3 samples. | ||
| 2 | 61.44 ± 0.67 | 12.28 ± 0.21 |
| 3 | 31.73 ± 0.83 | 24.07 ± 0.96 |
| 4 | 64.79 ± 0.70 | 11.64 ± 0.20 |
| 5 | 16.85 ± 0.73 | 44.97 ± 2.56 |
| 6 | 19.53 ± 0.90 | 38.83 ± 2.29 |
| 7 | 16.90 ± 0.70 | 44.83 ± 2.50 |
| 8 | 19.35 ± 0.79 | 39.14 ± 1.97 |
| 9 | 61.64 ± 0.67 | 12.24 ± 0.21 |
| Naproxen | 78.20 ± 0.83 | 9.65 ± 0.17 |
| Aspirin | 55.15 ± 0.76 | 13.68 ± 0.13 |
| Zileuton | 89.45 ± 1.49 | 8.43 ± 0.05 |
Furthermore, compound 2, characterized by 4-methoxybenzylidene, and compound 9, characterized by 2-oxo-1,2-dihydrobenzo[h]quinoline, demonstrated comparable inhibitory activities with IC50 values of 12.28 ± 0.21 and 12.24 ± 0.21 µg mL−1, respectively, in comparison to Zileuton (IC50 = 8.43 ± 0.05 µg mL−1). This suggests that the electron-donating methoxy group that is present in compound 2 may enhance resonant stability and increase electron density towards the azomethine group. In the case of compound 9, the presence of the benzo[h]quinoline core appears to augment the hydrophobic surface area and enhance planarity, thereby promoting van der Waals interactions. Conversely, the heterocyclic cores present in compounds 5, 6, 7, and 8 that characterized with inden-2-ylene, furan-2-yl methylene, 1,3-diphenyl-1H-pyrazole, and 5-(sulfonyl)inden-2-one exhibited weaker IC50 values ranging from 38.83 ± 2.29 to 44.97 ± 2.56 µg mL−1. This indicates that these derivatives may contribute to weaker interactions, as the indene or furan structures possess planar configurations and fewer interactive heteroatoms. Additionally, the diminished activity observed for compounds 7 and 8 may be attributed to steric hindrance. Furthermore, the presence of the 4-isopropylbenzylidene fragment in compound 3 exhibited moderate activity with IC50 value of 24.07 ± 0.96 µg mL−1. These lower activity levels may be associated with the bulkiness of the isopropyl group and the hydrophobic characteristics of this group, as well as its spatial orientation.
Finally, the most promising candidate was compound 4, which demonstrated potency comparable to Naproxen and was close to Zileuton. In contrast, bulky or sterically hindered structures exhibited reduced activity. Furthermore, the SAR analysis indicates that derivatives 2, 4, and 9 significantly enhance the inhibition of 5-LOX when substituted with electron-donating groups, fused aromatic scaffolds, and sulfonyl functionalities.
| Sample (µg mL−1) | Anti-arthritic activity | ||
|---|---|---|---|
| Protein denaturationa | Proteinasea | ||
| Inhibition (%) | Inhibition (%) | IC50 (µg mL−1) | |
| a All previous values were calculated as mean + SE from n = 3 samples. | |||
| 2 | 23.75 ± 0.31 | 19.58 ± 0.31 | 39.39 ± 1.19 |
| 3 | 25.29 ± 0.33 | 21.12 ± 0.33 | 36.51 ± 1.10 |
| 4 | 24.51 ± 0.45 | 20.34 ± 0.45 | 37.96 ± 1.48 |
| 5 | 22.11 ± 0.58 | 17.94 ± 0.58 | 43.03 ± 1.57 |
| 6 | 24.25 ± 0.72 | 20.08 ± 0.72 | 38.51 ± 2.00 |
| 7 | 22.15 ± 0.56 | 17.98 ± 0.56 | 42.93 ± 1.53 |
| 8 | 24.11 ± 0.64 | 19.94 ± 0.64 | 38.71 ± 1.23 |
| 9 | 22.11 ± 1.14 | 17.94 ± 1.14 | 43.37 ± 3.38 |
| Naproxen | 67.35 ± 0.71 | 63.18 ± 0.71 | 12.20 ± 0.33 |
| Aspirin | 18.20 ± 0.59 | 14.03 ± 0.59 | 55.10 ± 2.54 |
| Diclofenac sodium | 83.71 ± 0.89 | 79.54 ± 0.89 | 9.69 ± 0.10 |
Concerning the inhibition of protein denaturation, it was observed that all the synthesized derivatives exhibited the ability to prevent protein denaturation, which can function as autoantigens and provoke inflammatory responses by attacking joint tissue and exacerbating joint damage. The synthesized salicylic acid derivatives demonstrated moderate protective effects against protein denaturation, with inhibitory percentage values ranging from 22.11 ± 0.58% to 25.29 ± 0.33% at a concentration of 100 µg mL−1. Among the synthesized derivatives, salicylic acid conjugated with 4-isopropylbenzylidene via the azomethine group, referred to as compound 3, exhibited the highest inhibitory percentage (IP) of 25.29 ± 0.33%, followed by compound 4 (IP = 24.25 ± 0.72%), compound 6 (IP = 24.25 ± 0.72%), and compound 8 (IP = 24.11 ± 0.64%). All tested derivatives displayed lower activity compared to the positive control drugs, naproxen (IP = 67.35 ± 0.71%) and diclofenac sodium (IP = 83.71 ± 0.89%), but demonstrated higher activity than salicylic acid (IP = 18.20 ± 0.59%) (Table 1).
Furthermore, the inhibition of proteinases is posited to play a protective role in preserving cartilage and connective tissue from degradation in the context of arthritis. The azomethine salicylic acid derivatives 2–9 exhibited low to moderate inhibitory activity, with inhibition percentages ranging from 17.94 ± 0.58% (compound 5) to 21.12 ± 0.33% (compound 3), indicating that these synthesized derivatives were not effective in efficiently inhibiting proteinase enzymes. However, the azomethine salicylic acid derivatives 2–9 demonstrated higher inhibitory percentages compared to aspirin (IP = 14.03 ± 0.59%). In contrast, naproxen and diclofenac sodium exhibited markedly greater inhibition, with values of 63.18 ± 0.71% and 79.54 ± 0.89%, respectively. Furthermore, our study extended to determine the IC50 values for proteinase inhibition, expressed in (µg mL−1). The results indicated that these derivatives displayed moderate activity, with IC50 values ranging from 36.51 ± 1.10 to 43.03 ± 1.57 µg mL−1, in comparison to aspirin, which had an IC50 of 55.10 ± 2.54 µg mL−1. Notably, Naproxen (IC50 = 12.20 ± 0.33 µg mL−1) and Diclofenac sodium (IC50 = 9.69 ± 0.10 µg mL−1) exhibited significantly greater potency, achieving 50% inhibition at considerably lower concentrations (Table 1).
Finally, the previous results indicated that the synthesized compounds possess moderate antiarthritic properties, particularly in preventing protein denaturation and inhibiting proteinase activity. Additionally, the designed derivatives demonstrated lower activity than diclofenac sodium or naproxen, but they exhibited comparable or better results than Aspirin.
| Compound no. | Human normal fibroblast (BJ-1) cell line IC50 (µg mL−1) | Selectivity indicesa | ||
|---|---|---|---|---|
| SICOX-1 | SICOX-2 | SI5-LOX | ||
| a SI = Selectivity indices can be calculated by (IC50 BJ-1/IC50 of (COX-1 or COX-2 or 5-LOX) | ||||
| 2 | 169.05 ± 8.50 | 16.6 | 22.0 | 13.8 |
| 4 | 157.40 ± 13.17 | 16.3 | 21.5 | 13.5 |
| 9 | 121.49 ± 7.94 | 12.0 | 15.9 | 9.9 |
| Doxorubicin | 39.75 ± 2.86 | — | — | — |
2.4 Toxicity prediction analysis
One of the essential steps is preparing samples to conduct toxicity tests on newly designed molecules to determine whether they will enhance the therapeutic potential of future drugs.60,61 The in-silico toxicity prediction of the designed Schiff base derivatives 2–9 was conducted using Pro Tox 3.0 (https://tox.charite.de/protox3/index.php?site=home; last accessed 13/6/2025) as described previously.62,63Firstly, we found that 4-methoxybenzylidene derivative 2 and 4-isopropylbenzylidene derivative 3 exhibited a good toxicity profile, with an expected lethal dose (LD50) of 500 mg kg−1 indicated that the toxicity is moderate, placing it in toxicity class 4, which has a low toxicity rate and suggests limited use. The Topological Polar Surface Area (TPSA) for compounds 2 and 3 revealed 79.12 and 69.89 Å2, meanwhile they exhibited log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (lipophilicity) values of 2.85 and 3.96, respectively indicating moderate hydrophobicity. In terms of organ toxicity (active targets), compound 3 exhibited to have risk of nephrotoxicity (active, with a probability of 0.63), which may indicate potential kidney harm, and respiratory toxicity (active, with a probability of 0.66), suggesting possible harm to the lungs or airways. For organ toxicity (inactive targets), there is cardiotoxicity (inactive, with a probability of 0.56) (Fig. SI 25). Regarding toxicity endpoints, compound 3 exhibited notable ecotoxicity properties clinical toxicity with probabilities of 0.50 and 0.56, respectively. These findings indicate a potential risk to environmental organisms and suggest the possibility of adverse effects in humans, including drug side effects. In contrast, compound 2 displayed inactive properties concerning immunotoxicity, mutagenicity, cytotoxicity, blood–brain barrier permeability, nutritional toxicity, and ecotoxicity, with probability values ranging from 0.57 to 0.93. Conversely, the 4-methoxybenzylidene derivative 2 exhibited inactive properties concerning neurotoxicity, cardiotoxicity, and carcinogenicity, with associated probability values of 0.55, 0.57, and 0.58, respectively. Furthermore, this derivative was predicted to demonstrate active behavior regarding hepatotoxicity, nephrotoxicity, and respiratory toxicity, with probability values of 0.63, 0.55, and 0.52, respectively (Fig. SI 25).
P (lipophilicity) values of 2.85 and 3.96, respectively indicating moderate hydrophobicity. In terms of organ toxicity (active targets), compound 3 exhibited to have risk of nephrotoxicity (active, with a probability of 0.63), which may indicate potential kidney harm, and respiratory toxicity (active, with a probability of 0.66), suggesting possible harm to the lungs or airways. For organ toxicity (inactive targets), there is cardiotoxicity (inactive, with a probability of 0.56) (Fig. SI 25). Regarding toxicity endpoints, compound 3 exhibited notable ecotoxicity properties clinical toxicity with probabilities of 0.50 and 0.56, respectively. These findings indicate a potential risk to environmental organisms and suggest the possibility of adverse effects in humans, including drug side effects. In contrast, compound 2 displayed inactive properties concerning immunotoxicity, mutagenicity, cytotoxicity, blood–brain barrier permeability, nutritional toxicity, and ecotoxicity, with probability values ranging from 0.57 to 0.93. Conversely, the 4-methoxybenzylidene derivative 2 exhibited inactive properties concerning neurotoxicity, cardiotoxicity, and carcinogenicity, with associated probability values of 0.55, 0.57, and 0.58, respectively. Furthermore, this derivative was predicted to demonstrate active behavior regarding hepatotoxicity, nephrotoxicity, and respiratory toxicity, with probability values of 0.63, 0.55, and 0.52, respectively (Fig. SI 25).
For 2-hydroxy-4-((2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzylidene)amino)benzoic acid 4, the LD50 is 3000 mg kg−1, classifying it in toxicity class 5. It has a Topological Polar Surface Area (TPSA) of 135.88 Å2, indicating low acute oral toxicity and high polarity. The octanol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) is 3.35, classifying it as moderately lipophilic. Regarding organ toxicity, the active targets show nephrotoxicity (P ∼ 0.55) and respiratory toxicity (P ∼ 0.72), while inactive targets indicate hepatotoxicity (P ∼ 0.60) and neurotoxicity (P ∼ 0.74). For toxicity endpoints, the active targets display clinical toxicity (P ∼ 0.62), while the inactive targets show ecotoxicity (P ∼ 0.72) (Fig. SI 25). On the other hand, compound 5, named 4-((1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)amino)-2-hydroxybenzoic acid, exhibited LD50 with a favorable toxicity profile and the expected lethal dose (LD50) is 1500 mg kg−1, and it falls within toxicity class 4, indicating a low risk for acute poisoning. The topological polar surface area (TPSA) is 104.03 Å, indicating high polarity, while the octanol/water partition coefficient (log
P) is 3.35, classifying it as moderately lipophilic. Regarding organ toxicity, the active targets show nephrotoxicity (P ∼ 0.55) and respiratory toxicity (P ∼ 0.72), while inactive targets indicate hepatotoxicity (P ∼ 0.60) and neurotoxicity (P ∼ 0.74). For toxicity endpoints, the active targets display clinical toxicity (P ∼ 0.62), while the inactive targets show ecotoxicity (P ∼ 0.72) (Fig. SI 25). On the other hand, compound 5, named 4-((1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)amino)-2-hydroxybenzoic acid, exhibited LD50 with a favorable toxicity profile and the expected lethal dose (LD50) is 1500 mg kg−1, and it falls within toxicity class 4, indicating a low risk for acute poisoning. The topological polar surface area (TPSA) is 104.03 Å, indicating high polarity, while the octanol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) is 1.07, demonstrating balanced lipophilicity. In terms of organ toxicity (active targets), the probabilities are as follows: hepatotoxicity (0.56) and nephrotoxicity (0.66). For organ toxicity, it was found that compound 5 exhibited inactive targets with elevated probabilities against neurotoxicity (P ∼ 0.68) and cardiotoxicity (P ∼ 0.68). Additionally, for toxicity endpoints (active targets), the probability for clinical toxicity is 0.63, while for toxicity endpoints (inactive targets), the probability for immunotoxicity is 0.99 (Fig. SI 26).
P) is 1.07, demonstrating balanced lipophilicity. In terms of organ toxicity (active targets), the probabilities are as follows: hepatotoxicity (0.56) and nephrotoxicity (0.66). For organ toxicity, it was found that compound 5 exhibited inactive targets with elevated probabilities against neurotoxicity (P ∼ 0.68) and cardiotoxicity (P ∼ 0.68). Additionally, for toxicity endpoints (active targets), the probability for clinical toxicity is 0.63, while for toxicity endpoints (inactive targets), the probability for immunotoxicity is 0.99 (Fig. SI 26).
There are other derivatives, including derivatives 6 and 7, featuring heterocyclic cores of furan and pyrazole, respectively. These compounds exhibited a favorable toxicity profile, with an expected lethal dose (LD50) of 500 and 670 mg kg−1, respectively, categorizing them within medium toxicity (toxicity class 4). Besides, the topological polar surface area (TPSA) was predicted to be 83.03 and 87.71 Å2 for derivatives 6 and 7, respectively. Additionally, the Octanol/Water Partition Coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) was predicted to be 2.43 and 4.69 for derivatives 6 and 7, respectively. These derivatives exhibited probabilities for organ toxicity, with values of 0.62 and 0.65 for hepatotoxicity, while compound 7 displayed an active probability of neurotoxicity (P ∼ 0.60) compared to furan derivative 6, which demonstrated inactive neurotoxicity with a probability value of 0.59. Further, the tested derivatives 6 and 7 were found to be inactive regarding cardiotoxicity, with probability values of 0.70 and 0.76, respectively (see Fig. SI 26).
P) was predicted to be 2.43 and 4.69 for derivatives 6 and 7, respectively. These derivatives exhibited probabilities for organ toxicity, with values of 0.62 and 0.65 for hepatotoxicity, while compound 7 displayed an active probability of neurotoxicity (P ∼ 0.60) compared to furan derivative 6, which demonstrated inactive neurotoxicity with a probability value of 0.59. Further, the tested derivatives 6 and 7 were found to be inactive regarding cardiotoxicity, with probability values of 0.70 and 0.76, respectively (see Fig. SI 26).
Furthermore, the 2-hydroxy-4-((5-((4-methylpiperidin-1-yl)sulfonyl)-2-oxoindolin-3-ylidene) amino)benzoic acid (8) has an expected lethal dose (LD50) of 4000 mg kg−1, indicating low acute toxicity and placing it in Toxicity Class 5, which is considered practically non-toxic. The topological polar surface area (TPSA) measures 144.75 Å, suggesting poor membrane permeability with Octanol/Water Partition Coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) is 3.74. Additionally, compound 8 showed active properties for hepatotoxicity and neurotoxicity, with probabilities of 0.65 and 0.60, respectively. Conversely, the compound is predicted to have inactive properties for cardiotoxicity and respiratory toxicity, with probabilities values of 0.76 and 0.84. Additionally, it shows active toxicity endpoints for clinical toxicity, with a probability of 0.54, and inactive properties for immunotoxicity, with a probability of 0.99. Compound 9 that named as (2-hydroxy-4-(((2-oxo-1,2-dihydrobenzo[h]quinolin-3-yl)methylene)amino)benzoic acid), is predicted to have a lethal dose (LD50) of 2000 mg kg−1 and falls into toxicity class 4. This derivative demonstrated active properties for respiratory toxicity and nephrotoxicity, with probability values of 0.53 and 0.64, respectively. It is predicted to be inactive for hepatotoxicity and neurotoxicity, with probability values of 0.55 and 0.69, respectively. The topological polar surface area (TPSA) is 102.75 Å, and the octanol/water partition coefficient (log
P) is 3.74. Additionally, compound 8 showed active properties for hepatotoxicity and neurotoxicity, with probabilities of 0.65 and 0.60, respectively. Conversely, the compound is predicted to have inactive properties for cardiotoxicity and respiratory toxicity, with probabilities values of 0.76 and 0.84. Additionally, it shows active toxicity endpoints for clinical toxicity, with a probability of 0.54, and inactive properties for immunotoxicity, with a probability of 0.99. Compound 9 that named as (2-hydroxy-4-(((2-oxo-1,2-dihydrobenzo[h]quinolin-3-yl)methylene)amino)benzoic acid), is predicted to have a lethal dose (LD50) of 2000 mg kg−1 and falls into toxicity class 4. This derivative demonstrated active properties for respiratory toxicity and nephrotoxicity, with probability values of 0.53 and 0.64, respectively. It is predicted to be inactive for hepatotoxicity and neurotoxicity, with probability values of 0.55 and 0.69, respectively. The topological polar surface area (TPSA) is 102.75 Å, and the octanol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) is 3.84 (Fig. SI 27).
P) is 3.84 (Fig. SI 27).
All tested derivatives 2–9 were predicted, and the obtained data were compared to the positive control drugs utilized in this study, including aspirin, naproxen, and indomethacin. The results indicated an average similarity and prediction accuracy of 100% according to the model employed in Protex 3.0, with LD50 values of 250, 12, and 248 mg kg−1 and corresponding toxicity classes of 3, 2, and 3, respectively. For aspirin, the data revealed inactive properties concerning hepatotoxicity, neurotoxicity, respiratory toxicity, cardiotoxicity, carcinogenicity, immunotoxicity, mutagenicity, cytotoxicity, ecotoxicity, and neurotoxicity, with probability values ranging from 0.51 to 0.99. Conversely, aspirin exhibited active properties for nephrotoxicity, blood–brain barrier permeability, and clinical toxicity, with probability values of 0.69, 0.85, and 0.73, respectively. In contrast, indomethacin displayed a poor toxicity profile, being predominantly active for various toxicity endpoints, including hepatotoxicity, neurotoxicity, nephrotoxicity, respiratory toxicity, immunotoxicity, blood–brain barrier permeability, clinical toxicity, and nutritional toxicity, with probability values ranging from 0.59 to 0.90. However, it demonstrated inactive properties and a safe profile against cardiotoxicity, mutagenicity, cytotoxicity, and ecotoxicity. Additionally, naproxen exhibited a moderate toxicity profile, showing active behaviors towards hepatotoxicity, neurotoxicity, nephrotoxicity, respiratory toxicity, and blood–brain barrier permeability, while presenting an inactive profile for cardiotoxicity, carcinogenicity, mutagenicity, cytotoxicity, ecotoxicity, and clinical toxicity (Fig. SI 28).
Finally, the modification of salicylic acid derivatives with varying cores, including both aromatic and heterocyclic structures, via the azomethine group significantly improves the toxicity profile of these compounds. The synthesized derivatives generally exhibit favorable toxicity characteristics, demonstrating limited activity in terms of organ and endpoint toxicity when compared to established positive control drugs. Furthermore, the majority of the evaluated derivatives are classified within toxicity classes 4 and 5, suggesting that they possess minimal toxicity relative to indomethacin, which falls under toxicity class 2, and aspirin and naproxen, which are classified as toxicity class 3.
2.5 Computational molecular docking studies
The molecular docking simulation of the most active azomethine salicylic acid derivatives 2, 4, and 9 was conducted inside the active site of 5-lipoxygenase (PDB: 7TTJ) (https://www.rcsb.org/structure/7TTJ; last access 1/10/2025) (Fig. 3). The docking simulation results of the designed derivatives were compared with the approved FDA drugs as Aspirin and Zileuton. The binding energy of the designed frequently referred to as the “S score”, serves as an estimate of the free energy of binding between a ligand and its receptor. A noted decrease in this energy value (i.e., a more negative score) implies stronger and more favorable interactions, which are indicative of an enhanced binding affinity. This increase in binding strength is primarily ascribed to the specific molecular interactions that occur between the ligand and the residues within the receptor's binding pocket. The binding energy can be generated and related to different types of interaction as H-bonding, arene-cation (π–+) interaction, arene–arene (π–π), hydrophobic interaction, ionic bond interaction, and Vander wale force.The docking results indicated that the most promising derivatives, namely 2, 4, and 9, exhibited binding affinities of −13.48, −15.36, and −13.52 kcal mol−1, respectively. In comparison, Aspirin and Zileuton displayed binding affinities of −9.33 kcal mol−1 and −10.24 kcal mol−1, respectively. Notably, the 2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzylidene derivative (compound 4) exhibited the highest binding affinity at −15.36 kcal mol−1 and demonstrated the formation of six hydrogen bonds along with one arene-cation interaction. To elucidate the binding characteristics of the optimal conformation associated with compound 4 in relation to the active site, we observed that the residue Pro98 interacted with the phenolic (OH) group and the hydrogen of the carboxylic group of salicylic acid, establishing two backbone donor hydrogen bonds with distances of 3.2 Å and 1.8 Å, and bond strengths of 19% and 51%, respectively. Furthermore, the sulfonamide derivative of salicylaldehyde hybridized with salicylic acid via the azomethine group, revealing that the nitrogen of the azomethine moiety formed a bond with Arg101, characterized by a bond length of 2.1 Å and a bond strength of 11%. Additionally, two oxygen atoms from the SO2 group of the sulfonamide formed two side-chain acceptor hydrogen bonds with bond lengths of 2.3 Å and bond strengths of 17% and 23%. Similarly, the phenolic group of the sulfonated salicylimide was able to form a side-chain acceptor hydrogen bond with the residue Arg138, exhibiting a bond length of 1.9 Å and a bond strength of 51%. The arene-cation interaction was observed between the residue Arg101 and the phenyl group of the benzylidene core. Hydrophobic interactions were also identified between the conformation of compound 4 within the active site pocket and several residues, including Lys394, Arg112, Ala388, Leu111, Val107, Glu108, Val110, Thr137, Lys133, Val389, Glu134, and Cys99 (Fig. 4 and Table 5).
 | ||
| Fig. 4 2D and 3D structure of most active derivative 4 inside the active site of 5-lipoxygenase (PDB: 7TTJ). | ||
| Cpd no. | Binding energy (kcal mol−1) | Residues amino acid | Interacting group | Interaction | |
|---|---|---|---|---|---|
| Length of bond (Å) | Strength of bond (%) | ||||
| 2 | −13.48 | Pro98 | Proton of carboxylic-OH | 1.9 | 36% |
| Arg101 | Nitrogen of azomethine | 2.1 | 16 | ||
| Arg138 | Phenyl of 4-methoxybenzylidine | Arene-cation | |||
| Arg101 | Phenyl of 4-methoxybenzylidine | Arene-cation | |||
| 4 | −15.36 | Pro98 | Aromatic hydroxyl group | 3.2 | 19 |
| Pro98 | Carboxylic-OH | 1.8 | 51 | ||
| Arg101 | Nitrogen of azomethine | 2.1 | 11 | ||
| Arg101 | Oxygen of sulfonamide (SO2) | 2.3 | 17 | ||
| Arg101 | Oxygen of sulfonamide (SO2) | 2.3 | 23 | ||
| Arg138 | Phenolic hydroxyl group of sulfonated salicylimine | 1.9 | 51 | ||
| Arg101 | Phenolic hydroxyl group of sulfonated salicylimine | Arene-cation | |||
| 9 | −13.52 | Arg68 | Proton of carboxylic-OH | 1.9 | 67 |
| Gln129 | Oxygen of carbonyl of carboxylic group | 2.2 | 13 | ||
| Ile126 | Oxygen of carbonyl of carboxylic group | 3.0 | 19 | ||
| Arg101 | Phenyl of naphthalene | Arene-cation | |||
| Aspirin | −9.33 | Arg101 | Oxygen of acetoxy group | 2.3 | 18 |
| Arg101 | Proton of carboxylic-OH | 2.4 | 12 | ||
| Lys133 | Phenyl of acetyl salicylic acid | Arene-cation | |||
| Zileuton | −10.24 | Glu108 | Amino group | 2.3 | 11 |
| Arg101 | Hydroxyl group attached to nitrogen atom | 2.1 | 53 | ||
| Arg101 | Thiophene of benzo[b]thiophene | Arene-cation | |||
| Arg138 | Thiophene of benzo[b]thiophene | Arene-cation | |||
| Arg101 | Phenyl of benzo[b]thiophene | Arene-cation | |||
| Arg132 | Phenyl of benzo[b]thiophene | Arene-cation | |||
Furthermore, the 4-methoxybenzylidene derivative 2 exhibited a binding affinity of −13.48 kcal mol−1 and interacted with the active site through the formation of one hydrogen bond as a backbone donor with the residue Pro98, involving the proton of carboxylic-OH with a bond length of 1.9 Å and a strength of 36%. Additionally, the nitrogen of the azomethine group established a hydrogen bond as a sidechain acceptor with Arg101, characterized by a bond length of 2.1 Å and a strength of 16%. Furthermore, the residues Arg138 and Arg101 contributed to an arene-cation interaction with the phenyl group of 4-methoxybenzylidene. A hydrophobic interaction was also observed between compound 2 and the active site pocket, involving the residues Lys394, Asp113, Ala388, Arg112, Val389, Tyr142, Gln141, Asp166, Leu111, Glu134, and Cys99. In contrast, the 2-oxo-1,2-dihydrobenzo[h]quinoline derivative 9 demonstrated a binding affinity of −13.52 kcal mol−1. This compound exhibited hydrogen bond acceptors with the residue Arg68 through interaction with the proton of carboxylic-OH of salicylic acid, characterized by a bond length of 1.9 Å and a strength of 67%. Additionally, Gln129 formed a bond with the oxygen of the carbonyl group of the carboxylic acid, with a bond length of 2.2 Å and a strength of 13%. Moreover, the oxygen of the carbonyl group of the carboxylic acid could serve as a hydrogen bond backbone donor to Ile126, with a bond length of 3.0 Å and a strength of 19%. Moreover, Arg101 contributed to an arene-cation interaction with the phenyl group of naphthalene, alongside hydrophobic interactions observed with the residues Val129, Leu66, His130, Lys133, Glu134, Val110, Thr137, Val107, and Glu141 (Fig. 5 and Table 5).
 | ||
| Fig. 5 2D and 3D structure of (a) compound 2 and (b) compound 9 inside the active site of 5-lipoxygenase (PDB: 7TTJ). | ||
In terms of comparison, docking simulations were conducted for Aspirin, a derivative of salicylic acid, alongside Zileuton, which served as a positive control drug for 5-LOX. The results indicated that Aspirin exhibited a binding affinity of −9.33 kcal mol−1, characterized by two hydrogen bond interactions with sidechain acceptors. The first hydrogen bond involved the residue Arg101, which formed a bond with the oxygen of the acetoxy group, displaying a bond length of 2.3 Å and a bond strength of 18%. The second hydrogen bond was observed with the proton of carboxylic-OH, featuring a bond length of 2.4 Å and a bond strength of 12%. The conformation of Aspirin within the active site demonstrated an arene-cation interaction between the residue Lys133 and the phenyl group of acetylsalicylic acid. Additionally, Aspirin displayed hydrophobic interactions with the residues Val107, His130, Glu108, Val110, Glu134, and Thr137.
Conversely, Zileuton exhibited a binding energy of −10.24 kcal mol−1 and formed a hydrogen bond as a backbone donor between Glu108 and the amino group, with a bond length of 2.3 Å and a bond strength of 11%. Furthermore, another hydrogen bond was established as a sidechain acceptor between Arg101 and the hydroxyl group attached to the nitrogen atom of the hydroxyl urea fragment, characterized by a bond length of 2.1 Å and a bond strength of 53%. The conformation of Zileuton in the active site revealed four arene-cation interactions with the residues Arg101 and Arg138, which were linked to the thiophene ring and the phenyl group of benzothiophene, respectively. Zileuton also exhibited hydrophobic interactions with the residues Val107, Glu134, Asp166, Thr137, Tyr383, and Glu141 (Fig. 6 and Table 5).
 | ||
| Fig. 6 2D and 3D structure of (a) compound Aspirin and (b) Zileuton inside the active site of 5-lipoxygenase (PDB: 7TTJ). | ||
3 Conclusion
A series of azomethine salicylic acid derivatives 2–9 were designed and synthesized through a condensation reaction between 4-aminosalicylic acid and various formyl and ketone compounds, utilizing acetic acid as a solvent and catalyst. The synthesized derivatives were evaluated as potential anti-inflammatory agents targeting COX-1, COX-2, and 5-LOX. The results indicated moderate to good potency, with compounds 2, 4, and 9 identified as the most promising derivatives. Among these, compound 4 exhibited the highest potency, with IC50 values of 9.68 ± 0.17 µg mL−1 and 7.32 ± 0.04 µg mL−1 against COX-1 and COX-2, respectively, in comparison to Naproxen (IC50 = 6.18 ± 0.04 µg mL−1), Aspirin (IC50 = 8.45 ± 0.05 µg mL−1), and Indomethacin (IC50 = 5.47 ± 0.04 µg mL−1). Additionally, the designed derivatives were assessed for their anti-arthritic activity, including the determination of inhibition percentages against protein denaturation and protease activity. The findings revealed that these derivatives possess moderate anti-arthritic properties, particularly in preventing protein denaturation and inhibiting protease activity. The anti-arthritic activity demonstrated that the most active derivatives 2, 4, and 9 exhibited comparable or superior efficacy to Aspirin, although their activity is lower than that of Diclofenac sodium and Naproxen. In silico toxicity predictions were conducted for all the synthesized derivatives, and the results indicated favorable toxicity profiles, demonstrating limited organ and endpoint toxicity compared to established positive control drugs. Finally, docking simulations of the most active derivatives revealed that the presence of the azomethine group, benzylidene core, and phenolic group are critical for binding at the active site, facilitating the formation of hydrogen bonds and arene-cation interactions, thereby achieving excellent docking scores.4 Experimental section
The chemicals utilized in the research articles, including 4-amino-2-hydroxybenzoic acid, ninhydrin, 4-methoxybenzaldehyde, cuminaldehyde, and furfural, were procured from Sigma-Aldrich company. Meanwhile, methanol, piperidine, and acetic acid were sourced from Adwic Company, and all chemicals were employed without further purification. The synthesized derivatives were comprehensively characterized, with instrument details provided in the supplementary material. The elemental analysis data (reflecting the percentages of C, H, and N) for the synthesized derivatives were found to be within ±0.4% of the theoretically calculated values. The 1H NMR and 13C NMR spectra were recorded on a Bruker spectrometer operating at 400 MHz, utilizing tetramethylsilane (TMS) as the internal standard. All derivatives were dissolved in deuterated dimethyl sulfoxide, and chemical shifts were reported in δ ppm units. Additionally, elemental analysis was performed at the Regional Center for Mycology and Biotechnology at Al-Azhar University. Mass spectra were obtained using a Thermo Scientific ISQLT mass spectrometer at the Regional Center for Mycology and Biotechnology, Al-Azhar University. The in vitro cytotoxic activity on the normal human fibroblast cell line (BJ-1) was evaluated utilizing the MTT assay at VACSERA, Cairo, Egypt. Additionally, the synthesized Schiff base known as 2-hydroxy-4-((4-methoxybenzylidene)amino)benzoic acid (2) was prepared as previously reported,64 while 4-((furan-2-ylmethylene)amino)-2-hydroxybenzoic acid (6) was prepared according to a reported method.65 All spectra figures were represented in supplementary material figures (Fig. SI1–SI24)4.1 Synthesis of target Schiff bases
A mixture of 4-amino-2-hydroxybenzoic acid (1) and various formyl or ketone derivatives, including cuminaldehyde, 2-hydroxy-5-(pyrrolidin-1-ylsulfonyl)benzaldehyde, ninhydrin, 1,3-diphenyl-1H-pyrazole-4-carbaldehyde, 5-((4-methylpiperidin-1-yl)sulfonyl)indoline-2,3-dione, and 2-chlorobenzo[h]quinoline-3-carbaldehyde, was dissolved in 10 mL of acetic acid and subjected to reflux conditions with stirring for a duration of 4–12 h. Following this period, the solution was allowed to cool to room temperature, at which point the solid product that formed was washed with methanol. Subsequently, the solid product was recrystallized from methanol and dried at room temperature to yield the final product as a solid powder.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1605 (C
O), 1605 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 9.97, 9.76 (2 s, 2H, 2OH), 9.12 (s, 1H, CH = N), 7.87 (d, J = 8.0 Hz, 2H, Ar–H), 7.37 (d, J = 8.0 Hz, 2H, Ar–H), 7.19 (s, 1H, Ar–H), 7.02 (d, J = 8.4 Hz, 1H, Ar–H), 6.67 (d, J = 8.4 Hz, 1H, Ar–H), 2.96 (s, 1H, Isopropyl methine –CH), 1.22 (d, J = 6.8 Hz, 3H, CH3), 1.15 (d, J = 6.8 Hz, 3H, CH3); 13C NMR (101 MHz, DMSO) δ/ppm 172.50 (C
N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 9.97, 9.76 (2 s, 2H, 2OH), 9.12 (s, 1H, CH = N), 7.87 (d, J = 8.0 Hz, 2H, Ar–H), 7.37 (d, J = 8.0 Hz, 2H, Ar–H), 7.19 (s, 1H, Ar–H), 7.02 (d, J = 8.4 Hz, 1H, Ar–H), 6.67 (d, J = 8.4 Hz, 1H, Ar–H), 2.96 (s, 1H, Isopropyl methine –CH), 1.22 (d, J = 6.8 Hz, 3H, CH3), 1.15 (d, J = 6.8 Hz, 3H, CH3); 13C NMR (101 MHz, DMSO) δ/ppm 172.50 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.77 (C–OH), 154.08 (C
O), 167.77 (C–OH), 154.08 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 129.93, 128.93, 127.45, 126.98, 123.62, 119.48, 117.37, 115.42 (Ar–Cs), 33.93 (CH-), 24.02, 21.52 (CH3); MS (EI, 70 eV): m/z (%) = 283.23 [M+] (54.38%), 173 (100%); Analyt. Cal. C17H17NO3 (Mw.t. = 283.33) calculated C, 72.07; H, 6.05; N, 4.94; Found C, 72.21; H, 5.91; N, 5.06.
N), 129.93, 128.93, 127.45, 126.98, 123.62, 119.48, 117.37, 115.42 (Ar–Cs), 33.93 (CH-), 24.02, 21.52 (CH3); MS (EI, 70 eV): m/z (%) = 283.23 [M+] (54.38%), 173 (100%); Analyt. Cal. C17H17NO3 (Mw.t. = 283.33) calculated C, 72.07; H, 6.05; N, 4.94; Found C, 72.21; H, 5.91; N, 5.06.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1601 (C
O), 1601 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1328, 1149 (SO2); 1H NMR (400 MHz, DMSO-d6) δ/ppm 11.39, 10.32, 9.72 (3 s, 3H, 3OH), 8.19 (s, 1H, CH = N), 7.99 (s, 1H, Ar–H), 7.91 (d, J = 8.8 Hz, 1H, Ar–H), 7.41 (d, J = 8.4 Hz, 1H, Ar–H), 7.20 (d, J = 8.4 Hz, 1H, Ar–H), 6.08 (d, J = 10.8 Hz, 1H, Ar–H), 5.97 (s, 1H, Ar–H), 3.10 (s, 4H, N(CH2)2), 1.65 (s, 4H, 2CH2); 13C NMR (101 MHz, DMSO-d6) δ/ppm 172.55 (C
N) 1328, 1149 (SO2); 1H NMR (400 MHz, DMSO-d6) δ/ppm 11.39, 10.32, 9.72 (3 s, 3H, 3OH), 8.19 (s, 1H, CH = N), 7.99 (s, 1H, Ar–H), 7.91 (d, J = 8.8 Hz, 1H, Ar–H), 7.41 (d, J = 8.4 Hz, 1H, Ar–H), 7.20 (d, J = 8.4 Hz, 1H, Ar–H), 6.08 (d, J = 10.8 Hz, 1H, Ar–H), 5.97 (s, 1H, Ar–H), 3.10 (s, 4H, N(CH2)2), 1.65 (s, 4H, 2CH2); 13C NMR (101 MHz, DMSO-d6) δ/ppm 172.55 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.89, 156.29 (2C–OH), 150.62 (C
O), 163.89, 156.29 (2C–OH), 150.62 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 135.07, 131.92, 128.85, 127.46, 122.63, 118.97, 108.71, 106.70, 100.57, 98.98 (Ar–Cs), 48.30 (pyrrolidine N(CH2)2), 25.13 (pyrrolidine 2CH2); MS (EI, 70 eV): m/z (%) = 390.68 [M+] (13.38%), 263 (100%); Analyt. Cal. C18H18N2O6S (Mw.t. = 390.41) calculated C, 55.38; H, 4.65; N, 7.18; Found C, 55.54; H, 4.77; N, 7.01.
N), 135.07, 131.92, 128.85, 127.46, 122.63, 118.97, 108.71, 106.70, 100.57, 98.98 (Ar–Cs), 48.30 (pyrrolidine N(CH2)2), 25.13 (pyrrolidine 2CH2); MS (EI, 70 eV): m/z (%) = 390.68 [M+] (13.38%), 263 (100%); Analyt. Cal. C18H18N2O6S (Mw.t. = 390.41) calculated C, 55.38; H, 4.65; N, 7.18; Found C, 55.54; H, 4.77; N, 7.01.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1606 (C
O), 1606 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 12.42, 9.79 (2 s, 2H, 2OH), 8.07 (d, J = 7.2 Hz, 1H, Ar–H), 8.02 (d, J = 6.8 Hz, 1H, Ar–H), 7.97–7.91 (m, 1H, Ar–H), 7.82 (d, J = 14.0 Hz, 1H, Ar–H), 7.59 (d, J = 14.0 Hz, 1H, Ar–H), 7.25–7.20 (m, 1H, Ar–H), 7.11 (s, 1H, Ar–H); 13C NMR (101 MHz, DMSO) δ/ppm 174.06, 172.50 (C
N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 12.42, 9.79 (2 s, 2H, 2OH), 8.07 (d, J = 7.2 Hz, 1H, Ar–H), 8.02 (d, J = 6.8 Hz, 1H, Ar–H), 7.97–7.91 (m, 1H, Ar–H), 7.82 (d, J = 14.0 Hz, 1H, Ar–H), 7.59 (d, J = 14.0 Hz, 1H, Ar–H), 7.25–7.20 (m, 1H, Ar–H), 7.11 (s, 1H, Ar–H); 13C NMR (101 MHz, DMSO) δ/ppm 174.06, 172.50 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 158.00 (C–OH), 140.80 (C
O), 158.00 (C–OH), 140.80 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 137.08, 135.29, 131.45, 129.73, 124.75, 123.83, 110.57, 110.21, 106.62 (Ar–Cs); MS (EI, 70 eV): m/z (%) = 295.95 [M+] (37.18%), 69 (100%); Analyt. Cal. C16H9NO5 (Mw.t. = 295.25) calculated C, 65.09; H, 3.07; N, 4.74; Found C, 65.24; H, 3.21; N, 4.61.
N), 137.08, 135.29, 131.45, 129.73, 124.75, 123.83, 110.57, 110.21, 106.62 (Ar–Cs); MS (EI, 70 eV): m/z (%) = 295.95 [M+] (37.18%), 69 (100%); Analyt. Cal. C16H9NO5 (Mw.t. = 295.25) calculated C, 65.09; H, 3.07; N, 4.74; Found C, 65.24; H, 3.21; N, 4.61.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 10.03, 9.34 (2 s, 2H, 2 OH), 8.01 (d, J = 8.4 Hz, 1H, Ar–H), 7.94 (d, J = 9.2 Hz, 1H, Ar–H), 7.85 (t, J = 8.4 Hz, 1H, Ar–H), 7.73–7.68 (m, 1H, Ar–H), 7.62 (d, J = 8.0 Hz, 1H, Ar–H), 7.59 (s, 1H, CH = N), 7.56 (d, J = 8.4 Hz, 1H, Ar–H), 7.53 (s, 1H, pyrazole-H), 7.51 (d, J = 6.8 Hz, 1H, Ar–H), 7.47–7.44 (m, 1H, Ar–H), 7.42–7.37 (m, 1H, Ar–H), 7.37–7.33 (m, 1H, Ar–H), 7.24 (s, 1H, Ar–H), 7.19 (t, J = 6.8 Hz, 1H, Ar–H), 7.13–7.10 (m, 1H, Ar–H); 13C NMR (101 MHz, DMSO) δ/ppm 172.50 (C
N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 10.03, 9.34 (2 s, 2H, 2 OH), 8.01 (d, J = 8.4 Hz, 1H, Ar–H), 7.94 (d, J = 9.2 Hz, 1H, Ar–H), 7.85 (t, J = 8.4 Hz, 1H, Ar–H), 7.73–7.68 (m, 1H, Ar–H), 7.62 (d, J = 8.0 Hz, 1H, Ar–H), 7.59 (s, 1H, CH = N), 7.56 (d, J = 8.4 Hz, 1H, Ar–H), 7.53 (s, 1H, pyrazole-H), 7.51 (d, J = 6.8 Hz, 1H, Ar–H), 7.47–7.44 (m, 1H, Ar–H), 7.42–7.37 (m, 1H, Ar–H), 7.37–7.33 (m, 1H, Ar–H), 7.24 (s, 1H, Ar–H), 7.19 (t, J = 6.8 Hz, 1H, Ar–H), 7.13–7.10 (m, 1H, Ar–H); 13C NMR (101 MHz, DMSO) δ/ppm 172.50 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 166.11 (C–OH), 157.18, 153.57 (C
O), 166.11 (C–OH), 157.18, 153.57 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 145.71, 140.57, 139.69, 136.69, 135.33, 133.96, 131.87, 130.89, 129.68, 128.79, 127.84, 125.89, 119.74, 118.53, 113.71 (Ar–Cs); MS (EI, 70 eV): m/z (%) = 383.01 [M+] (27.44%), 239 (100%); Analyt. Cal. C23H17N3O3 (Mw.t. = 383.41) calculated C, 72.05; H, 4.47; N, 10.96; Found C, 72.13; H, 4.62; N, 10.84.
N), 145.71, 140.57, 139.69, 136.69, 135.33, 133.96, 131.87, 130.89, 129.68, 128.79, 127.84, 125.89, 119.74, 118.53, 113.71 (Ar–Cs); MS (EI, 70 eV): m/z (%) = 383.01 [M+] (27.44%), 239 (100%); Analyt. Cal. C23H17N3O3 (Mw.t. = 383.41) calculated C, 72.05; H, 4.47; N, 10.96; Found C, 72.13; H, 4.62; N, 10.84.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1618 (C
O), 1618 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1355, 1148 (SO2); 1H NMR (400 MHz, DMSO-d6) δ/ppm 10.71, 10.39, 9.46 (3 s, 3H, 2OH +NH), 7.67 (s, 1H, Ar–H), 7.36 (d, J = 8.4 Hz, 1H, Ar–H), 7.02–6.90 (m, 1H, Ar–H), 6.81–6.64 (m, 1H, Ar–H), 6.21 (m, 1H, Ar–H), 5.92 (d, J = 8.4 Hz, 1H, Ar–H), 3.00 (s, 2H, N-(CH2)), 2.04–2.06 (m, 2H, N-(CH2)), 1.64 (s, 2H, CH2), 1.54 (s, 1H, CH–Me), 1.25 (s, 1H, CH), 1.07 (s, 1H, CH), 0.83 (s, 3H, N–CH3); 13C NMR (101 MHz, DMSO) δ/ppm 173.54, 164.14 (2C
N) 1355, 1148 (SO2); 1H NMR (400 MHz, DMSO-d6) δ/ppm 10.71, 10.39, 9.46 (3 s, 3H, 2OH +NH), 7.67 (s, 1H, Ar–H), 7.36 (d, J = 8.4 Hz, 1H, Ar–H), 7.02–6.90 (m, 1H, Ar–H), 6.81–6.64 (m, 1H, Ar–H), 6.21 (m, 1H, Ar–H), 5.92 (d, J = 8.4 Hz, 1H, Ar–H), 3.00 (s, 2H, N-(CH2)), 2.04–2.06 (m, 2H, N-(CH2)), 1.64 (s, 2H, CH2), 1.54 (s, 1H, CH–Me), 1.25 (s, 1H, CH), 1.07 (s, 1H, CH), 0.83 (s, 3H, N–CH3); 13C NMR (101 MHz, DMSO) δ/ppm 173.54, 164.14 (2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.78 (C–OH), 153.53 (C
O), 161.78 (C–OH), 153.53 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 145.72, 132.88, 131.57, 128.41, 127.97, 121.43, 115.73, 114.32, 104.87, 99.67 (Ar–Cs), 46.45, 44.07 (N(CH2)2), 33.30 (2CH2), 29.91 (CH–CH3), 22.72 (CH3); MS (EI, 70 eV): m/z (%) = 443.13 [M+] (15.15%), 74 (100%); Analyt. Cal. C21H21N3O6S (Mw.t. = 443.47) calculated C, 56.88; H, 4.77; N, 9.48; Found C, 56.96; H, 4.54; N, 9.56.
N), 145.72, 132.88, 131.57, 128.41, 127.97, 121.43, 115.73, 114.32, 104.87, 99.67 (Ar–Cs), 46.45, 44.07 (N(CH2)2), 33.30 (2CH2), 29.91 (CH–CH3), 22.72 (CH3); MS (EI, 70 eV): m/z (%) = 443.13 [M+] (15.15%), 74 (100%); Analyt. Cal. C21H21N3O6S (Mw.t. = 443.47) calculated C, 56.88; H, 4.77; N, 9.48; Found C, 56.96; H, 4.54; N, 9.56.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1607 (C
O), 1607 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 10.42 and 10.26 (2 s, 2H, 2OH), 9.03 (d, J = 8.0 Hz, 1H, Ar–H), 8.96 (s, 1H, quinoline CH = N), 8.09 (d, J = 7.4 Hz, 2H, Ar–H), 8.08 (d, J = 6.8 Hz, 2H, Ar–H), 7.98 (d, J = 7.6 Hz, 1H, Ar–H), 7.86 (s, 1H, quinoline-H4), 7.85–7.81 (m, 2H, 1 Ar–H+ NH), 7.73 (d, J = 8.4 Hz, 1H, Ar–H), 7.67 (d, J = 8.0 Hz, 1H, Ar–H); 13C NMR (101 MHz, DMSO) δ/ppm 172.06 (C
N); 1H NMR (400 MHz, DMSO-d6) δ/ppm 10.42 and 10.26 (2 s, 2H, 2OH), 9.03 (d, J = 8.0 Hz, 1H, Ar–H), 8.96 (s, 1H, quinoline CH = N), 8.09 (d, J = 7.4 Hz, 2H, Ar–H), 8.08 (d, J = 6.8 Hz, 2H, Ar–H), 7.98 (d, J = 7.6 Hz, 1H, Ar–H), 7.86 (s, 1H, quinoline-H4), 7.85–7.81 (m, 2H, 1 Ar–H+ NH), 7.73 (d, J = 8.4 Hz, 1H, Ar–H), 7.67 (d, J = 8.0 Hz, 1H, Ar–H); 13C NMR (101 MHz, DMSO) δ/ppm 172.06 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 165.16 (C–OH), 162.05 (C
O), 165.16 (C–OH), 162.05 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 152.99 (C
O), 152.99 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 148.06, 141.25, 140.15, 138.28, 135.40, 134.47, 130.11, 129.51, 125.39, 124.25, 117.07, 114.67, 111.50, 108.50 (Ar–Cs); MS (EI, 70 eV): m/z (%) = 358.34 [M+] (31.13%), 305 (100%); Analyt. Cal. C21H14N2O4 (Mw.t. = 358.35) calculated C, 70.39; H, 3.94; N, 7.82; Found C, 70.19; H, 4.12; N, 7.62.
N), 148.06, 141.25, 140.15, 138.28, 135.40, 134.47, 130.11, 129.51, 125.39, 124.25, 117.07, 114.67, 111.50, 108.50 (Ar–Cs); MS (EI, 70 eV): m/z (%) = 358.34 [M+] (31.13%), 305 (100%); Analyt. Cal. C21H14N2O4 (Mw.t. = 358.35) calculated C, 70.39; H, 3.94; N, 7.82; Found C, 70.19; H, 4.12; N, 7.62.4.2 Biological activity
The 5-lipoxygenase inhibition assay was conducted utilizing the 5-LOX kit (no. 437996, Sigma-Aldrich), in accordance with the protocol established by Huang et al. (2019).67 Various concentrations of the extracts were combined with 90 µL of 5-LOX and 100 µL of the chromogen. Subsequently, 10 µL of arachidonic acid was added, and the mixture was agitated for 10 minutes. The absorbance was subsequently measured at 490 nm and compared with blank control. The IC50 value was calculated employing linear regression analysis.
4.3 Docking simulation
The molecular docking simulation of the most active azomethine salicylic acid derivatives 2, 4, and 9, alongside positive control drugs (Aspirin and Zileuton), was conducted within the active site of 5-lipoxygenase (PDB: 7TTJ) (https://www.rcsb.org/; last accessed 1/10/2025). The docking and visualization of docking pose was executed using the Molecular Operating Environment (MOE) software version 10.2009. The structures of the most active derivatives and the positive controls were exported to MOE, where they were saved as an mdb file after the addition of hydrogen atoms and subsequent minimization based on the MMFF94x force field, in accordance with previously established methodologies.71,72 The active site of 5-lipoxygenase (PDB: 7TTJ) was generated following a previously reported protocol, which involved the use of a dummy atom and only one chain (chain A). The standard procedure included employing the Triangle Matcher for placement and London dG for rescoring, with a retention of 30 poses as described previously.73Conflicts of interest
The authors declare that there are no conflicts of interest.Data availability
The data supporting this article are included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra07622f.Acknowledgements
The authors gratefully acknowledge the Faculty of Science, Galala University, for providing the laboratory facilities, technical support, and research environment that significantly contributed to the completion of this work.References
- J. K. Sinha, K. Jorwal, K. K. Singh, S. S. Han, R. Bhaskar and S. Ghosh, Mol. Neurobiol., 2025, 62, 6748–6763 Search PubMed.
- P. Choppara, Y. V Prasad, C. V Rao, K. Hari Krishna, G. Trimoorthulu, G. U. Maheswara Rao, J. Venkateswara Rao, M. S. Bethu and Y. L. N. Murthy, Arab. J. Chem., 2019, 12, 2328–2335 CrossRef CAS.
- S. Viveka, Dinesha, P. Shama, G. K. Nagaraja, S. Ballav and S. Kerkar, Eur. J. Med. Chem., 2015, 101, 442–451 CrossRef CAS PubMed.
- H. S. F. Salem, M. N. Raafat, I. M. El Mancy and I. M. Bayomi, Al-Azhar Int. Med. J., 2024, 5, 16 Search PubMed.
- Y. Ali, M. S. Alam, H. Hamid, A. Husain, S. Bano, A. Dhulap, C. Kharbanda, S. Nazreen and S. Haider, Eur. J. Med. Chem., 2015, 92, 490–500 Search PubMed.
- D. Ribeiro, M. Freitas, S. M. Tomé, A. M. S. Silva, G. Porto, E. J. Cabrita, M. M. B. Marques and E. Fernandes, Eur. J. Med. Chem., 2014, 72, 137–145 Search PubMed.
- E. Aitella, C. Romano, L. Ginaldi and D. Cozzolino, 2025, DOI:10.3390/ijms26030927.
- N. P. Reddy, T. Chandramohan Reddy, P. Aparoy, C. Achari, P. R. Sridhar and P. Reddanna, Eur. J. Med. Chem., 2012, 47, 351–359 CrossRef CAS PubMed.
- M. Zaki, Al-Azhar J. Pharm. Sci., 2019, 59, 88–106 Search PubMed.
- P. Srivastava, V. K. Vyas, B. Variya, P. Patel, G. Qureshi and M. Ghate, Bioorg. Chem., 2016, 67, 130–138 Search PubMed.
- T. Crosson, N. Bretaud and S. Ugolini, Immunity, 2025, 58, 1161–1174 Search PubMed.
- K. Bishayee and A. R. Khuda-Bukhsh, Acta Biochim. Biophys. Sin., 2013, 45, 709–719 Search PubMed.
- B. Jawabrah Al-Hourani, S. K. Sharma, J. Kaur and F. Wuest, Med. Chem. Res., 2015, 24, 78–85 CrossRef CAS.
- R. Ayman, A. M. Radwan, A. M. Elmetwally, Y. A. Ammar and A. Ragab, Arch. Pharm., 2023, 356, e2200395 Search PubMed.
- R. Ayman, M. S. Abusaif, A. M. Radwan, A. M. Elmetwally and A. Ragab, Eur. J. Med. Chem., 2023, 249, 115138 CrossRef CAS PubMed.
- H. A. Abd El Razik, M. H. Badr, A. H. Atta, S. M. Mouneir and M. M. Abu-Serie, Arch. Pharm., 2017, 350, 1700026 Search PubMed.
- M. Arora, S. Choudhary, P. K. Singh, B. Sapra and O. Silakari, Life Sci., 2020, 251, 117631 Search PubMed.
- M. A. Abdelgawad, M. B. Labib, W. A. M. Ali, G. Kamel, A. A. Azouz and E.-S. EL-Nahass, Bioorg. Chem., 2018, 78, 103–114 Search PubMed.
- S. Hussein, E. A. Fayed, A. Ragab, M. S. Abusaif, Y. A. Ammar, R. El-Sayed Mansou, A. Musa and T. M. Ramsis, BMC Chem., 2025, 19, 271 CrossRef CAS PubMed.
- A. Rauf, A. Olatunde, M. R. Islam, Z. Ahmad, N. Hafeez, H. A. Hemeg, M. Imran, M. S. Mubarak, G. Ribaudo and N. Schmiedebergs, Arch. Pharmacol., 2025, 398, 9387–9403 CrossRef CAS.
- M. A. Chowdhury, K. R. A. Abdellatif, Y. Dong, D. Das, G. Yu, C. A. Velázquez, M. R. Suresh and E. E. Knaus, Bioorg. Med. Chem. Lett., 2009, 19, 6855–6861 Search PubMed.
- P. Michel, Int. J. Mol. Sci., 2025, 26, 7280 Search PubMed.
- A. Bashir I.J., C, S. Kankipati, S. Jones, R. Newman, S. Safrany T., C. Perry J. and I. Nicholl, Int J Oncol, 2019, 54, 1256–1270 Search PubMed.
- M. Abdalla, A. E. Elmasry, M. T. M. Nemr, M. M. Alanazi, N. G. M. Attallah and Y. A. A. M. Elshaier, ChemistrySelect, 2025, 10, e202500175 CrossRef CAS.
- L. H. Al-Wahaibi, M. A. Mahmoud, H. A. Alzahrani, H. A. Abou-Zied, A. Abdelmoez, B. G. M. Youssif, S. Bräse and S. M. Rabea, Front. Chem., 2024, 12, 1493906 Search PubMed.
- E. M. Sarshira, N. M. Hamada, Y. M. Moghazi and M. M. Abdelrahman, J. Heterocycl. Chem., 2016, 53, 1970–1982 Search PubMed.
- G. Paraskevopoulos, M. Krátký, J. Mandíková, F. Trejtnar, J. Stolaříková, P. Pávek, G. Besra and J. Vinšová, Bioorg. Med. Chem., 2015, 23, 7292–7301 CrossRef CAS PubMed.
- H. C. Sweany, G. C. Turner, M. Lichtenstein and S. Entin, Dis. Chest, 1949, 16, 633–656 Search PubMed.
- J. A. Forbes, W. T. Beaver, E. H. White, R. W. White, G. B. Neilson and R. W. Shackleford, Jama, 1982, 248, 2139–2142 Search PubMed.
- J. Hannah, W. V Ruyle, H. Jones, A. R. Matzuk, K. W. Kelly, B. E. Witzel, W. J. Holtz, R. A. Houser and T. Y. Shen, J. Med. Chem., 1978, 21, 1093–1100 Search PubMed.
- Y. A. Ammar, J. A. Micky, D. S. Aboul-Magd, S. M. A. Abd El-Hafez, S. A. Hessein, A. M. Ali and A. Ragab, Chem. Biol. Drug Des., 2023, 101, 245–270 Search PubMed.
- Y. A. Ammar, A. Ragab, M. A. Migahed, S. Al-Sharbasy, M. A. Salem, O. K. M. Riad, H. M. R. M. Selim, G. A. Abd-elmaksoud and M. S. Abusaif, RSC Adv., 2023, 13, 27363–27384 Search PubMed.
- Y. M. Abdel-Baky, A. M. Omer, E. M. El-Fakharany, Y. A. Ammar, M. S. Abusaif and A. Ragab, Sci. Rep., 2023, 13, 22792 Search PubMed.
- E. Raczuk, B. Dmochowska, J. Samaszko-Fiertek and J. Madaj, Molecules, 2022, 27(3), 787 CrossRef CAS PubMed.
- T. Biswas, R. K. Mittal, V. Sharma, Kanupriya and I. Mishra, Lett. Org. Chem., 2024, 21(6), 505–519 CrossRef CAS.
- Z. Yang, L. Lv, Y. Wang, X. Wang, W. Wu, Q. Shen, C. Zhang, J. Qin, Z. Jiang, F. Sun, J. Liu and H. Lu, Colloids Surfaces B Biointerfaces, 2025, 254, 114866 CrossRef CAS PubMed.
- Ö. Güngör and P. Gürkan, J. Mol. Struct., 2014, 1074, 62–70 CrossRef.
- P. Misra, B. K. Mishra and G. B. Behera, Int. J. Chem. Kinet., 1991, 23, 639–654 Search PubMed.
- M. A. Vázquez, F. Muñoz, J. Donoso and F. Garcia Blanco, Amino Acids, 1992, 3, 81–94 CrossRef PubMed.
- F. Benachenhou, N. Mimouni, Y. Mederbel and R. K. Slimane, Arab. J. Chem., 2012, 5, 245–250 Search PubMed.
- R. F. Martínez Vázquez, E. Matamoros Castellano, P. Cintas Moreno and J. C. Palacios Albarrán, J. Org. Chem., 2020, 85, 5838–5862 Search PubMed.
- Z. Feng, S. Jia, H. Chen and L. You, Tetrahedron, 2020, 76, 131128 Search PubMed.
- A. A. Almehizia, A. M. Naglah, S. S. Aljafen, A. S. Hassan and W. M. Aboulthana, Pharmaceutics, 2025, 17(2), 180 Search PubMed.
- A. Ragab, A. Minarčíková, A. Kleinová, J. Kronek and Z. Kroneková, J. Mol. Struct., 2025, 1341, 142584 Search PubMed.
- M. E. Hassan, T. M. Tamer, K. Valachová, A. Ragab and L. Šoltés, Int. J. Biol. Macromol., 2025, 318, 145054 Search PubMed.
- A. S. Hassan, N. M. Morsy, W. M. Aboulthana and A. Ragab, RSC Adv., 2023, 13, 9281–9303 RSC.
- Y. M. Abdel-Baky, A. Ragab, M. S. Abusaif, Y. A. Ammar and A. M. Omer, Carbohydr. Polym., 2025, 124049 Search PubMed.
- M. H. Helal, A. Ragab, M. S. Abusaif, E. M. Ahmed, S. M. Gomha, Y. A. Ammar and S. I. Eissa, J. Mol. Struct., 2026, 1349, 143703 Search PubMed.
- M. S. Abusaif, A. M. Sh El-Sharief, Y. A. Mohamed, Y. A. Ammar, M. A. Ismail, W. M. Aboulthana, M. S. A. El-Gaby and A. Ragab, Sci. Rep., 2025, 15, 19409 CrossRef CAS PubMed.
- A. Ragab, RSC Adv., 2025, 15, 3607–3645 RSC.
- M. M. S. Wassel, W. M. Gamal Eldin, A. Ragab, G. A. M. Elhag Ali, Y. A. Ammar and J. Appl, Vet. Sci., 2020, 5, 37–46 Search PubMed.
- K. E. Saadon, A. Ragab, N. M. H. Taha, N. A. Mahmoud, A. K. Khalil and G. A. M. Elhagali, Drug Dev. Res., 2025, 86, e70189 CrossRef CAS PubMed.
- Y. A. Ammar, A. M. El-Sharief, Y. A. Mohamed, A. B. Mehany and A. Ragab, Al-Azhar Bull. Sci., 2018, 29, 25–37 Search PubMed.
- H. Khamees Thabet, Y. A. Ammar, M. Imran, M. Hamdy Helal, S. Ibrahim Alaqel, A. Alshehri, A. Ash Mohd, M. S. Abusaif and A. Ragab, Bioorg. Chem., 2024, 151, 107671 Search PubMed.
- H. K. Thabet, M. S. Abusaif, M. Imran, M. H. Helal, S. I. Alaqel, A. Alshehri, A. A. Mohd, Y. A. Ammar and A. Ragab, Comput. Biol. Chem., 2024, 111, 108097 CrossRef CAS PubMed.
- O. Rådmark, O. Werz, D. Steinhilber and B. Samuelsson, Biochim. Biophys. Acta - Mol. Cell Biol. Lipids, 2015, 1851, 331–339 Search PubMed.
- A. A. Silva, L. L. Oliveira, C. M. Queiroz-Junior, B. F. C. V. Caldeira, A. C. B. R. Soares, G. C. Mattos, A. D. Braga, P. L. Bosi, A. M. S. Reis, M. M. Teixeira and F. A. Amaral, Inflammopharmacology, 2025, 33, 3909–3918 CrossRef CAS PubMed.
- A. Ragab, R. R. Raslan, M. S. Abusaif, H. K. Thabet, Y. A. Ammar and N. A. Gohar, Eur. J. Med. Chem., 2025, 294, 117751 Search PubMed.
- M. A. Salem, M. S. Abusaif, N. A. Gohar, Y. A. Ammar and A. Ragab, Drug Dev. Res., 2025, 86, e70085 Search PubMed.
- R. R. Raslan, Y. A. Ammar, S. A. Fouad, S. A. Hessein, N. A. M. Shmiess and A. Ragab, RSC Adv., 2023, 13, 10440–10458 Search PubMed.
- H. A. Mohamed, Y. A. Ammar, G. A. M. Elhagali, H. A. Eyada, D. S. Aboul-Magd and A. Ragab, J. Mol. Struct., 2023, 1287, 135671 CrossRef CAS.
- H. Khamees Thabet, A. Ragab, M. Imran, M. H. Helal, S. Ibrahim Alaqel, A. Alshehri, A. Ash Mohd, M. Rakan Alshammari, M. S. Abusaif and Y. A. Ammar, Eur. J. Med. Chem., 2024, 275, 116589 Search PubMed.
- M. A. Ismail, M. S. Abusaif, M. S. A. El-Gaby, Y. A. Ammar and A. Ragab, RSC Adv., 2023, 13, 12589–12608 RSC.
- A. A. Fahmy, Phosphorus. Sulfur. Silicon Relat. Elem., 1991, 63, 81–86 CrossRef CAS.
- Y. H. Y. Almalky and O. M. Alatawi, Int. J. Biol. Macromol., 2025, 316, 144353 Search PubMed.
- A. A.-M. Abdel-Aziz, A. S. El-Azab, L. A. Abou-Zeid, K. E. H. ElTahir, N. I. Abdel-Aziz, R. R. Ayyad and A. M. Al-Obaid, Eur. J. Med. Chem., 2016, 115, 121–131 Search PubMed.
- Y. Huang, B. Zhang, J. Li, H. Liu, Y. Zhang, Z. Yang and W. Liu, Eur. J. Med. Chem., 2019, 180, 41–50 Search PubMed.
- D. Soni and P. Sureshkumar, Res. J. Biotechnol., 2016, 11, 4 Search PubMed.
- O. O. Oyedapo and A. J. Famurewa, Int. J. Pharmacogn., 1995, 33, 65–69 CrossRef.
- V. Vichai and K. Kirtikara, Nat. Protoc., 2006, 1, 1112–1116 Search PubMed.
- H. F. Rizk, M. A. El-Borai, A. Ragab, S. A. Ibrahim and M. E. Sadek, Polycycl. Aromat. Compd., 2023, 43, 500–522 Search PubMed.
- H. K. Thabet, A. Ragab, M. Imran, M. Hamdy Helal, S. Ibrahim Alaqel, A. Alshehri, A. Ash Mohd, S. S. Alshammari, Y. A. Ammar and M. S. Abusaif, RSC Adv., 2024, 14, 15691–15705 Search PubMed.
- A. Ragab, R. Ayman, M. A. Salem, Y. A. Ammar and M. S. Abusaif, Eur. J. Med. Chem., 2025, 290, 117499 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |



