Open Access Article
Open Access ArticleEmerging trends and structure–activity insights of Schiff base–lanthanide complexes as antibacterial agents
Sikandar Paswan
 a,
Nitesh Jaiswal
a,
Nitesh Jaiswal
 b,
Urvashi
b,
Urvashi
 c,
Amar Nath
c,
Amar Nath
 a,
Manoj Kumar
a,
Manoj Kumar
 d,
Divya Mishra
d,
Divya Mishra
 e and
Navneet Yadav
e and
Navneet Yadav
 *c
*c
aDepartment of Chemistry, Baba Raghav Das Post Graduate College, Deoria, DDU Gorakhpur University, Gorakhpur, U. P. 274001, India
bDepartment of Chemistry, Prof. Rajendra Singh (Rajju Bhaiya) Institute of Physical Sciences for Study & Research, Veer Bahadur Singh Purvanchal University Jaunpur, U. P. 222003, India
cDepartment of Chemical Engineering, Swansea University, Bay Campus, Fabian Way, Swansea SA1 8EN, UK. E-mail: navaneetyadav840@gmail.com; Navneet.yadav@swansea.ac.uk
dDepartment of Chemistry, Government Degree College, Dhadha Bujurg-Hata, Kushinagar, DDU Gorakhpur University, Gorakhpur, U. P. 274207, India
eTumor Microenvironment Unit, Humanitas University, Via Reti Levi Montalcini, 4, 20090 Pieve Emanuele, Milan, Italy
First published on 15th January 2026
Abstract
The rapid escalation of microbial resistance has accelerated the search for next-generation antibacterial agents with novel mechanisms of action. Schiff bases and their lanthanide complexes have emerged as promising candidates due to their tunable structures, coordination versatility and enhanced bioactivity upon metal binding. While earlier reviews laid the foundational understanding of antibacterial activity of Schiff bases and their metal complexes, this review advances the field by offering a data-driven, mechanistically informed and structure–activity-focused perspective on the most recent antibacterial findings (2018–2025) of Schiff bases and their Ln(III) complexes. In this review article, we critically compare the antibacterial performance of diverse ligand frameworks with lanthanide ions, under varied experimental conditions against both Gram-negative and Gram-positive bacterial strains highlighting trends in structure–activity correlations. We also identify clear trends: Schiff base ligands alone usually show modest or weak antibacterial activity, which is significantly enhanced upon complexation with lanthanide ions. Additionally, unresolved challenges, including toxicity, bioavailability and resistance modulation are discussed alongside proposed future research pathways. This perspective aims to guide the rational design of Schiff base–lanthanide complexes toward clinical translation as potent antibacterial agents.
1. Introduction
Antimicrobial resistance (AMR) arises when microorganisms including bacteria, fungi, parasites, and viruses adapt and become resistant to the antimicrobial agents used to treat them. Today, AMR is a major global health threat and is projected to cause up to 10 million deaths annually by 2050. Its rapid rise is largely driven by the misuse and overuse of antibiotics in human medicine, agriculture, animal care, and the food industry.1a,b Known as the “Silent Pandemic,” AMR is already undermining the effectiveness of existing treatments. Multidrug-resistant pathogens such as MRSA (Methicillin-Resistant Staphylococcus aureus), VRE (Vancomycin-Resistant Enterococci), and CRE (carbapenem-resistant Enterobacteriaceae) are causing increased treatment failures, prolonged illnesses, and higher mortality rates. Since the early antibiotic era, it has been recognized that inappropriate or excessive use accelerates resistance, a problem that has now escalated globally. To combat AMR, international organisations and governments have adopted coordinated strategies, including the “One Health Approach” and the WHO's Global Action Plan and GLASS surveillance system, which aim to strengthen monitoring and promote responsible antimicrobial use worldwide.1c,d The advancement in sanitation, vaccine development and the supplementary public health procedures are insufficient to address the widespread morbidity and mortality caused by infectious diseases. The infections by old diseases and the emergence of new ones have been facilitated due to growing environmental changes, increased transport and human movement, and rising in global warming.1e,2 Over the past three decades, more than 30 new infectious agents have been discovered worldwide; 60% of which are zoonotic, and more than two-thirds of these have their origins in wildlife.3 A literature review reported 1407 species of human pathogens, including 208 viruses, 538 bacterial species, 317 fungal species, 57 protozoan species, and 287 helminths. Of these, 177 species (13%) were identified as emerging or re-emerging pathogens, including 77 viruses (37%), 54 bacteria (10%), 22 fungi (7%), 14 protozoa (25%), and 10 helminths (3%). Emerging and re-emerging infectious diseases account for 26% of annual deaths worldwide.4–8 Antimicrobial susceptibility screening is widely studied in the drug discovery, epidemiology and prediction of therapeutic outcomes.9,10 Over the last few decades, scientists achieved great success by reporting several different types of antimicrobial drugs to counter the global public health emergency arising from various G(+) and G(−) bacterial strains.11–13 The above resistance crisis has not only undermined the efficacy of frontline therapeutic agents but also emphasized the more research for alternate antimicrobial strategies with novel mechanism of action. The metallo-organic compounds have emerged as main candidates in this search, proposing unique physicochemical properties that often transform into enhanced biological activities.14 An important organic ligand called Schiff bases, generated by the condensation reaction of primary amines with aldehydes or ketones have attracted significant attention because of their structural diversity, ease of synthesis and flexible donor atom sets. Lanthanide–Schiff base complexes exhibit enhanced stability relative to the uncoordinated ligand, due to chelation and electron donation from N O donors.15 In particular, the incorporation of lanthanide (Ln(III)) ions into Schiff base frameworks has yielded a variety of coordination architectures with notable antibacterial activity.16 The azomethine (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) or an imine (>C
N–) or an imine (>C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) groups present in Schiff bases elucidate the mechanism of transamination and racemisation reaction in biological systems, which makes Schiff bases an important fragment for medical purpose.17–21 The large ionic radii and unique shielding of 4f orbitals of lanthanide ions offer range of coordination number for metal which allowed the generation of stable mono and poly-nuclear metal complexes. These properties induce superior lipophilicity, and enhanced interaction with bacterial biomolecules compared to the free ligands.22 Compounds containing lanthanide(III) ions (Ln3+) have been of interest since the 19th century. In the 1960s, complexes of the Ln(III) ion exhibiting pharmacological properties such as anticoagulation, anti-inflammatory, antibacterial, anti-allergic and anticancer effects were discovered.23–26 The antibacterial properties of lanthanide metals were first recognized when Ce(III) salts exhibited antibacterial activity and the oxidizing properties of Ce(IV) led to the use of Ce(IV) sulphate as an antiseptic powder.27 Consequently, the coordination chemistry of lanthanides and their complexes has attracted significant interest among researchers. The biological properties of Ln(III) ions arise from their similarity to calcium ions, which sparked studies into their potential medical application because Ln3+ can substitute for Ca2+ proteins and can influence calcium-dependent enzymes either by inhibiting their function or by enhancing their activation.28 Additionally, Ln(III) ions such as Nd(III), Sm(III) and Yb(III) ions, are ideal factors for luminescence imaging in vivo due to their emission in the NIR region, which can be detected through animal tissue of great thickness.29–32 Although the potential of Schiff bases and their lanthanide complexes in antibacterial research is well established, the literature still reveals a gap in systematic investigation. An examination of earlier reports reveals a notable gap, suggesting that more comprehensive investigations into structure–activity relationships are required.33 In particular; limited attention has been devoted to how donor atom sets, substituent effects and coordination numbers influence antibacterial potency. Consequently, the field still lacks a systematic synthesis that links the chemistry of Schiff base–lanthanide complexes with their biological significance.34–37
N–) groups present in Schiff bases elucidate the mechanism of transamination and racemisation reaction in biological systems, which makes Schiff bases an important fragment for medical purpose.17–21 The large ionic radii and unique shielding of 4f orbitals of lanthanide ions offer range of coordination number for metal which allowed the generation of stable mono and poly-nuclear metal complexes. These properties induce superior lipophilicity, and enhanced interaction with bacterial biomolecules compared to the free ligands.22 Compounds containing lanthanide(III) ions (Ln3+) have been of interest since the 19th century. In the 1960s, complexes of the Ln(III) ion exhibiting pharmacological properties such as anticoagulation, anti-inflammatory, antibacterial, anti-allergic and anticancer effects were discovered.23–26 The antibacterial properties of lanthanide metals were first recognized when Ce(III) salts exhibited antibacterial activity and the oxidizing properties of Ce(IV) led to the use of Ce(IV) sulphate as an antiseptic powder.27 Consequently, the coordination chemistry of lanthanides and their complexes has attracted significant interest among researchers. The biological properties of Ln(III) ions arise from their similarity to calcium ions, which sparked studies into their potential medical application because Ln3+ can substitute for Ca2+ proteins and can influence calcium-dependent enzymes either by inhibiting their function or by enhancing their activation.28 Additionally, Ln(III) ions such as Nd(III), Sm(III) and Yb(III) ions, are ideal factors for luminescence imaging in vivo due to their emission in the NIR region, which can be detected through animal tissue of great thickness.29–32 Although the potential of Schiff bases and their lanthanide complexes in antibacterial research is well established, the literature still reveals a gap in systematic investigation. An examination of earlier reports reveals a notable gap, suggesting that more comprehensive investigations into structure–activity relationships are required.33 In particular; limited attention has been devoted to how donor atom sets, substituent effects and coordination numbers influence antibacterial potency. Consequently, the field still lacks a systematic synthesis that links the chemistry of Schiff base–lanthanide complexes with their biological significance.34–37
In the present review, we aim to address this gap by providing a comprehensive and focused account of the antibacterial properties of Schiff bases and their lanthanide complexes reported between 2018 and 2025. In contrast to earlier surveys, this work provide new advances in synthesis, coordination features of metal ions, mechanistic insights and antibacterial evaluation into a unified framework that delivers the connection between coordination chemistry and biological outcome. Special attention is given to the comparison of Schiff bases with their corresponding lanthanide complexes, revealing general trends in activity. In addition, the authors have also expressed structure–activity relationships across a wide range of ligands and its Ln(III) complexes which identifies specific structural motifs and coordination environments associated with strong antibacterial activity. Through this approach, the review not only summarizes progress in the field but also clarifies design principles that can inform the rational development of next-generation antibacterial agents. By combining recent literature with comparative and mechanistic analysis, it aims to bridge the disciplines of coordination chemistry and microbiology, thereby offering insights that are both timely and urgently needed in the broader context of combating antibiotic resistance.
2. Schiff bases and lanthanide complexes: overview
Schiff bases, first described by Hugo Schiff in 1864, are formed by the condensation of primary amines with aldehydes or ketones and are characterized by the presence of the azomethine (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) functional group.38 Their structural versatility arises from the wide variety of amine and carbonyl precursors, which allows fine-tuning of steric and electronic environments. While the imine nitrogen atom serves as the principal coordination site, additional substituents such as –OH, –COOH, –SH and –OCH3 increase the denticity of the ligands and enhance the stability of metal complexes.39 Lanthanide ions, typically in the +II, +III, and +IV oxidation states, are highly electropositive and classified as hard Lewis acids. According to the hard and soft acids and bases (HSAB) principle, they preferentially coordinate with donor atoms such as O, N and S, making Schiff bases ideal ligands for their stabilization. Unlike transition metals, lanthanides possess shielded 4f orbitals, resulting in weak crystal field effects and predominantly ionic bonding. Nevertheless, Schiff bases stabilize lanthanide center effectively, affording robust and thermally stable complexes that can be mononuclear, binuclear or polynuclear in nature.
N–) functional group.38 Their structural versatility arises from the wide variety of amine and carbonyl precursors, which allows fine-tuning of steric and electronic environments. While the imine nitrogen atom serves as the principal coordination site, additional substituents such as –OH, –COOH, –SH and –OCH3 increase the denticity of the ligands and enhance the stability of metal complexes.39 Lanthanide ions, typically in the +II, +III, and +IV oxidation states, are highly electropositive and classified as hard Lewis acids. According to the hard and soft acids and bases (HSAB) principle, they preferentially coordinate with donor atoms such as O, N and S, making Schiff bases ideal ligands for their stabilization. Unlike transition metals, lanthanides possess shielded 4f orbitals, resulting in weak crystal field effects and predominantly ionic bonding. Nevertheless, Schiff bases stabilize lanthanide center effectively, affording robust and thermally stable complexes that can be mononuclear, binuclear or polynuclear in nature.
Lanthanide coordination chemistry is largely dictated by the large ionic radii of Ln3+ ions and the steric demands of the coordinating ligands. Because the 4f electrons are effectively shielded and do not participate significantly in bonding, lanthanide–ligand interactions are primarily ionic, leading to high and flexible coordination numbers. With respect to complex formation, lanthanide ions are classified as “Class A” or hard Lewis acids and therefore preferentially bind to hard donor ligands, following the general order O > N > S and F > Cl. In aqueous solution, Ln3+ ions are strongly hydrated and form well-defined hydration shells. The number of coordinated water molecules depends on ionic size: the lighter lanthanides typically exhibit higher coordination numbers, commonly forming [Ln(H2O)9]3+ species (Ln = La–Eu), while the heavier lanthanides favor lower hydration numbers, most often [Ln(H2O)8]3+ (Ln = Dy–Lu). Eu3+ and Gd3+ frequently exist as mixed populations with both 8 and 9 coordinate hydration states. This reduction in hydration number across the series is attributed to increasing Ln–O bond strength and decreasing ionic radius, which limit the number of water molecules that can be accommodated. Because of this strong hydration, only ligands capable of forming highly stable chelates can effectively compete with water for coordination in aqueous media.40a Bulky ligands such as bis(trimethylsilyl)amine may enforce lower coordination, whereas small, multidentate ligands such as nitrates or macrocycles can stabilize coordination numbers up to 12. A gradual decrease in average coordination number is observed from La to Lu due to the lanthanide contraction, which also shortens Ln–donor distances and influences complex stability, preferred geometries, and ligand selectivity. Oxygen donors dominate the first coordination sphere, followed by carbon donors (mainly from cyclopentadienyl ligands) and nitrogen donors, together comprising roughly 95% of all donor atoms. High-hapticity ligands such as Cp significantly impact both coordination numbers and their variability. Unlike transition metals, lanthanides exhibit minimal ligand field splitting, resulting in limited stereochemical control and a tendency to form irregular coordination geometries such as bicapped trigonal prisms, square antiprisms, and dodecahedra.40b–d Their weak ligand field also renders f–f transitions Laporte-forbidden, producing the sharp emission bands characteristic of lanthanide-based luminescent materials. Consistent with HSAB principles, lanthanide ions show strong affinity toward hard donor atoms; thus, oxygen-containing ligands-including carboxylates, β-diketonates, phosphates, and polyoxometalates-form particularly stable complexes. Nitrogen donors such as amines and Schiff bases also coordinate effectively, though generally with slightly reduced stability. These preferences are critical for designing lanthanide-based MOFs, luminescent probes, and antibacterial coordination compounds. Increasing attention is now focused on tuning ligand denticity, steric constraints, and solvent interactions to modulate magnetic anisotropy, energy-transfer efficiency, and biological activity. Such structural tunability underpins the versatile applications of lanthanide complexes in catalysis, photophysics, and medicinal chemistry, especially in the development of next-generation antibacterial agents.
Synthesis of Schiff base and its lanthanide complexes typically involve a two-step process: initial condensation of aldehydes and amines, usually in alcoholic solvents such as ethanol or methanol, followed by coordination with hydrated lanthanide salts (nitrates or chlorides). Conventional methods generally employ reflux conditions, which are effective but can be time-consuming and occasionally result in reduced selectivity.41 In recent years, green chemistry strategies such as solvent-free mechanochemical grinding, microwave-assisted synthesis and ionic liquid-mediated reactions have been explored to reduce reaction times, minimize chemical waste and improve yields. Ethanol remains the most widely used solvent due to its benign and eco-friendly nature; however, greener alternatives are increasingly being adopted in Schiff base and their lanthanide chemistry. In general, the synthetic strategies commonly employed for the preparation of Schiff bases and their lanthanide complexes as reported in various scientific studies are summarized in the following flow chart in Scheme 1.42–47
Structurally, Schiff base ligands are particularly suitable for stabilizing lanthanide ions, which owing to their large ionic radii, usually adopt high coordination numbers ranging from 8 to 10 are most common but values of 7 and 11–12 are also observed. The resulting geometries include bicapped trigonal prisms, dodecahedra, and monocapped square antiprisms, with specific arrangements governed mainly by steric and solvation effects rather than crystal field stabilization energy.48–51 These geometrical features significantly influence the physicochemical properties of the complexes, such as electronic behaviour, lipophilicity, and biological activity. Importantly, the enhanced membrane permeability and lipophilic balance of Schiff base–lanthanide complexes are consistently associated with superior antibacterial activity against both G(+) and G(−) pathogens. In addition, certain systems exhibit dual luminescent and theranostic properties, extending their potential beyond antimicrobial applications to bio-imaging and multifunctional drug design. The structure–activity relationship (SAR) of lanthanide Schiff base complexes, correlating coordination number, donor atom set and antibacterial response against selected pathogens were given in Table 1.52–55,66,67,92,107,111,115,122,123 The structure–activity relationship (SAR) analysis highlights that Schiff base ligands with amine-derived donor atoms (NOS) play a crucial role in modulating antibacterial potency of Ln(III) complexes. A clear trend is observed where higher coordination numbers (8–10) often correlate with enhanced activity, likely due to increased stability and lipophilicity. Lanthanide identity strongly influences selectivity with Ce(III), Pr(III), Nd(III), Gd(III) and Dy(III) showing broader antibacterial spectra. Overall, the interplay of coordination number, donor environment and Ln(III) ions nature governs antibacterial efficiency against both G(+) and G(−) pathogens.
| Sr. no. | Ln(III) ions | Observed coord. no. | Ligand donor features | Amine derived fragments | Pathogens showing enhanced activity |
|---|---|---|---|---|---|
| 1 | Ce(III) | 8–10 | NO; NN | Aromatic hydrazones; thiazole based amines | SA, LM, BC, PBA |
| 2 | Pr(III) | 6–10 | NO; NN | Hydrazones, ethoxy, pyridyl/phenyl; thiazole, pyridyl | SE, SA, MRSA, EF, PBA, KP, EC, PA |
| 3 | Nd(III) | 6–10 | NO; NN; NOS; NS | Hydrazones, pyridyls, amino phenol; alkyls, thiazoles; benzothiazoles | SA, SE, EC, BS, KP, EF, MRSA, PBA,PV |
| 4 | Sm(III) | 6–10 | NO; NS | Hydrazones, aminophenol, pyridyl, phenyl; benzothiazole | SA, EC, EF, MRSA, KP, SE |
| 5 | Eu(III) | 7–9 | NO; NOS | Hydrazones, phenyls; amino phenols | SE, EC, KP, PA, SA, EF |
| 6 | Gd(III) | 7–10 | NO; NN; NOS; NS | Hydrazones, amino phenols, phenyls; alkyls; benzothiazoles | SA, EC, KP, PA, EF, PV, BS, MRSA, SE |
| 7 | Tb(III) | 7–9 | NO; NOS | Hydrazones, phenyls; amino phenols | SA, SE, KP, PA, EC, EF, MRSA |
| 8 | Dy(III) | 7–10 | NO; NN; NOS | Hydrazones, amino phenols, phenyls; alkyls; benzothiazolres | EC, SA, SAl, BS, ML, EF, MRSA, KP, SE |
| 9 | Er(III) | 6–10 | NO; NN; NOS | Hydrazones, alanines, alkyls; pyridyls, naphthyls, alkyls; benzothiazoles | EC, SA, BS, PA, EF, MRSA, KP, SE, SF |
| 10 | Yb(III) | 6 | NO; NN | Alanine; pyridyl, naphthyls | SA, PA, EC, BS, NG, SF |
3. Summary of reported antibacterial activity of Schiff bases and Ln(III) complexes
In recent decades, the pursuit of new antibacterial agents has been an active area of research in medicine and pharmacy. Lanthanide complexes, owing to their distinctive electronic configuration, capacity for high coordination number, flexibility in ligand binding and unique physicochemical characteristics have emerged as promising candidates for therapeutic use.56 Representative examples of lanthanide ions with different Schiff base ligands and their antibacterial activity are presented SI in Table 1S.Sogukomerogullari et al. (2025)57–59 synthesized a tridentate NOS donor Schiff base, (H2L1) by condensing 2,5-thiophenedicarboxaldehyde with 2-aminophenol in MeOH. This ligand was subsequently used to prepare three Ln(III) complexes of the general formula [Ln(L1)Cl(H2O)]·nH2O (Ln = Eu, Tb, Gd). Spectroscopic analyses confirmed a coordination number of seven, involving two nitrogen, two oxygen and one sulfur donor atoms from the ligand, in addition to one aqua and from one chlorido ligand as shown in Fig. 1a. The antibacterial activities of these complexes were evaluated against G(+) strains (SA-ATCC25923, EF-ATCC29212) and G(−) strains (EC-ATCC25922, KP-ATCC700603, PA-ATCC27853) using disk diffusion and broth micro-dilution assays. The complexes consistently displayed higher antibacterial activity compared to the free H2L1 and in some cases were comparable to the standard drug amoxicillin–clavulanic acid (30 µg). Among them, the Eu(III) complex showed the most potent activity, with a 13 mm inhibition zone, MIC of 97.7 µM and MBC of 195.3 µM against SA. In contrast, the Tb(III) complex demonstrated lower potency (10 mm inhibition against EC; MIC/MBC of 390.7/781.3 µM). The zone of inhibition plots of theses complexes against above selected bacterial strains were given in Fig. 8. The enhanced antimicrobial activity of these complexes has been attributed to their increased lipophilicity, resulting from electron delocalization across the chelate ring. This facilitates penetration of bacterial lipid membranes and interference with essential metallo-enzymes by blocking their active sites.60,61
Abdel-Fatah et al. (2025)54 prepared a tridentate Schiff base H2L2 by the reaction of 5-bromosalicylaldehyde and 2-amino-3-hydroxypyridine in EtOH and its three lanthanide complexes having general formula [Ln(L2)Cl(H2O)2]·nH2O (Ln = Pr, Nd, Sm) as shown in Fig. 2a. Spectral analysis confirm coordination number six of Ln(III) ions involving two oxygen, one nitrogen atoms from Schiff base L2, two oxygen from two aqua and one chlorido ligands. The antibacterial activities of these complexes were examined against EC in DMSO solvent by using disk diffusion and dilution methods and results of this study were expressed in diameters of zone of inhibition and in MICs.27 The metal complexes displayed higher activity compare to free H2L2 and comparable with standard drug ofloxacin. The antibacterial efficacy of the these complexes followed the order ofloxacin > Nd(III) > Pr(III) > Sm(III) > H2L2.
Zhang et al. (2025)55 using a template-assisted cyclo-condensation procedure, prepared two Schiff base macro-cyclic mononuclear Ln(III) complexes with a ‘lasso-type’ architecture, having the general formula [Ln(HL3)(NO3)3] (Ln = Pr, Nd). The coordination number of the metal ions in these complexes is ten, arising from four oxygen and two nitrogen donor atoms of the ligand, along with six oxygen atoms from three bidentate nitrate ligands. The antibacterial activity was evaluated against six bacterial strains, including G(+) bacteria (SA, MRSA, EF) and G(−) bacteria (EC, PA, SE), using DMSO as the solvent, and the MIC values were determined by the dilution method.62–64 The results indicated that, due to the synergistic antibacterial effect of the Schiff base and the Ln3+ center, the Pr(III) and Nd(III) complexes exhibited pronounced antibacterial activity against SA, PA, and MRSA, with low MIC values, including 3.91 µg mL−1 for MRSA.
Abo-Rehab and co-workers (2024)65 synthesized a Schiff base (L4) through the condensation of 2-aminobenzothiazole with coumarin and this ligand was further used in the preparation of four novel Ln(III) complexes of general formula [LnL4Cl3]·2H2O (Ln = La, Ce, Nd, Dy). Structural analysis indicated that the Ln(III) centers adopt a coordination number of six, involving one nitrogen, one sulfur, and one oxygen donor atom from the Schiff base ligand, along with three chloride ions as shown in Fig. 3a. The antibacterial activities of the free L4 and its Ln(III) complexes were evaluated using the agar diffusion method against two G(−) (EC-ATCC10536, KP-ATCC10031) and two G(+) (SA-ATCC13565, BS-DSM1088). The results, expressed as diameters of zone of inhibition (mm), were compared with the standard antibiotic gentamicin. The complexes generally exhibited stronger antibacterial activity than the free ligand, except for the La(III) complex, which displayed comparable activity. Among the series, the Ce(III) complex showed the highest activity, producing inhibition zones of 21.3 mm (KP) and 20.3 mm (BS). However, all complexes were less potent than gentamicin. The observed antibacterial activity followed the order: gentamicin > Ce(III) > Nd(III) > Dy(III) > La(III) > free ligand (L4).
 | ||
| Fig. 3 General structure of aminobenzothiazole derived Schiff bases and their Ln(III) complexes (a–d). | ||
Miao et al. (2024)66 published an article dealing with synthesis of H2L5 ligand from 2-(hydroxyimino)propanehydrazide and 5-bromosalicylaldehyde in MeOH and its one cluster compound [Dy4(CO3)(H2L5)4(acac)2(CH3OH)2(CH2OH)2]2CH3OH. The structures of ligand are given in Fig. 2b. Single crystal X-ray analysis confirmed coordination numbers eight and nine with geometries triangular dodecahedron and spherical capped square antiprism, respectively. The antibacterial activities of the Dy(acac)3·2H2O, H2L5 and Dy(III)–H2L5 complexes against the five bacteria (EC, SA, SAl, BS, ML) were evaluated by paper disc diffusion and nutrient broth dilution methods, and results expressed as diameter of zone of inhibition (mm) and MIC, respectively as shown in Fig. 9. The findings demonstrate that Dy(III) complex exhibits markedly higher antibacterial activity than both the free ligand H2L5 and Dy(acac)3·2H2O salt. Among the five tested bacteria, Dy(III) complex exhibits relatively strong antibacterial activity, with notable inhibition zone diameters of 26.8 mm and 25.5 mm against SAl and ML, respectively, which is better than 22.7 mm against EC, 23.5 mm against SA, and 21.5 mm against BS. This superior activity is likely due to the Schiff base's complexation with Dy(III) ions, which reduces the polarity of the Dy(III) ion through partial sharing of its positive charge with the donor groups.67 The proposed antibacterial mechanism involves disruption of the microbial cell membrane, resulting in intracellular protein denaturation. This process likely inhibits the function of electron-transport enzymes and cell respiration enzymes, ultimately compromising bacterial viability.68,69
Taha et al. (2024)70,71 synthesized a Schiff base ligand, (H2L6) via the condensation of 3,5-di-tert-butyl-2-hydroxybenzaldehyde with picolinohydrazide in EtOH, and prepared a series of nine lanthanide complexes of the general composition [NEt3]2[Ln2(L6)2(NO3)4]·nH2O (Ln = La, Pr, Nd, Sm, Eu, Gd, Tb, Dy, Er). Single-crystal structural studies revealed a coordination number of nine for the Ln(III) ions, achieved through three oxygen and two nitrogen donor atoms from the Schiff base and four oxygen atoms from two bidentate nitrate ligands as shown in Fig. 2c and representative ORTEP diagram of Dy(III) complex shown in Fig. 11a. The antibacterial activity of the H2L6 and its Ln(III) complexes was investigated against three G(+) (SA, MRSA, EF) and three G(−) (KP, EC, SE) using both the micro-broth dilution and disk diffusion methods, with amoxicillin as the reference drug. The free ligand H2L6 and most complexes showed moderate antibacterial activity, predominantly against G(−) bacteria. Notably, Eu(III), Nd(III) and Pr(III) complexes demonstrated enhanced activity against SE with a MIC of 64 µg mL−1, compared to the free ligand (MIC 128 µg mL−1). However, none of the compounds were effective against G(+) strains, and all exhibited lower activity than the standard antibiotic. The authors attributed the improved activity of selected Ln(III) complexes to Overtone's concept of cell permeability, where chelation increases lipophilicity, facilitating transport across bacterial membranes. The differential response between G(+) and G(−) strains was explained by differences in cell wall architecture: G(+) bacteria possess a thick peptidoglycan wall, while G(−) bacteria have a thinner peptidoglycan layer combined with an outer lipopolysaccharide membrane, which influences permeability and susceptibility to antimicrobial agents.
Hussein et al. (2024)72 synthesized a Schiff base, (L7) through the condensation of 4-antipyrinecarboxaldehyde with 2-aminobenzothiazole and a series of lanthanide complexes of general formula [Ln2(L7)2(NO3)6]·6H2O (Ln = La, Nd, Er, Gd, Dy). Spectroscopic analysis confirmed that the Ln(III) ions adopt a nine-coordinate geometry, with donor atoms contributed by one nitrogen, one sulfur, and one oxygen from the ligand, along with six oxygen atoms from four nitrate ligands (Fig. 3b). The antibacterial activity of the ligand and its complexes was evaluated against two G(+) (SA, BS) and two G(−) (EC, KP) strains using the disk diffusion method. Against SA, Er(III) (30 mm), Gd(III) (33 mm) and Dy(III) (31 mm) complexes exhibited higher activity than the free ligand (26 mm), while Nd(III) (26 mm) was comparable and La(III) (20 mm) was less active. For BS, Er(III) (29 mm), Gd(III) (30 mm), and Nd(III) (25 mm) showed improved activity relative to the ligand (24 mm), whereas Dy(III) (20 mm) and La(III) (22 mm) were less effective. Against EC, most complexes demonstrated activity similar to the free ligand, except Gd(III) and Dy(III), which were more active. Remarkably, both the ligand and all complexes exhibited strong antibacterial effects against KP, with the Nd(III) complex showing the highest inhibition zone of 34 mm. The authors attributed the enhanced antibacterial activity of Ln(III)–Schiff base complexes to their ability to disrupt bacterial cell membranes, leading to increased permeability, leakage of intracellular components, and ultimately cell death.73
Alqasaimeh et al. (2023)74 synthesized a Schiff base, (HL8) by condensation reaction of salicylaldehyde with p-toluidine and its three Ln(III) complexes of the general formula [Ln(HL8)3(NO3)3] (Ln = Nd, Tb Dy) (Fig. 2d). The coordination number of Ln(III) ions is nine, six from three bidentate nitrate ions and three from three HL8. The representative ORTEP diagram of Nd(III) complex shown in Fig. 11b. The authors investigated antibacterial properties of theses complexes against G(+) (SA-ATCC29213, SA-ATCC33591) and G(−) (EC-ATCC25922, PA-ATCC27853) using the disk diffusion; diameter of zone of inhibition (mm) and micro-dilution methods; MIC with gentamicin and amikacin as reference drugs. The results showed that HL8 exhibits no zone of inhibition against all bacteria strains whereas its Ln(III) complexes shows some sensitivity against SA-ATCC29213 and EC-ATCC25922 except other bacterial strains SA-ATCC33591 and PA-ATCC27853. All the Ln(III) complexes with MIC 0.75 mg mL−1 against SA-ATCC29213 is very efficient than free Schiff base.
Andiappan et al. (2023)75 reported a Schiff base (L9) through the condensation reaction of 9-anthraldehyde with 2,6-diaminopyridine in MeOH and their three Ln(III) complexes (Ln = Yb, Pr, Er) as shown in Fig. 1b. Antibacterial performance was evaluated against G(+) (SA) and G(−) (PA) using the well diffusion method (zone of inhibition, mm) and microdilution analysis (MIC, µg mL−1) at concentrations of 20, 40, 60, and 80 µg mL−1. Against PA, the free ligand showed inhibition zones of 10, 12, 12, and 14 mm across the tested concentrations, while the Pr(III) complex exhibited markedly enhanced activity with zones of 18, 22, 24, and 24 mm. This activity was comparable to the standard antibiotic streptomycin, which produced a 25 mm inhibition zone. Against SA, the free ligand (20 mm) demonstrated moderate activity, which was further improved in its complexes: Pr(III) (24 mm), Er(III) (21 mm), and Yb(III) (22 mm). The antibacterial activity data of theses complexes at various concentrations against both strains were given in Fig. 10. The results highlight that complexation with Ln(III) ions significantly enhances the antibacterial properties of the Schiff base ligand, with the Pr(III) complex emerging as the most potent candidate, approaching the activity of the standard drug. The improved bioactivity was attributed to increased lipophilicity of the complexes, facilitating greater bacterial membrane penetration and interaction with intracellular targets.76,77
Xue et al. (2023)78 synthesized a Schiff base ligand, H2L10 via the condensation of 5-hydroxypyridine-2-carbohydrazide with 6-methoxypyridine-2-carbaldehyde in MeOH, and subsequently prepared three trinuclear Ln(III) complexes of the general formula [Ln3(dbm)5(L10)2(CH3OH)]CH2Cl2 (Ln = Nd, Sm, Tb) through reaction with dibenzoylmethane (dbm) salts of lanthanides, Ln(dbm)3·2H2O (Fig. 4a). Single-crystal X-ray diffraction confirmed that the complexes adopt multinuclear architectures, with coordination numbers of 8 and 9 around the Ln(III) centres. The representative ORTEP diagrams of Sm(III) and Tb(III) complexes are shown in Fig. 11c and d. Antibacterial activity was evaluated against G(−) (PSy) using both agar diffusion (zone of inhibition) and broth dilution (MIC) methods, with DMF as solvent.79–82 The results demonstrated that the complexes displayed enhanced antibacterial efficacy (24–27 mm inhibition zone), particularly for Tb(III) (27.5 mm), compared to the free ligand H2L10 (22 mm) and the parent metal salts Ln(dbm)3·2H2O (16–17.5 mm). These findings indicate that complexation with Ln(III) ions significantly improves the antibacterial potency of Schiff bases, with Tb(III) showing the strongest effect, likely due to favourable coordination geometry and higher redox activity facilitating bacterial membrane disruption.
Khalil and Mohamed (2023),83 synthesized a Schiff base (L11) via condensation of benzophenone with naphthalene-1,8-diamine and its mixed-ligand lanthanide complexes through coordination with o-aminophenol and Ln(III) salts, affording compounds of the type [Ln(L11)(o-AP)Cl2]Cl nH2O (Ln = La, Er, Yb). Structural analysis revealed a coordination number of six, with Ln(III) centers bound by two nitrogen donors from L11, one oxygen, one nitrogen atom from o-AP, and from two chlorido ligands (Fig. 1c). The antibacterial activities were evaluated against G(+) (BS) and G(−) (EC) strains using the disc diffusion method, with gentamicin and ampicillin as standard ref. 84 The results demonstrated that all Ln(III) complexes displayed higher antibacterial activity than the free Schiff base (L11), though slightly weaker than o-aminophenol alone. The order of potency was: ampicillin > o-AP > La(III) = Er(III) = Yb(III) > L11 (against BS), and o-AP > gentamicin > La(III) = Er(III) = Yb(III) > L11 (against EC). These findings indicate that while the Ln–Schiff base complexes exhibit moderate enhancement over the parent ligand, their activity remains below that of standard antibiotics, highlighting scope for further ligand modification to optimize antibacterial performance.
Abu-Yamin and co-workers (2022)85 prepared a Schiff base, (L12) by the condensation reaction of 3-(2-furyl)acrolein with 2-amino-6-ethoxybenzothiazole in EtOH solvent and their three lanthanide complexes of composition [Ln(L12)2(NO3)3] (Ln = Gd, Sm, Nd). They were tested their antibacterial activity against both G(+) (SA-ATCC25923, BS-RCMB015) and G(−) (EC-ATCC25922, PV-RCMB004) bacterial strain. The lanthanide ions posses ten coordination number by six oxygen atoms from three bidentate nitrate ions and two oxygen, two nitrogen atoms from two L12 ligands as shown in Fig. 3c. The results antibacterial activity of these complexes was expressed in MIC by using broth micro-dilution method and compared these results with standard drug gentamycin. The results showed that lanthanide complexes expressed higher antibacterial activity than free Schiff base and lower than gentamycin. The results showed that Nd(III) complex possess greater activity than free Schiff base and other two La(III) and Sm(III) complexes and these value comprised with previous antibacterial results. The authors concluded that the enhanced activity of lanthanide complexes than free Schiff base is due to higher lipophilicity of the Ln(III) ions and also by inhibition of growth of microbes by blocking their active mechanisms.86–89
Taha et al. (2022)90 were synthesized two Schiff base ligands, H2L13 and H2L14 through condensation of 5-chlorosalicylaldehyde and 5-bromosalicylaldehyde, respectively with 2-fluorobenzoic hydrazide. Each ligand was further used to prepare a series of nine Ln(III) complexes with the general composition [Ln(H2L13–14)(NO3)2(H2O)] (Ln = La, Pr, Nd, Sm, Eu, Gd, Tb, Dy, Er). Spectroscopic characterization revealed that the Ln(III) ions adopt an eight-coordinate environment, involving two oxygen and one nitrogen donor atoms from the Schiff base ligand, four oxygen atoms from two bidentate nitrate ligands, and one oxygen from a coordinated aqua as shown in Fig. 2e and f. The antibacterial activities of the free ligands H2L13 and H2L14 and their Ln(III) complexes were evaluated against G(+) (EF, SA, MRSA) and G(−) (EC, KP, SE) bacterial strains using the micro-broth dilution method, with amoxicillin as the reference drug.91 The results demonstrated that the Ln(III) complexes generally exhibited stronger antibacterial activity than the free Schiff bases and, notably, showed superior inhibitory effects compared to the reference drug against EF. Furthermore, the H2L14-derived lanthanide complexes displayed higher antibacterial potency than their H2L13 analogues, indicating that the introduction of a bromine substituent on the aromatic ring enhances antimicrobial efficacy.
Mishra et al. (2020)92 reported the synthesis of two new tetradentate Schiff base ligands, L15 and L16, derived from the condensation of 5-nitrobenzothiazole and 2-amino-5-methylthiazole, respectively, with 1,3-benzenedicarboxaldehyde in EtOH. These ligands were further employed to prepare six Ln(III) complexes of the general compositions [Ln(L15)2] and [Ln(L16)2] (Ln = Ce, Nd, Pr) by reacting the ligands with the respective lanthanide nitrate salts in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio. Spectroscopic analyses confirmed that the Ln(III) ions adopt an eight-coordinate environment involving eight nitrogen atoms from two ligands in both series of complexes as shown in Fig. 3d and 1d. The antibacterial activities of the free ligands and their complexes were evaluated using the agar diffusion method against two G(+) bacteria (SA, PBA). The results revealed that the Ln(III) complexes exhibited higher antibacterial potency than the corresponding Schiff bases, particularly against SA (responsible for skin infections and food poisoning) and PBA (a major cause of acne). These findings suggest that coordination with Ln(III) ions enhances the antimicrobial efficiency of the Schiff bases by improving lipophilicity and membrane permeability.93–95
2 molar ratio. Spectroscopic analyses confirmed that the Ln(III) ions adopt an eight-coordinate environment involving eight nitrogen atoms from two ligands in both series of complexes as shown in Fig. 3d and 1d. The antibacterial activities of the free ligands and their complexes were evaluated using the agar diffusion method against two G(+) bacteria (SA, PBA). The results revealed that the Ln(III) complexes exhibited higher antibacterial potency than the corresponding Schiff bases, particularly against SA (responsible for skin infections and food poisoning) and PBA (a major cause of acne). These findings suggest that coordination with Ln(III) ions enhances the antimicrobial efficiency of the Schiff bases by improving lipophilicity and membrane permeability.93–95
Deghadi et al. (2022)96–98 synthesized two Schiff base ligands, L17 and HL18 obtained by the reaction of 1,8-naphthalenediamine and alanine, respectively with 2-acetylferrocene as shown in Fig. 5a. These Schiff bases were further used for the preparation of six new lanthanide(III) complexes of general formula [Ln(L17)(H2O)2Cl2] and [Ln(HL18)(H2O)(1–3)Cl(1–3)] (Ln = La, Er, Yb). All the synthesized Ln(III) complexes adopt octahedral geometry. The authors evaluated the antibacterial activity of these complexes against four G(+) (SA, BC, BS, SF) and four G(−) (EC, PA, NG, ST) bacterial strains in term of zone of inhibition by using disc diffusion method,99 with DMF as negative control and gentamicin as a positive standard. The results showed that Yb(III) complexes of L17 Schiff base exhibited enhanced activity compare to free ligand L17 against NG and SF bacterial strains.
Hussein and Shaalan (2022)100 reported a Schiff base, L19 by the reaction of 4,4-methylenediantipyrine with ethylenediamine in EtOH and their five new lanthanide complexes of the general formula [Ln(L19)2(NO3)3] (Ln = Nd, La, Er, Gd, Dy). Four nitrogen atoms from two Schiff base ligands and six oxygen atoms from three bidentate nitrate ions provide coordination number ten for the lanthanide ions in complexes (Fig. 5b). The authors reported the antibacterial activity of theses complexes against two G(−) bacteria (EC, KP) and two G(+) bacteria (SA, BS) by utilizing disc diffusion method. The activity of complexes reveals that the majority of lanthanide complexes exhibited higher activity as compared to free L19. This research indicates that most of lanthanide complexes are more effective against SA followed by BS than G(−) bacteria. The lanthanide complexes showed more powerful activity against KP than EC. Although all bacteria have an inner cell membrane, G(−) bacteria have a distinct outer membrane, which is responsible for the increased resistance to antibacterial treatments in these pathogen's cell. This outer layer keeps a number of medications and antibiotics from penetrating the cell.101–103
Khalil and his co-workers (2021)104 also uses Schiff base, L11 for the synthesis of another three lanthanide complexes of composition [Ln(L11)(H2O)2Cl2]Cl (Ln = La, Yb, Er). These Ln(III) complexes were adopt octahedral geometry involving two chlorido, two oxygen from two aqua molecules and two nitrogen atoms from L11 as shown in Fig. 5c. The antibacterial activity of ligand and its Ln(III) complexes against G(+) (BS) and G(−) (EC) strains were evaluated by using disc diffusion method with gentamycin and ampicillin as standard drugs.99 The results showed that Ln(III) complexes as compared to free L11 displayed higher activity against both strains according to this order as ampicillin > Er(III) > La(III) > Yb(III) = L11> for BS and gentamicin > Er(III) > La(III) = Yb(III) = L11 for EC. Overtone's concept and Chelation theory might be used to explain why metal complexes have increased activity.105,106
Another series of lanthanide Schiff base complexes have been published by Taha et al. (2021),107 by using an important organic compound (H2L20) which have composition [Ln(L20)(NO3)(H2O)]·nH2O (Ln = La, Pr, Nd, Sm, Eu, Gd, Tb, Dy). The tetradentate ligand (H2L20) obtained by the reaction of phenylenediamine and 5-nitrosalicylaldehyde in EtOH. The coordination number of Ln(III) ions in their complexes is seven achieved by two nitrogen, two oxygen atoms from H2L20, two oxygen atoms from one bidentate nitrate ions and one oxygen atom from one aqua ligand (Fig. 2g). The antibacterial investigation of these complexes was performed against G(−) (SE, KP, EC) and G(+) (SA, MRSA, EF) strains using micro-broth dilution method and results expressed in term of MIC108 with amoxicillin as a reference drug. On the basis of MIC values, the free H2L20 showed less activity against all tested bacterial strains except SA and MRSA for which H2L20 alone showed approximately higher activity than amoxicillin. The most noticeable inhibitory activity was observed in case of La(III) and Pr(III) complexes against KP, SA and MRSA. The Dy(III) complex had higher activity than amoxicillin for EF and MRSA. Finally the authors concluded that with few exceptions, lanthanide(III) complexes were more active on G(+) bacterial strains than on G(−) bacterial strains. The enhanced activity against G(+) bacteria may be due to their increased susceptibility to antibiotics due to the lack of an outer low-permeability barrier.109
Hussein (2021),110 synthesized a tetradentate Schiff base (H2L21) by the reaction of salicylaldehyde and ethylenediamine in EtOH solvent which is further used in the preparation of europium complex of composition [Er(H2L21)(NO3)2]NO3. The reported coordination number of Er(III) ion is eight involving two nitrogen, two oxygen atoms from Schiff base and four oxygen atoms from two bidentate nitrate ions (Fig. 2h). The author evaluated the antibacterial activity of complexes by using EC and SA bacterial strains in term of zone of inhibition. The results showed that antibacterial activity of erbium complex is greater than free Schiff base.
Muriel et al. (2021)111 reported four bidentate Schiff base ligands HL22, HL23, HL24, HL25 derived from 2-(m-aminophenyl)benzimidazole, 2-(p-aminophenyl)benzimidazole, salicylaldehyde, 2,4-dihydroxybenzaldehyde and its eight lanthanide complexes having composition [Ln(L22–25)2Cl(H2O)m]·nH2O (Ln = La, Ce) (Fig. 7). The antibacterial investigation of Schiff bases and their Ln(III) compounds were completed against two G(+) (SA-ATCC25923, LM-ATCC19115) and two G(−) (EC-ATCC25922, PA-ATCC27583) bacterial strains by using micro-broth dilution assays,112,113 with ciprofloxacin as standard drug. These complexes showed more activity against G(+) as compare to against G(−) which is due to outer membrane act like permeability barriers that holds the compound not reach at the targets.114 La(III) complex of L22 ligand with MIC125 µg mL−1 showed highest activity as compare to all other complexes. The current investigation concludes that bacteriostatic effect of free Schiff bases increases upon complexation with Ln(III) ions.
Mahmoud and co-workers (2020)115 synthesized another ligand H2L26, through the condensation reaction of dibenzoylmethane with anthranilic acid and their three Ln(III) complexes of composition [Ln(L26)(H2O)(NO3)]H2O (Ln = Gd, Pr). The six coordination of Ln(III) ions obeyed by two nitrogen, two oxygen atoms from L26, one oxygen atom from water molecule and one oxygen from one nitrate ion (Fig. 5d). The antibacterial investigation was measured against two G(+) (SA, BS) and two G(−) (ST, EC) pathogens employing disc diffusion method.99 The activity of Schiff base H2L26 and its Gd(III) and Pr(III) complexes was evaluated using DMSO as solvent, with results expressed as inhibition zone diameters (mm). The free ligand exhibited inhibition zones of 10–16 mm, while the Gd(III) and Pr(III) complexes showed slightly enhanced activity with inhibition zones of 13–15 mm and 12–14 mm, respectively. The authors concluded that both complexes displayed greater potency against G(−) bacteria, whereas their activity against G(+) bacteria was slightly lower compared to the free ligand H2L26.
Taha et al. (2020)116 published an article dealing with the synthesis ligand, H2L27 through the condensation reaction of picolinoylhydrazide and salicylaldehyde in EtOH and their six lanthanide(III) complexes of the general formula [Ln(H2L27)2(NO3)2]NO3 (Ln = La, Sm, Eu, Gd, Tb, Dy). The coordination number of Ln(III) ions in their complexes is ten involving four oxygen, two nitrogen atoms from L27, four oxygen atoms from two bidentate nitrate ions as shown in Fig. 4b. The antibacterial activities were evaluated against three G(−) (SE, KP, EC) and three G(+) (SA, MRSA, EF) bacterial strains using broth-dilution technique (MIC) with oxytetracycline as standard drug. The results showed that Ln(III) complexes exhibited more activity against G(−) bacteria than G(+) bacteria and authors concluded that the activity of Schiff base enhance upon complexes with Ln(III) ions which is comparable with the activity of standard drug oxytetracycline.
In another study, Paswan et al. (2020)117 prepared two Schiff bases, H2L28 and H2L29 through the condensation reaction o-vanillin and 2-hydroxy-1-naphthaldehyde, respectively with diethylenetriamine (DETA) and their six lanthanide(III) complexes of compositions [Ln(NO3)2(H2L28)]NO3·xCHCl3.nH2O and [Ln(NO3)2(H2L29)]NO3·xCHCl3 (Ln = Nd, Yb, Dy). The reported coordination number of lanthanide ions was as eight occupied by two nitrogen, two oxygen atoms from ligands and four oxygen atoms from two bidentate nitrate ions as depicted in Fig. 2i and 6a. The activity of lanthanide nitrate salts, Schiff bases and lanthanide complexes were investigated against two G(−) (EC, ST) and two G(+) (SA, EF) bacterial strains by agar disc diffusion method; diameter of zone of inhibition and micro-broth dilution method; MIC.118 The antibacterial evaluation of Schiff bases H2L28, H2L29 and their Ln(III) complexes revealed selective activity against both G(−) and G(+) strains. The Yb(III) and Dy(III) complexes of H2L28 exhibited notable inhibition against ST, with inhibition zones of 17 and 18 mm and MIC values of >500 and 500 µg mL−1, respectively. In contrast, Nd(III) and Dy(III) complexes with H2L29 demonstrated enhanced activity against EF and SA, showing inhibition zones of 16 mm and MIC values <500 µg mL−1. These findings suggest that Dy(III) complexes, in particular, display broad antibacterial potential across multiple strains, with activity surpassing the corresponding free Schiff bases.
In another study (2020), a bidentate ligand (HL30) produced by the reaction of sulfanilamide and 2-hydroxy-1-naphthaldehyde and utilized for the synthesis of six lanthanide complexes having composition [Ln(NO3)2(L30)(H2O)2]nH2O (Ln = Nd, Dy, Tb Eu Yb).119 The coordination number of lanthanide ions in their metal complexes is eight by one nitrogen atom, one oxygen atom from HL30, four oxygen atoms from two bidentate nitrate ions and two oxygen atoms from two aqua molecules (Fig. 6b). The antibacterial activity of ligand and its lanthanide complexes was assessed against EC, SA, KP, AB, PA strains using micro-broth dilution method (MICs) with levofloxacin as reference drug. The free ligand showed only moderate activity against SA, KP, and PA (MIC 32 µg mL−1) and no effect on EC or AB (MIC > 64 µg mL−1). Among the complexes, Dy(III) and Tb(III) displayed the most potent effects against SA (MIC 16 µg mL−1), while Dy(III) also exhibited moderate inhibition of EC, AB, and PA (MIC 32 µg mL−1). Tb(III) showed weaker activity against PA and no effect on EC, KP, or AB, whereas Eu(III) displayed minimal activity limited to SA. Although less effective than the reference drug levofloxacin, these results highlight Dy(III) and Tb(III) complexes as the most promising candidates, particularly against SA.
Andiappan et al. (2019)76,120 prepared Schiff base (L31) by the condensation reaction between 3,4-diaminopyridine and 9-anthraldehyde in MeOH and its three new lanthanide complexes of composition [Ln(L31)2] (Ln = Er, Pr, Yb) (Fig. 6c). These complexes were screened for antibacterial activity against G(+) (SA, BS) and G(−) (PA, EC) strains using well diffusion and micro dilution method and results expressed in term of size of inhibition zones and in MICs values with gentamycin as standard drug. The Er(III) complex demonstrated strong inhibition against EC and BS, but only weak activity toward SA and PA. A concentration-dependent enhancement of antibacterial activity was observed, with higher doses leading to larger inhibition zones. The Pr(III) complex showed the most pronounced effects, yielding inhibition zones of 20 mm (BS), 17 mm (SA), 20 mm (EC), and 23 mm (PA) at 100 µg mL−1. Moreover, all the complexes displayed low MIC values, confirming their promising antibacterial potential. Among them, the Pr(III) complex exhibited the strongest activity, correlating with its broader inhibition spectrum and lower MIC values. Overall, all the synthesized lanthanide Schiff base complexes were effective against the tested bacterial pathogens.
Mosquera and collaborators (2018)121 described the synthesis of two Schiff base ligands, HL32 HL33 and its six novel lanthanide complexes having compositions[Ln(L32)2(Cl)]nH2O and [Ln(L33)2(Cl)]·nH2O (Ln = La, Nd, Eu). The stoichiometric ratio of metal to ligand in all the synthesized complexes is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The coordination number of lanthanide ions in their complexes is nine involving two nitrogen atoms, two sulphur atoms, four oxygen atoms from Schiff bases and from one chlorido ligand as shown in Fig. 4c and d. The antibacterial activity of ligands and its lanthanide complexes were evaluated by micro-dilution technique in term of MIC against one G(+) bacteria (SA-ATCC25923) and four G(−) bacteria (EC-ATCC25922, ST-ATCC14028, KP-ATCC2146, KP-ATCC1705).112 The results demonstrated that all lanthanide–Schiff base complexes exhibited comparatively lower antibacterial activity against G(−) strains, while showing enhanced activity against G(+) strains, particularly SA. Notably, the Eu(III) complex displayed the strongest effect with a MIC value of 63 µg mL−1. These findings highlight that coordination with lanthanide ions significantly improves the antibacterial potency of Schiff base ligands, likely due to increased lipophilicity and facilitated penetration of the bacterial cell wall.
2. The coordination number of lanthanide ions in their complexes is nine involving two nitrogen atoms, two sulphur atoms, four oxygen atoms from Schiff bases and from one chlorido ligand as shown in Fig. 4c and d. The antibacterial activity of ligands and its lanthanide complexes were evaluated by micro-dilution technique in term of MIC against one G(+) bacteria (SA-ATCC25923) and four G(−) bacteria (EC-ATCC25922, ST-ATCC14028, KP-ATCC2146, KP-ATCC1705).112 The results demonstrated that all lanthanide–Schiff base complexes exhibited comparatively lower antibacterial activity against G(−) strains, while showing enhanced activity against G(+) strains, particularly SA. Notably, the Eu(III) complex displayed the strongest effect with a MIC value of 63 µg mL−1. These findings highlight that coordination with lanthanide ions significantly improves the antibacterial potency of Schiff base ligands, likely due to increased lipophilicity and facilitated penetration of the bacterial cell wall.
Ajlouni et al. (2018)122,123 synthesized Schiff base (HL34), via condensation of furan-2-carbaldehyde with 2-fluorobenzohydrazide and its eight lanthanide complexes with the general formula [Ln(L34)2(NO3)2]NO3 (Ln = La, Pr, Nd, Sm, Eu, Gd, Dy, Er). The complexes exhibited a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal-to-ligand stoichiometry, with the Ln(III) ions attaining a coordination number of ten through interactions with two nitrogen atoms, four oxygen atoms from HL34, and four oxygen atoms from two bidentate nitrate ligands (Fig. 4e). Antibacterial activity, evaluated by the agar well diffusion method, revealed weak to negligible effects against G(+) (SA, SPy, EF), but comparatively higher activity against G(−) (EC, KP, PM, SE, PA) bacterial strains. PM and SE were the most susceptible, with MIC values ranging from 16–250 µg mL−1, while EC and KP were also inhibited within the same range. Among G(+) strains, SPy showed moderate susceptibility to five complexes (MIC 125–250 µg mL−1). Overall, the activity of the ligand and its complexes was significantly lower than that of the reference drug oxytetracycline. The authors concluded that Ln(III)–hydrazone complexes act as selective inhibitors of G(−) pathogens, likely due to their interaction with structural features of the bacterial cell wall.124a
2 metal-to-ligand stoichiometry, with the Ln(III) ions attaining a coordination number of ten through interactions with two nitrogen atoms, four oxygen atoms from HL34, and four oxygen atoms from two bidentate nitrate ligands (Fig. 4e). Antibacterial activity, evaluated by the agar well diffusion method, revealed weak to negligible effects against G(+) (SA, SPy, EF), but comparatively higher activity against G(−) (EC, KP, PM, SE, PA) bacterial strains. PM and SE were the most susceptible, with MIC values ranging from 16–250 µg mL−1, while EC and KP were also inhibited within the same range. Among G(+) strains, SPy showed moderate susceptibility to five complexes (MIC 125–250 µg mL−1). Overall, the activity of the ligand and its complexes was significantly lower than that of the reference drug oxytetracycline. The authors concluded that Ln(III)–hydrazone complexes act as selective inhibitors of G(−) pathogens, likely due to their interaction with structural features of the bacterial cell wall.124a
We have observed that most of synthesized Schiff bases generally contain both N and O donor atoms, to avoid dechelation they prepared in the absence of water. The hard Lewis acid nature of Ln(III) ions enables this ions that they preferentially coordinate oxygen donors, whereas Schiff bases primarily bind through azomethine nitrogen atoms. Consequently, many Ln–Schiff base complexes exhibit comparatively weaker metal–ligand interactions than oxygen-rich chelators, making them susceptible to dechelation and hydrolytic ligand exchange, particularly in aqueous and biologically relevant environments.124b,c Recent potentiometric studies on Eu(III), Gd(III), and Tb(III) complexes with salicylidene-based Schiff bases demonstrate the formation of hydrolysed species, Ln–SB(OH)x, at higher pH, highlighting partial disruption of the coordination sphere.124d Such instability has direct implications for luminescent applications, as partial ligand dissociation allows water coordination, which quenches lanthanide-centered emission through O–H vibrational relaxation. Dechelation also plays a pivotal role in biological systems. Schiff base complexes of Ln(III) ions may release aqua Ln(III) ions in solution, which readily interact with biomolecules such as amino acids, nucleotides, and phosphate-containing metabolites due to the ions' high kinetic lability.124e,f Amino acids like aspartate, glutamate, cysteine, and histidine, as well as nucleotide phosphates, can displace coordinated water to form Ln(III)-biomolecular adducts, leading to dynamic speciation in physiological conditions. This competitive coordination can modulate metal ion uptake, enzymatic activity, and nucleic acid interactions, while also facilitating metal-mediated oxidative stress or catalytic processes.124g Importantly, dechelation is not an inherent limitation of all Schiff base systems. Modern ligand designs incorporating phenolate oxygen donors, increased denticity, rigid backbones, and mixed N,O-donor sets significantly improve thermodynamic stability and kinetic inertness.124d,h Mixed-ligand complexes, such as those combining Schiff bases with strong β-diketonate co-ligands, demonstrate robust structural integrity in solution and preserve luminescence, underscoring the critical role of ligand design in mitigating dechelation. Overall, the biological activity and solution behavior of Ln(III) ions are intimately connected to their kinetic lability and coordination environment. In both Schiff base complexes and biological media, Ln(III) ions predominantly exist as dynamic aqua or biomolecularly coordinated species. Their interactions with amino acids, nucleotides, and other biomolecules govern their luminescence, pharmacological activity, and potential toxicity. Therefore, consideration of both dechelation and biomolecular adduct formation is essential for evaluating the functional and biological implications of lanthanide complexes.
The majority of reported lanthanide–Schiff base complexes exhibit good solubility in polar aprotic solvents such as DMSO and DMF, limited solubility in alcohols and moderately polar organic solvents, and negligible solubility in non-polar media and water.107,110,122–124a These solvent-dependent solubility trends are consistent with their ionic and coordination-driven structures. Representative examples reported by Taha et al.90 further confirm that such complexes are air-stable, non-hygroscopic, and remain intact in solution. The solution stability is corroborated by well-defined UV-Vis spectral signatures and the consistent enhancement of antibacterial activity relative to the free Schiff bases. The clear differences in spectroscopic profiles and MIC values between ligands and their corresponding complexes strongly support the persistence of intact metal–ligand coordination in solution, which is essential for reliable biological evaluation. The limited solubility of these complexes in common solvent may also indicate the presence of aggregate or nanoparticulate species, which could contribute into the biological activity.
3.1 Characterization techniques for lanthanide complexes
A comprehensive characterization is essential for establishing the structure, purity, and stability of Schiff base lanthanide complexes prior to evaluating their biological properties. A set of complementary tools are required to characterize lanthanide complexes due to their high coordination numbers and flexible geometries.Fourier transform infrared (FT-IR) spectroscopy is typically the first line of evidence for metal coordination. Upon chelation, the azomethine HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching band (usually 1610–1640 cm−1) undergoes a characteristic shift to lower wavenumbers, indicating donation of electron density from the imine nitrogen to Ln(III) ions.54,57,70,74,85,90 Concurrently, disappearance or broadening of phenolic O–H stretching signals in lanthanide complexes compare to ligands confirms deprotonation and O-coordination.57,107 Shifts in C–O, C
N stretching band (usually 1610–1640 cm−1) undergoes a characteristic shift to lower wavenumbers, indicating donation of electron density from the imine nitrogen to Ln(III) ions.54,57,70,74,85,90 Concurrently, disappearance or broadening of phenolic O–H stretching signals in lanthanide complexes compare to ligands confirms deprotonation and O-coordination.57,107 Shifts in C–O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, and sometimes N–N vibrations provide additional diagnostic information and help differentiate mono-, bi-, or tridentate binding modes.107
N, and sometimes N–N vibrations provide additional diagnostic information and help differentiate mono-, bi-, or tridentate binding modes.107
Elemental analysis (CHN) and high-resolution ESI mass spectrometry (HR-MS) provide indispensable confirmation of stoichiometry, purity, and the presence of intact complex ions. HR-MS often reveals ligand-loss fragments or solvated species, allowing insight into solution speciation-especially important for Ln(III) complexes with labile coordination spheres.83
Single-crystal X-ray diffraction remains the authoritative method for elucidating coordination geometry, structural topology, and ligand denticity around Ln(III) centre. Due to the large ionic radii and flexible coordination environments, diffraction often reveals 7–10 coordination numbers, distorted polyhedral geometries, and intramolecular or intermolecular hydrogen-bond networks that influence solution stability and biological function.74 Lanthanide contraction trends, such as decreasing Ln–O/N bond distances across the series, have been extensively documented using crystallographic data.24 Thus, even a small number of representative ORTEP structures significantly strengthen structural arguments in any investigation.
UV-Vis absorption spectroscopy is widely used to monitor ligand centered (π–π*, n–π*) transitions and ligand-to-metal charge transfer (LMCT) bands. Time-dependent UV-Vis measurements also allow qualitative assessment of solution stability under physiological pH ranges. For many Schiff base ligands, dramatic spectral differences upon complexation with metal ions serve as additional evidence for chelation. Electronic transitions in the 4f orbitals of lanthanide ions give rise to characteristic absorption spectra. Only Y3+, La3+ and Lu3+ do not absorb in the UV or visible domain; all the other lanthanides have characteristic bands in this region, but the absorptivities have low values. The literature indicates that Schiff bases typically display two major absorption peaks: one below 300 nm attributed to π–π* transitions and another between 300–450 nm corresponding to n–π* transitions. Upon complexation with Ln(III) ions, these bands shift, confirming coordination between the ligand and the metal center. Additionally, the Ln(III) complexes exhibit new absorption features in the 410–475 nm range, which are absent in the free ligands. These newly appeared bands are assigned to intra-ligand charge–transfer transitions, consistent with quantum-chemical calculations.54,57,66,70,74,83
Lanthanide ions such as Sm(III), Eu(III), Tb(III), and Dy(III) exhibit strong characteristic luminescence because their accepting 4f excited states lie slightly below the excited energy levels of coordinated ligands, enabling efficient ligand-to-metal energy transfer (LMET). Detectable luminescence is typically achieved upon excitation by ultraviolet light, X-rays, or other high-energy stimuli. Complexation with suitable chromophoric ligands markedly enhances lanthanide emission through the well-known antenna effect, in which the ligand absorbs energy and transfers it non-radiatively to the metal center. Luminescence spectroscopy is therefore a powerful tool for probing Eu(III), Tb(III), Dy(III) and Sm(III) complexes. Schiff base ligands commonly act as effective sensitizers in these systems, and photophysical parameters such as emission intensity, excitation profiles, and excited-state lifetimes provide critical information on energy-transfer efficiency, coordination environment, and dominant quenching pathways. Numerous studies have demonstrated that Sm(III) and Tb(III) Schiff base complexes display intense, metal-centered emissions, confirming efficient ligand-sensitized energy transfer, whereas in some Eu(III) and Gd(III) systems weak or ligand-based emission indicates inefficient antenna behaviour.40b,57,124a,b
Magnetic susceptibility measurements and electron paramagnetic resonance (EPR) are valuable for paramagnetic lanthanides such as Gd(III), Dy(III), and Tb(III). These techniques provide insight into electronic environments, spin–orbit coupling, and coordination anisotropy. Although less frequently reported than IR or UV-Vis spectroscopy, magnetic data enhance understanding of structure–property relationships, particularly in mixed-metal or high-spin systems. Alquasaimeh et al.74 measured magnetic moment of Nd, Tb, Dy complexes with HL8 Schiff base by measuring magnetic susceptibility by the Curie–Weis (C–W) law:
| χT = χdia + C/T − TCW |
Together, these characterization techniques ensure that Schiff base lanthanide complexes are structurally well-defined, chemically pure, and stable under relevant conditions.
4. Mechanistic insights and structure–activity relationships (SAR)
4.1 Structure–activity relationships (SAR)
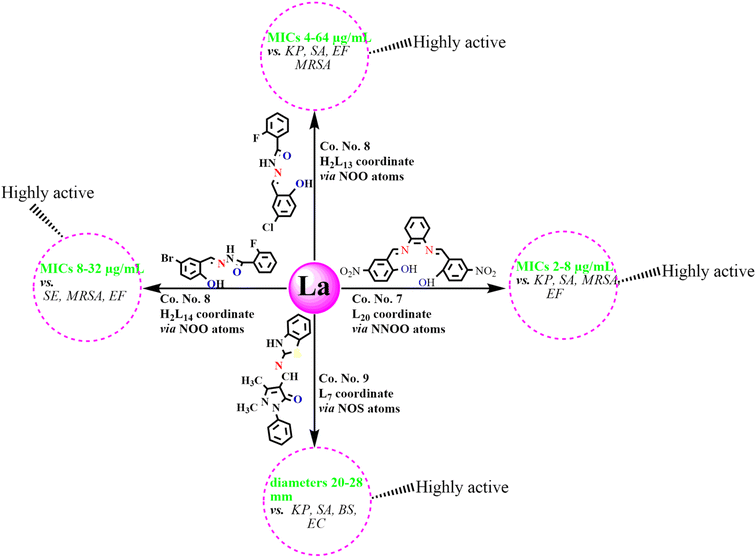
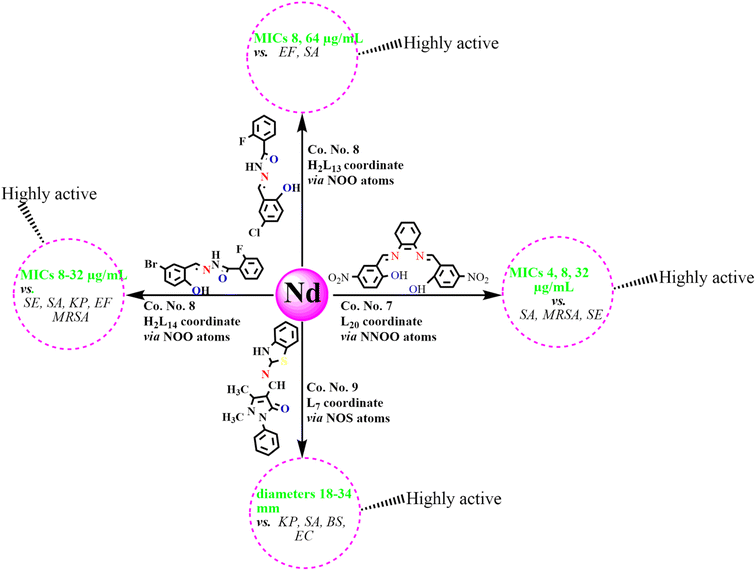
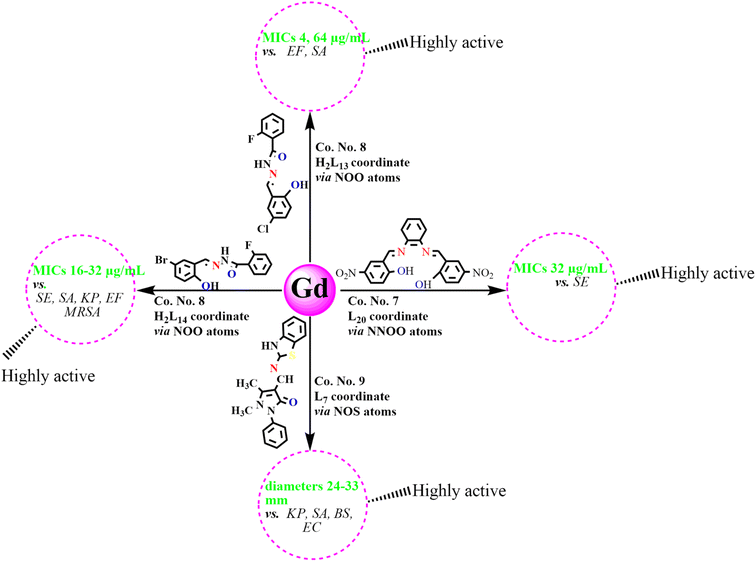
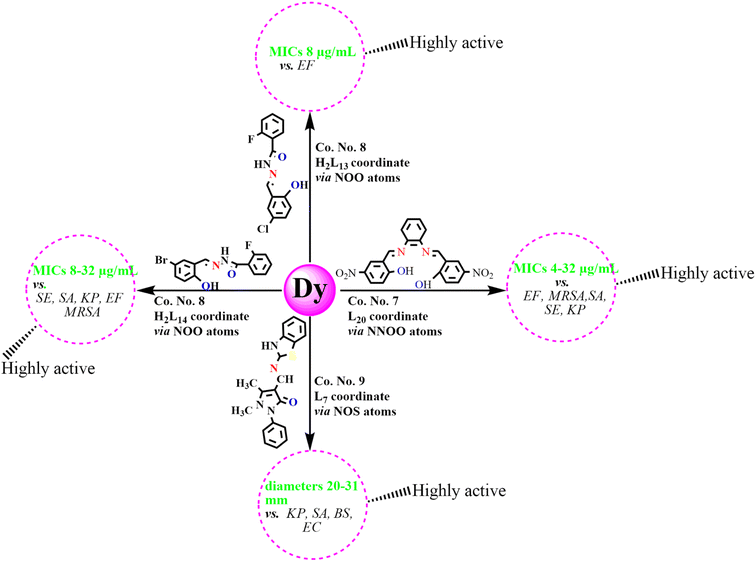
An analysis of antibacterial studies published between 2018 and 2025 indicates that Schiff bases and their Ln(III) complexes may be broadly classified into three categories of activity: highly active, moderately active and weakly active agents. Ligands such as L7, H2L13–14, and H2L20, along with their Ln(III) complexes displayed the most promising activity with MIC values of 2–64 µg mL−1 and inhibition zones ≥20 mm, particularly against SA, EF, MRSA, KP, SE, and EC. By contrast, ligands including L4, L12, L15–16, HL30, HL32–34 and their complexes were only moderately active, with MICs of 64–195 µg mL−1 and inhibition zones of 10–20 mm, mainly effective against SA, BS, and EC. Finally, Schiff bases such as H2L6, L22–25, H2L27 and their corresponding complexes exhibited weak or negligible antibacterial activity (MIC > 195 µg mL−1; inhibition zones <10 mm), showing limited effectiveness against most bacterial strains (Fig. 12). Among the studied compounds, La(III), Nd(III), Gd(III) and Dy(III) complexes with highly active tridentate and tetradentate Schiff bases (L7, H2L13–14, L20), incorporating electron-withdrawing substituents such as Cl, Br, and –NO2, were found to possess coordination numbers of 7–9 along with their notable antibacterial activities, as reflected by MIC values and inhibition zone diameters, are summarized in Schemes 2–5. These observations emphasize that ligand substituents, donor atom sets (N, O, S), and coordination numbers (6–9) play key roles in modulating activity. The halogen and heteroatom-substituted Schiff bases generally enhancing antibacterial potency. The biological activity consistently follows the trend: free Schiff base < Ln(III)–Schiff base complex < conventional antibiotics-although, top-performing Ln(III) complexes occasionally approach standard drug efficacy. These structure–activity insights underscore the importance of both the identity of the lanthanide ion and the coordination environment in determining antibacterial potency. | ||
| Fig. 8 Agar disc diffusion assay results of Eu(III), Tb(III) and Gd(III) complexes of H2L1 ligand against (a) EC, (b) KP, (c) PA, (d) SA, (e) EF, bacterial strains. | ||
 | ||
| Fig. 10 The diameters of zone of inhibition of L9 ligand and its Ln(III) (Er, Pr, Yb) complexes against PA and SA strains at various concentrations 20, 40, 60, 80 µg mL−1. | ||
4.2 Mechanistic insights
An antimicrobial agent is a compound that can kill bacteria by binding to essential metabolic components and preventing the formation of functioning biomolecules, which interferes with normal cellular functions.125 The antibacterial activity of drugs is primarily achieved through two strategies, which entail interfering chemically with the synthesis or functioning of crucial bacterial components and/or dodging known antibacterial resistance mechanism. The bacterial cell contains many targets for antibacterial drugs that include (a) bacterial protein biosynthesis (b) bacterial cell-wall biosynthesis (c) bacterial cell membrane destruction (d) bacterial DNA replication and repair, and (e) inhibition of a metabolic pathway.126 It is very crucial to find a solution for antibacterial resistance mechanisms because bacteria can exhibit resistance through a variety of mechanisms, which primarily include activating the efflux pumps, destroying antibacterial agents with the help of destruction enzymes, modifying antibiotics with the help of modifying enzymes, and altering target structures in the bacterium that have lower affinities for an antibiotic.127 Some types of antibiotics need metal ions to work properly. These metal ions can either be important to the structure of the antibiotic, and therefore their removal renders the antibiotic structure inactive, or bind to the antibiotic molecules and cause important chemical and biochemical reactions without changing the antibiotic structure.128 The lanthanide complexes as antibacterial drugs have a benefit that Ln3+ and Ca2+ have similar ionic radii, the lanthanide ions have the potential to block calcium channels because of their greater charge and strong affinity for the Ca2+ sites on biological molecules. Thus, even though the lanthanide ions cannot get through cell membranes on their own, they can still affect calcium channel's external face by blocking them. The lanthanides have the potential to substitute for the calcium in proteins, which can either inhibit or significantly enhance the activity of calcium-dependent enzymes.25 The higher antibacterial activity of metal complexes can be explained on the basis of two theories that comprise (i) Tweedy's chelation theory129 (ii) Overtone's concept.130 The presence of the imine (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) group in the Schiff base ligands, which provides them their biological activity, facilitated the rationalisation the antibacterial activity and offered a mechanism of transamination and resamination reactions in biological systems work.121,124a,131,132 The ligands have demonstrated to form hydrogen bonds with bacterial strain through azomethine moiety, thereby resulting in enhanced cell inhibition. Furthermore, it has been hypothesised that Schiff bases with nitrogen, oxygen and sulphur donor atoms may also be capable of inhibiting bacterial cell growth.133–135 According to extant literature in the preceding discussion; most research has demonstrated that lanthanide complexes with Schiff bases exhibit higher activity than free Schiff bases and metal salts. This enhancement in activity can be attributed to the increased chelation exhibited by lanthanide complexes, a phenomenon that may be explained by Tweedy's chelation theory. This theory facilitates the delocalization of π-electrons across the whole chelate ring; resulting the positive charge on the metal ions is reduced and an enhancement of the lipophilicity of the metal chelate, favouring its permeation across the lipid barrier inside bacterial membranes.136,137 According to Overtone's concept of cell permeability, the lipid membrane that envelops the cell selectively permits the passage of lipid-soluble compounds; resulting liposolubility has been identified as a crucial variable that regulates antimicrobial action. It has been established that the overlap of the ligand orbital and the partial sharing of the positive charge of the metal ion with donor groups during chelation result in a lowering of the polarity of the metal ion. This, in turn, favours the lipophilicity of metal complexes. As the lipophilicity of the complexes increases, there is concomitant increase in penetration of the complexes through the lipid membranes of the cell. This is due to the metal binding sites in the bacterial enzymes being blocked, which in turn causes disturbance to the normal functioning of the cell. While moderate lipophilicity facilitates membrane penetration and enhances antimicrobial potency, excessive hydrophobicity in lanthanide–Schiff base complexes can promote self-aggregation and nanoparticle formation, which significantly alters their biological behavior. Such nanostructures may interact with bacterial membranes via surface adhesion, membrane destabilization, and mechanical perturbation. Moreover, their cellular uptake pathways can differ substantially from those of discrete molecular chelates, thereby influencing the observed antibacterial activity.
N) group in the Schiff base ligands, which provides them their biological activity, facilitated the rationalisation the antibacterial activity and offered a mechanism of transamination and resamination reactions in biological systems work.121,124a,131,132 The ligands have demonstrated to form hydrogen bonds with bacterial strain through azomethine moiety, thereby resulting in enhanced cell inhibition. Furthermore, it has been hypothesised that Schiff bases with nitrogen, oxygen and sulphur donor atoms may also be capable of inhibiting bacterial cell growth.133–135 According to extant literature in the preceding discussion; most research has demonstrated that lanthanide complexes with Schiff bases exhibit higher activity than free Schiff bases and metal salts. This enhancement in activity can be attributed to the increased chelation exhibited by lanthanide complexes, a phenomenon that may be explained by Tweedy's chelation theory. This theory facilitates the delocalization of π-electrons across the whole chelate ring; resulting the positive charge on the metal ions is reduced and an enhancement of the lipophilicity of the metal chelate, favouring its permeation across the lipid barrier inside bacterial membranes.136,137 According to Overtone's concept of cell permeability, the lipid membrane that envelops the cell selectively permits the passage of lipid-soluble compounds; resulting liposolubility has been identified as a crucial variable that regulates antimicrobial action. It has been established that the overlap of the ligand orbital and the partial sharing of the positive charge of the metal ion with donor groups during chelation result in a lowering of the polarity of the metal ion. This, in turn, favours the lipophilicity of metal complexes. As the lipophilicity of the complexes increases, there is concomitant increase in penetration of the complexes through the lipid membranes of the cell. This is due to the metal binding sites in the bacterial enzymes being blocked, which in turn causes disturbance to the normal functioning of the cell. While moderate lipophilicity facilitates membrane penetration and enhances antimicrobial potency, excessive hydrophobicity in lanthanide–Schiff base complexes can promote self-aggregation and nanoparticle formation, which significantly alters their biological behavior. Such nanostructures may interact with bacterial membranes via surface adhesion, membrane destabilization, and mechanical perturbation. Moreover, their cellular uptake pathways can differ substantially from those of discrete molecular chelates, thereby influencing the observed antibacterial activity.
5. Comparative analysis with previous reviews
Most of the earlier reviews on Schiff base and their Ln(III) complexes, largely published before 2020, mainly focused on synthetic strategies, structural characterization and general coordination chemistry with antibacterial application often mentioned only as a secondary aspect. For instance, Hameed et al. (2017)138 presented a patent-based general overview of Schiff bases and their metal complexes, highlighting numerous therapeutic applications (2010–2015), including antibacterial effects. However, systematic quantitative comparisons, mechanistic insights, and detailed structure–activity relationships of antibacterial investigation were lacking. The review “Metal complexes driven from Schiff bases and semicarbazones for biomedical and allied applications: a review” by More et al. (2019)139 examined a broad set of biological activities of Schiff base and its metal complexes, including antibacterial, but mostly focusing on transition metals, with fewer details for lanthanide systems. X. Leu and J. Hamon (2018),140 provided a broad overview of synthesis and structure of multidentate Schiff bases and its coordination with transition and inner transition metal ions to form metal complexes with catalytic, magnetic and few biological applications, while K. Prajapati (2018)141 highlighted several ligands and its Ln(III) complexes for antibacterial activities with general importance of these complexes in biological fields. In addition, Kaczmarek et al. (2018)33 highlighted synthetic method, several structure of Schiff base and their Ln(III) complexes, role of theses complexes in different biological fields such as cancer diagnosis, therapy and biological activity. I. Cota et al. (2019)2 published a comprehensive review that extensively compiled studies on Ln(III) Schiff base complexes as antibacterial and antifungal agents, covering the period from 1995 to 2019. This review emphasizes interaction different Schiff bases and lanthanide complexes against both G(+) and G(−) bacterial strains, their antibacterial results and mechanistic interpretations. Another review, ‘Advanced and Biomedical Applications of Schiff-Base Complexes and their metal complexes: A Review’ (2022),142 summarised recent work since about 2015, with some attention to antibacterial activity, but again the structure–activity relationships, coordination numbers, and systematic comparisons among Ln(III) ions were not thoroughly analysed. The work by Rakshit et al. (2023)143 mainly provides antimicrobial behaviours, some techniques of antimicrobial measurement and structure of most of the transition metal complexes derived from only N, O donor ligands but it did not provide proper information about structure–activity relationships, mechanism for higher activity, classification of ligands and their metal complexes into different activity categories.Unlike these prior works that emphasized synthesis, catalytic, photophysical or broad biological applications, our review mainly focused on data driven, mechanistic perspectives and structure–activity relationship (SAR) analysis of Schiff bases and their lanthanide complexes based on the 2018–2025 literature. Specifically, earlier reviews often lacked bulk of investigation on: (a) detailed MIC and inhibition zone data for different bacteria (b) comparisons of donor atom types, substituent effects (such as halogen or nitro groups) and coordination geometries (c) clear classification of compounds by potency (e.g. highly active vs. moderate vs. weak). By compiling recent studies, our review identifies clear trends: Schiff base ligands alone usually show modest or weak antibacterial activity, which is significantly enhanced upon complexation with lanthanides. We find that certain Ln(III) ions (La, Nd, Eu, Gd Dy) with ligand features (NOS donor sets, halogen and –NO2 substituents) correlate with higher potency. Moreover, our classification into highly, moderately, and weakly active agents as given in Fig. 12, provides a framework that was generally not present in previous reviews. We also tried to integrate mechanistic insights like enhanced lipophilicity, improved membrane permeability and enzyme inhibition. These mechanistic angles are systemically linked to structural parameters. Finally, our coverage includes direct comparisons of Ln–Schiff base complexes to standard antibiotics (in many 2018–2025 studies) in terms of MIC and inhibition zones, which few prior reviews addressed in detail.
In conclusion, while previous reviews laid the foundational understanding of Schiff bases and their metal complexes, our review advances the field by offering a data-driven, mechanistically informed and structure–activity focused perspective of the most recent antibacterial findings (2018–2025), which can guide rational design of new compounds.
6. Techniques for evaluating antibacterial activity of complexes
Some of important techniques used for evaluating the antibacterial activity of chemical compounds.6.1 Diffusion methods
6.2 Dilution methods
This approach is widely used for determining Minimum Inhibitory Concentration (MIC) values, offering flexibility to calculate the concentration of the tested antimicrobial agent in either the agar (agar dilution) or broth medium (macro dilution or micro dilution). The quantitative measurement of antimicrobial activity against bacterial and fungal strains is achieved through either the broth or agar dilution method.1577. Challenges and future directions
7.1 Current challenges
Despite considerable advances in the synthesis and evaluation of Schiff base–lanthanide complexes during 2018–2025, their transition from laboratory curiosity to practical antibacterial agents remains constrained by multiple challenges. While in vitro results are promising, significant hurdles regarding solubility, toxicity, stability and translational applicability continue to limit their scope. A consistent limitation across reported studies is poor solubility and bioavailability. Most the Schiff bases, particularly L7, H2L13, and H2L14, feature extended aromatic scaffolds that confer lipophilicity but reduce solubility in aqueous solvent, limiting their biological applicability.72,90 Even though these ligands and their complexes demonstrated strong activity against SA and KP, their hydrophobicity complicates formulation for in vivo delivery. The toxicity and selectivity shown by these complexes are still unresolved. Taha et al. (2024) reported that H2L6 and its Ln(III) complexes exhibited moderate activity against EC and KP, but no effect against G(+) strains, suggesting that broad-spectrum selectivity is still lacking.70 Stability under physiological conditions also poses a problem. Schiff base ligands are prone to hydrolysis and lanthanide complexes often undergo ligand exchange reactions. For instance, in the work of Mishra et al. (2022), Ln(III) complexes of L15–L16 showed antibacterial effects against skin pathogens, yet their long term stability in biological systems was not reported.92 Perhaps the most critical limitation is the absence of translational data. Most studies rely solely on agar diffusion or MIC assays. No reports have advanced into in vivo infection models, pharmacokinetics or preclinical trials. This gap between bench chemistry and biomedical validation remains the largest bottleneck for clinical adoption.7.2 Future perspectives
Research into Schiff bases and their lanthanide (Ln(III)) complexes has increasingly shown their promising antibacterial properties. However, despite these advancements, the transition from laboratory research to clinically applicable treatments remains limited. This highlights the need for a forward looking approach that addresses both the challenges and opportunities in this area. The future of antibacterial research involving Schiff base–lanthanide complexes will likely be driven by progress in sustainable synthesis methods,109 targeted structural design, mechanistic understanding and pharmacological validation.162,163 While Schiff bases and its Ln(III) complexes consistently showed activity mainly against G(+) bacteria, G(−) bacteria tend to be more resistant. The future work should be the selective targeting of G(−) bacteria by structural modifications of Schiff bases, such as adding amphiphilic groups or cationic moieties, to improve bacterial membrane penetration.109,163 The existing literature suggest that lanthanide–Schiff base complexes may exert antibacterial effects through multiple mechanisms, including DNA binding, membrane disruption and enzyme inhibition.85,96 The rational design of theses complexes with mechanistic guidance is very important for future works. The integration of biochemical assays, molecular docking and omics-based profiling can provide deeper insights into precise molecular targets of these complexes. The structural studies like X-ray crystallographic of ligand–protein interaction could further elucidate how specific coordination environment drive antibacterial activity.164 The pharmacokinetic and toxicological profiles of Schiff base and its lanthanide complexes remain insufficiently studied. For clinical translation, future research must assess solubility, stability, protein binding and bio-distribution.162,163 In vivo validation is also essential as many studies are limited to in vitro assays such as diameters of zone of inhibition, MIC and MBC tests, which do not fully capture biological complexity.37,165 Another promising direction is the development of mixed-metal and hetero-bimetallic complexes. The combination of lanthanides with transition metals or other bioactive ions may provide synergistic effects by combining redox activity with unique coordination chemistry.166 The pronounced ROS activity exhibited by Eu(III) and Dy(III) complexes indicates that the integration of redox-active centres with lanthanides could further promote ROS generation and thereby enhance antibacterial efficacy.167,168 The computational approaches including density functional theory (DFT) and molecular docking can greatly accelerate drug discovery. However, these methods are rarely applied to correlate observed MIC values with mechanistic insights. For instance, Hussein et al. (2024) proposed that Ln(III) complexes with the L7 Schiff base disrupt bacterial membranes, but this was inferred rather than confirmed through computational studies.72 In silico predictions of stability, permeability and enzyme inhibition could shift research from trial-and-error synthesis toward rational design.169 Additionally, scaling green and sustainable synthesis methods is important. Microwave-assisted and solvent-minimized approaches have been shown to produce Schiff bases rapidly with improved yields and lower environmental impact.170 Adoption of microwave irradiation, solvent-free grinding, ultrasonic methods, or biomass-derived starting materials could improve sustainability in ligand preparation and complexation.In conclusion, future research on Schiff bases and their lanthanide complexes is expected to focus primarily on the key aspects illustrated in Scheme 6.
8. Conclusion
Over the last seven years (2018–2025), the design and investigation of Schiff base ligands and their Ln(III) complexes as antibacterial drugs have progressed considerably. A consistent pattern appeared across this study, while free Schiff bases generally display only moderate antibacterial activity; their coordination with Ln(III) ions enhances lipophilicity, improves membrane penetration, and introduces additional mechanisms such as enzyme inhibition and reactive oxygen species (ROS) generation. As a result, many Ln(III)–Schiff base complexes exhibit greater antibacterial activity than the corresponding free ligands, although their activity still frequently lower than conventional standard antibiotics. The parent Schiff base's activity is increased after complexation with Ln(III) ions because of increased lipophilicity, which enables lanthanide(III) complexes to pass through the cell's lipid membranes and disrupt its normal function, ultimately resulting in cell death. Comparative studies reveals that Schiff bases like L7, H2L14, L20, HL30, HL32 with Ln(III) ions particularly La(III), Nd(III), Gd(III), Tb(III) and Dy(III) are more consistently associated with strong antibacterial activity. The most active Ln(III) complexes tend to feature coordination number range 7–9 with donor sets involving N, O and S atoms that create stable and biologically reach architectures. Schiff bases bearing electron donating substituents, extended aromatic system and hydrophilic groups have further shown to influence activity profiles. Despite these promising advances, the practical translation of Schiff base–lanthanide complexes remains constrained by key limitations, including poor aqueous solubility, limited stability under physiological conditions and the absence of comprehensive translational data beyond in vitro assay. Furthermore, activity against Gram-negative bacteria remains a bottleneck, likely due to permeability barriers in the outer membrane. Future developments in the subject would include the creation of mixed-metal or multinuclear systems, the use of green synthetic techniques to improve scalability and biocompatibility, and logical ligand design informed by computational modelling. Above all, converting these complexes into effective antibacterial medicines will require combining in vitro tests with mechanistic research and in vivo validation. Schiff base–lanthanide complexes, in summary, are a dynamic and adaptable class of substances having definite antibacterial potential. They provide new approaches to treating multidrug-resistant infections, even though they are not yet ready to take the place of traditional antibiotics. This is especially true if future studies continue to increase their selectivity, refine their structures, and close the crucial gap between bench and bedside.Author contributions
S. P.: investigation, conceptualization, writing original draft, methodology, formal analysis, data collection, review & editing. N. J.: validations, methodology, review & editing. A. N.: visualization, methodology, and reviewing. Urvashi: formal analysis, review & editing. M. K.: visualization, methodology, and reviewing. D. M.: formal analysis, visualization, review and editing. N. Y.: supervision, review & editing modification.Conflicts of interest
The authors declare no competing interests.Abbreviations
| G(+) | Gram positive |
| G(−) | Gram negative |
| SA | Staphylococcus aureus |
| EF | Enterococcus faecalis |
| EC | Escherichia coli |
| PA | Pseudomonas aeruginosa |
| KP | Klebsiella pneumonia |
| BS | Bacillus subtilis |
| SAl | Staphylococcus albus |
| ML | Micrococcus luteus |
| MRSA | Methicillin resistant staphylococcus aureus |
| PV | Proteus vulgaris |
| ST | Salmonella typhi |
| SE | Salmonella enteritidis |
| PBA | Propionibacterium acnes |
| BC | Bacillus cereus |
| SF | Shigella flexneri |
| NG | Neisseria gonorrhoeae |
| AB | Acinetobacter baumannii |
| PM | Proteus mirabilis |
| SPy | Streptococcus pyogenes |
| BM | Bacillus megaterium |
| SAg | Streptococcus agalactiae |
| SM | Serratia Marcescens |
| SD | Shigella dysenteriae |
| KPA | Klebsiella pseudomonas aeruginosa |
| SF | Streptococcus faecalis |
| LM | Listeria monocytogenes |
| SP | Streptococcus pneumonia |
| PC | Pseudomonas cepacicola |
| PSy | Pseudomonas syringae |
| Ln | Lanthanide |
| DMF | Dimethylformamide |
| DMSO | Dimethyl sulfoxide |
| dbm | dibenzoylmethane |
| SAR | Structure–activity relationships |
Data availability
The data presented in this review are derived from previously published studies and have been extracted and summarized for inclusion in this manuscript. Relevant information is organized within the tables and figures and no new data were generated or analysed as part of this review. The original datasets can be accessed through the cited sources in the reference list, and readers are encouraged to consult those original publications for further details.Supplementary information (SI): Table S1, summarizing reported Schiff bases and their Ln(III) complexes, the bacterial strains used for antibacterial evaluation, and relevant references. See DOI: https://doi.org/10.1039/d5ra08181e.
References
- (a) World Health Organization, Antimicrobial Resistance, 2021, available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance, accessed March 21, 2023; (b) K. W. K. Tang, B. C. Millar and J. E. Moore, Br. J. Biomed. Sci., 2023, 80, 11387 Search PubMed; (c) C. Llor and L. Bjerrum, Ther. Adv. Drug Saf., 2014, 5, 229–241 CrossRef; (d) I. H. Ifijen, P. Agyemang, E. Faderin, L. O. Odo, E. I. Okeke, D. I. Onugha, S. E. Obuba, O. Oluwafunke and C. Ogochukwu, Next Mater., 2025, 8, 100719 CrossRef; (e) K. L. Y. Christensen, R. C. Holman, C. A. Steiner, J. J. Sejvar, B. J. Stoll and L. B. Schonberger, Clin. Infect. Dis., 2009, 49, 1025 CrossRef PubMed.
- I. Cota, V. Marturano and B. Tylkowski, Coord. Chem. Rev., 2019, 396, 49 CrossRef CAS.
- T. Dikid, S. K. Jain, A. Sharma, A. Kumar and J. P. Narain, Indian J. Med. Res., 2013, 138, 19 CAS.
- R. K. Obi, N. M. Orji, F. C. Nwanebu, C. C. Okangba and U. U. Ndubuisi, Asian J. Exp. Biol. Sci., 2010, 1, 271 Search PubMed.
- S. Mohammed, J. Vet. Med. Anim. Sci., 2023, 6, 1132 Search PubMed.
- L. H. Taylor, S. M. Latham and M. E. Woolhouse, Philos. Trans. R. Soc. London, Ser. B, 2001, 356, 983 CrossRef CAS.
- National Institute of Allergy and Infectious Diseases, Emerging and Re-emerging Infectious Diseases, 2008, available at: http://www.naid.nih.gov/factsheet/infectiousdiseases.htm.
- M. E. Woolhouse and S. G. Sequeria, Emerging Infect. Dis., 2005, 11, 1842 CrossRef PubMed.
- B. A. Brown-Elliott, K. A. Nash and R. J. Wallace Jr, Clin. Microbiol. Rev., 2012, 25, 545 CrossRef CAS.
- T. M. Uddin, A. J. Chakraborty, A. Khusro, B. R. M. Zidan, S. Mitra, T. B. Emran, K. Dhama, M. K. H. Ripon, M. Gajdacs, M. U. K. Sahibzada and M. J. Hossain, J. Infect. Public Health, 2021, 14, 1750 CrossRef PubMed.
- K. E. Jones, N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman and P. Daszak, Nature, 2008, 451, 990 CrossRef CAS.
- S. K. Fathalla, H. A. El-Ghamry and M. Gaber, Inorg. Chem. Commun., 2021, 129, 108616 CrossRef CAS.
- H. R. van Doorn, Medicine, 2017, 45, 798 CrossRef.
- S. Ahmed, M. Z. Ahmed, S. Rafique, S. E. Almasoudi, M. Shah, N. A. Che Jalil and S. C. Ojha, BioMed Res. Int., 2023, 2023, 5250040 CrossRef PubMed.
- N. Mishra, R. Yadav, K. Kumar, H. Pandey and R. Pandey, J. Phys.: Conf. Ser., 2020, 1504, 012002 CrossRef CAS.
- A. F. Wady, M. B. Hussein and M. M. Mohammed, Sch. Int. J. Chem. Mater. Sci., 2021, 4, 46 Search PubMed.
- I. B. Amali, M. P. Kesavan, V. Vijayakumar, N. I. Gandhi, J. Rajesh and G. Rajagopal, J. Mol. Struct., 2019, 1183, 342 CrossRef.
- E. Ritter, P. Przybylski, B. Brzezinski and F. Bartl, Curr. Org. Chem., 2009, 13, 241 CrossRef CAS.
- N. H. Yarkandi, H. A. El-Ghamry and M. Gaber, Mater. Sci. Eng., C, 2017, 75, 1059 CrossRef CAS.
- R. El-Sharkawy and H. A. El-Ghamry, J. Mol. Liq., 2019, 282, 515 CrossRef CAS.
- N. Channa, R. H. Khan, A. M. Moina, A. Bano, I. Saghir, C. Umedan, L. R. Nasira and Q. Samra, J. Chem. Lett., 2024, 4, 232 Search PubMed.
- C. Alexander, Z. Guo, P. B. Glover, S. Faulkner and Z. Pikramenou, Chem. Rev., 2025, 125, 2269–2370 CrossRef CAS PubMed.
- A. L. Gassner, C. Duhot, J.-C. G. Bünzli and A. S. Chauvin, Inorg. Chem., 2008, 47, 7802 Search PubMed.
- S. V. Eliseeva and J.-C. G. Bünzli, Chem. Soc. Rev., 2010, 39, 189 Search PubMed.
- P. Hermann, J. Kotek, V. Kubicek and I. Lukes, Dalton Trans., 2008, 23, 3027 RSC.
- E. Kusrini, F. Hashim, M. I. Saleh, R. Adnan, A. Usman, I. N. Zakaria, W. W. Prihandini, N. Putra and E. A. Prasetyanto, J. Mater. Sci., 2020, 55, 9795 CrossRef CAS.
- R. Kumar, K. Seema, D. K. Singh, P. Jain, N. Manav, B. Gautam and S. N. Kumar, J. Coord. Chem., 2023, 76, 1065 CrossRef CAS.
- S. P. Fricker and S. Fricker, Chem. Soc. Rev., 2006, 35, 524 RSC.
- K. L. Peterson, J. V. Dang, E. A. Weitz, C. Lewandowski and V. C. Pierre, Inorg. Chem., 2014, 53, 6013 Search PubMed.
- L. Lekha, K. K. Raja, G. Rajagopal and D. Easwaramoorthy, J. Organomet. Chem., 2014, 753, 72 CrossRef CAS.
- L. Zhu, B. Zhu, J. Luo and B. Liu, ACS Mater. Lett., 2021, 3, 77 CrossRef CAS.
- T. Zhang, X. Zhu, W. K. Wong, H. L. Tam and W. Y. Wong, Chem.–Eur. J., 2013, 19, 739 CrossRef CAS.
- M. T. Kaczmarek, M. Zabiszak, M. Nowak and R. Jastrząb, Coord. Chem. Rev., 2018, 370, 42 CrossRef CAS.
- C. S. Cutler, C. J. Smith, G. J. Ehrhardt, T. T. Tyler, S. S. Jurisson and E. Deutsch, Cancer Biother. Radiopharm., 2000, 15, 531 Search PubMed.
- A. H. Pathan, G. N. Naik, R. P. Bakale, S. S. Machakanur and K. B. Gudasi, Appl. Organomet. Chem., 2012, 26, 148 CrossRef CAS.
- V. Amani and F. Norouzi, Chem. Pap., 2024, 78, 1699 CrossRef CAS.
- J. Ceramella, D. Iacopetta, A. Catalano, F. Cirillo, R. Lappano and M. S. Sinicropi, Antibiotics, 2022, 11, 191 CrossRef CAS PubMed.
- H. Schiff, Ann. Chem. Pharm., 1864, 131, 118–119 CrossRef.
- P. A. Vigato and S. Tamburini, Coord. Chem. Rev., 2004, 248, 1717–2128 CrossRef CAS.
- (a) S. Cotton, in Lanthanide and Actinide Chemistry, John Wiley & Sons, Hoboken, NJ, 2006, pp. 35–60 CrossRef; (b) K. Binnemans, Chem. Rev., 2009, 109, 4283–4374 Search PubMed; (c) R. E. Cramer, J. M. Rimsza and T. J. Boyle, Inorg. Chem., 2022, 61, 6120–6127 CrossRef CAS; (d) S. Li, S. Jansone-Popova and D.-E. Jiang, Sci. Rep., 2024, 14, 11301 CrossRef CAS.
- Z. H. Chohan, Appl. Organomet. Chem., 2006, 20, 112–116 CrossRef CAS.
- J. E. Muraleedharan and K. P. Viswanathan, Orient. J. Chem., 2025, 41(1), 254 CAS.
- S. Nagar, S. Raizada and N. Tripathee, Results Chem., 2023, 6, 101153 CrossRef CAS.
- M. T. Baig, M. T. Sayed, R. Aledamat, S. Hassan, A. AlReyashi, N. Sidiq and M. F. Mady, BMC Chem., 2025, 19, 201 CrossRef CAS PubMed.
- M. Draye, G. Chatel and R. Duwald, Pharmaceuticals, 2020, 13, 23 CrossRef CAS PubMed.
- A. M. Hassan, A. O. Said, B. H. Heakal, A. Younis, W. M. Aboulthana and M. F. Mady, ACS Omega, 2022, 7, 32418–32431 CrossRef CAS PubMed.
- A. Abd-El-Aziz, Z. Li, X. Zhang, S. Elnagdy, M. S. Mansour, A. ElSherif and A. S. Abd-El-Aziz, Top. Curr. Chem., 2025, 383, 8 CrossRef CAS PubMed.
- E. R. Trivedi, S. V. Eliseeva, J. Jankolovits, M. M. Olmstead, S. Petoud and V. L. Pecoraro, J. Am. Chem. Soc., 2014, 136, 1526 CrossRef CAS PubMed.
- T. Zhang, X. Zhu, C. C. Cheng, W. M. Kwok, H. L. Tam, J. Hao and K. L. Wong, J. Am. Chem. Soc., 2011, 133, 20120 CrossRef CAS PubMed.
- M. A. Katkova and M. N. Bochkarev, Dalton Trans., 2010, 39, 6599 RSC.
- J. Jin, S. Niu, Q. Han and Y. Chi, New J. Chem., 2010, 34, 1176 RSC.
- N. P. Ebosie, J. Chem. Soc. Niger., 2025, 50, 373–388 Search PubMed.
- H. E. Saad, G. M. Abu El Reash, M. Gaber, M. A. Hashem, Y. G. A. El-Reash, N. Y. Elamin and Y. S. El-Sayed, BMC Genomics, 2024, 25, 162 Search PubMed.
- S. M. Abdel-Fatah, M. T. Basha, A. I. Al-Sulami, M. R. Shehata and L. H. Abdel-Rahman, Appl. Organomet. Chem., 2025, 39, e70192 CrossRef CAS.
- K. Zhang, D. Wang, S. Wu, C. Wang, Z. Yu and L. Zhang, Struct. Chem., 2025, 36, 213–221 CrossRef CAS.
- M. A. Subhan, M. S. Rahman, K. Alam and M. M. Hasan, Spectrochim. Acta, Part A, 2014, 118, 944 CrossRef PubMed.
- H. G. Sogukomerogullari, E. Basaran, R. A. Kepekçi, B. Turkmenoglu, A. O. Sarıoglu and M. Kose, Polyhedron, 2025, 265, 117275 CrossRef CAS.
- A. A. Abdel Aziz and S. H. Seda, J. Fluoresc., 2015, 25, 1711 CrossRef CAS.
- D. Sahin, R. A. Kepekçi, B. Turkmenoglu and S. Akkoc, J. Biomol. Struct. Dyn., 2025, 43, 3375 CrossRef CAS PubMed.
- B. G. Tweedy, Phytopathology, 1964, 55, 910 Search PubMed.
- F. Chioma, O. Okpareke, S. N. Okafor and C. I. Ezugwu, J. Mol. Struct., 2023, 1291, 136070 CrossRef CAS.
- N. Raman, S. J. Raja and A. Sakthivel, J. Coord. Chem., 2009, 62, 691 CrossRef CAS.
- A. A. Emara, Spectrochim. Acta, Part A, 2010, 77, 117 CrossRef PubMed.
- S. H. Mahdi and L. K. A. Karem, Inorg. Chem. Commun., 2024, 165, 112524 CrossRef CAS.
- R. S. Abo-Rehab, E. A. Kasim, N. Farhan, M. S. Tolba, M. R. Shehata and E. M. Abdalla, Appl. Organomet. Chem., 2024, 38, e7622 CrossRef CAS.
- C.-Q. Miao, Y.-N. Ling, X.-Q. Ma, Y.-X. Chen, X.-J. Zhu, S.-R. Zhang and M. Fang, Inorg. Chim. Acta, 2024, 559, 121782 CrossRef CAS.
- T. Meng, T. Liu, Q.-P. Qin, Z.-L. Chen, H.-H. Zou, K. Wang and F.-P. Liang, Dalton Trans., 2020, 49, 4404 Search PubMed.
- Y.-L. Song, Y.-T. Li and Z.-Y. Wu, J. Inorg. Biochem., 2008, 102, 1691 CrossRef CAS.
- T. Ahamad, N. Nishat and S. Parveen, J. Coord. Chem., 2008, 61, 1963 CrossRef CAS.
- Z. A. Taha, A. K. Hijazi, H. Abul-Futouh, W. Al-Momani, B. M. Alziqili, P. Liebing and W. Weigand, J. Mol. Struct., 2024, 1316, 139054 Search PubMed.
- Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing, Wayne, PA, 2011, p. 100 Search PubMed.
- K. A. Hussein, N. Shaalan, A. K. Lafta and J. M. Al Akeedi, Indones. J. Chem., 2024, 24, 358 Search PubMed.
- K. A. Hussein and N. Shaalan, Chem. Methodol., 2021, 6, 103 Search PubMed.
- M. Alqasaimeh, A.-A. Abu-Yamin, S. Matar, K. Al Khalyfeh, T. Rüffer, H. Lang and D. Taher, J. Mol. Struct., 2023, 1274, 134458 Search PubMed.
- K. Andiappan, A. Sanmugam, E. Deivanayagam, K. Karuppasamy, H. S. Kim and D. Vikraman, Inorg. Chem. Commun., 2023, 150, 110510 CrossRef CAS.
- K. Andiappan, A. Sanmugam, E. Deivanayagam, K. Karuppasamy, H. S. Kim and D. Vikraman, Int. J. Biol. Macromol., 2019, 124, 403 CrossRef CAS PubMed.
- K. Andiappan, A. Sanmugam, E. Deivanayagam, K. Karuppasamy, H. S. Kim and D. Vikraman, Sci. Rep., 2018, 8, 3054 CrossRef PubMed.
- C. Xue, X. Xin, Y. Chang, Y.-J. Gao, J.-Y. Huang, H.-X. Qi and W.-M. Wang, J. Mol. Struct., 2023, 1291, 136058 CrossRef CAS.
- Z.-H. Shen, D.-F. Xu, N.-N. Cheng, X. Zhou, X. Chen, Y. Xu and Q. He, J. Coord. Chem., 2011, 64, 2342 CrossRef CAS.
- L. J. K. Boerner and J. M. Zaleski, Curr. Opin. Chem. Biol., 2005, 9, 135 CrossRef CAS PubMed.
- D.-F. Xu, S.-Z. Ma, G.-Y. Du, Q. He and D. Sun, J. Rare Earths, 2008, 26, 643 CrossRef.
- X.-Y. Xin, N. Qiao, C.-S. Cao, F.-J. Chen, W.-Y. Li, Y.-N. Ling and W.-M. Wang, Inorg. Chim. Acta, 2022, 541, 121092 CrossRef CAS.
- E. A. M. Khalil and G. G. Mohamed, Inorg. Chem. Commun., 2023, 153, 110825 Search PubMed.
- H. F. Abd El-Halim, M. M. Omar and G. G. Mohamed, Spectrochim. Acta, Part A, 2011, 78, 36 CrossRef PubMed.
- A.-A. Abu-Yamin, M. S. Abduh, S. A. Saghir and N. Al-Gabri, Pharmaceuticals, 2022, 15, 454 CrossRef CAS.
- I. P. Ejidike and P. A. Ajibade, Bioinorg. Chem. Appl., 2015, 2015, 1 CrossRef.
- M. F. Abo-Ashour, W. M. Eldehna, R. F. George, M. M. Abdel-Aziz, M. M. Elaasser, N. M. A. Gawad and S. M. Abou-Seri, Eur. J. Med. Chem., 2018, 160, 49 CrossRef CAS.
- P. L. Ho, K. H. Chow, W. U. Lo and V. C. Cheng, Int. J. Antimicrob. Agents, 2011, 37, 270 CrossRef CAS.
- H. S. Ibrahim, W. M. Eldehna, H. A. Abdel-Aziz, M. M. Elaasser and M. M. Abdel-Aziz, Eur. J. Med. Chem., 2014, 85, 480 CrossRef CAS PubMed.
- Z. A. Taha, T. S. Ababneh, A. K. Hijazi, S. M. Al-Aqtash, W. A. Al-Momani and I. Mhaidat, J. Saudi Chem. Soc., 2022, 26, 101400 Search PubMed.
- M. T. Kaczmarek, M. Zabiszak, M. Nowak and R. Jastrzab, Coord. Chem. Rev., 2018, 370, 42 CrossRef CAS.
- N. Mishra, K. Kumar, H. Pandey, S. R. Anand, R. Yadav, S. P. Srivastava and R. Pandey, J. Saudi Chem. Soc., 2020, 24, 925 CrossRef CAS.
- P. Ganapathi and K. Ganesan, J. Mol. Liq., 2017, 233, 452 Search PubMed.
- N. E. El-Gamel and K. A. Ali, J. Mol. Struct., 2017, 1147, 167 CrossRef CAS.
- K. M. Docherty Jr and C. F. Kulpa, Green Chem., 2005, 7, 185 Search PubMed.
- R. G. Deghadi and G. G. Mohamed, Comments Inorg. Chem., 2022, 42, 368 CrossRef.
- W. H. Mahmoud, R. G. Deghadi and G. G. Mohamed, J. Organomet. Chem., 2020, 917, 121113 Search PubMed.
- W. H. Mahmoud, R. G. Deghadi, M. M. El Desssouky and G. G. Mohamed, Appl. Organomet. Chem., 2019, 33, e4556 CrossRef.
- A. Albert, J. Pharm. Sci., 1979, 68, 662 CrossRef PubMed.
- K. A. Hussein and N. Shaalan, Indones. J. Chem., 2022, 22, 1365 Search PubMed.
- S. M. Obaid, A. J. Jarad and A. A. Al-Hamdani, J. Phys.: Conf. Ser., 2020, 1660, 012028 Search PubMed.
- T. D. Tavares, J. C. Antunes, J. Padrao, A. I. Ribeiro, A. Zille, M. T. P. Amorim and H. P. Felgueiras, Antibiotics, 2020, 9, 314 Search PubMed.
- A. H. Delcour, Biochim. Biophys. Acta, 2009, 1794, 808 CrossRef CAS.
- E. Khalil, W. Mahmoud and G. Mohamed, Egypt. J. Chem., 2021, 64, 3555 Search PubMed.
- L. Shi, R.-Q. Fang, J.-Y. Xue, Z.-P. Xiao, S.-H. Tan and H.-L. Zhu, Aust. J. Chem., 2008, 61, 28 Search PubMed.
- S. V. Kumar, W. K. Lo, H. J. Brooks, L. R. Hanton and J. D. Crowley, Aust. J. Chem., 2015, 69, 489 CrossRef.
- Z. A. Taha, A. K. Hijazi, A. Y. Al-Smadi, W. M. Al-Momani and F. Wedian, Russ. J. Gen. Chem., 2021, 91, 2292 CrossRef CAS.
- A. K. Hijazi, Z. A. Taha, A. Ibdah, I. M. Idris and W. M. Al-Momani, Chem. Pap., 2021, 75, 4611 Search PubMed.
- W. M. Al-Momani, Z. A. Taha, A. M. Ajlouni, Q. M. Shagra and M. A. Zouby, Asian Pac. J. Trop. Biomed., 2013, 3, 367 CrossRef CAS PubMed.
- K. B. Hussein and J. Zanco, Pure Appl. Sci., 2021, 33, 73 Search PubMed.
- J. D. L. Mosquera, A. A. Muriel, Y. Upegui, S. M. Robledo, M. T. Ramirez-Apan, D. Morales-Morales and D. Polo-Cerón, Antibiotics, 2021, 10, 728 CrossRef.
- J. M. Andrews, J. Antimicrob. Chemother., 2001, 48, 5 Search PubMed.
- J. M. Andrews, J. Antimicrob. Chemother., 2002, 49, 1049 Search PubMed.
- M. Arzanlou, W. C. Chai and H. Venter, Essays Biochem., 2017, 61, 49 CrossRef.
- N. F. Mahmoud, W. H. Mahmoud and G. G. Mohamed, Appl. Organomet. Chem., 2020, 34, e5801 Search PubMed.
- Z. A. Taha, A. K. Hijazi and W. M. Al-Momani, J. Mol. Struct., 2020, 1220, 128712 CrossRef CAS.
- S. Paswan, A. Anjum, N. Yadav, N. Jaiswal and R. K. Singh, J. Coord. Chem., 2020, 73, 686 Search PubMed.
- N. Yadav, A. K. Jaiswal, K. K. Dey, V. B. Yadav, G. Nath, A. K. Srivastava and R. R. Yadav, Mater. Chem. Phys., 2018, 218, 10 CrossRef CAS.
- S. Paswan, N. Jaiswal, V. K. Modanawal, M. K. Patel and R. K. Singh, Inorg. Chim. Acta, 2020, 513, 119955 CrossRef CAS.
- C. Nastasa, D. C. Vodnar, I. Ionuţ, A. Stana, D. Benedec, R. Tămaian, O. Oniga and B. Tiperciuc, Int. J. Mol. Sci., 2018, 19, 222 CrossRef PubMed.
- J. D. L. Mosquera, A. A. Muriel and D. P. Cerón, Univ. Sci., 2018, 23, 141 CrossRef.
- A. M. Ajlouni, Z. A. Taha, A. K. Hijazi and W. M. Al-Momani, Appl. Organomet. Chem., 2018, 32, e4536 Search PubMed.
- Z. A. Taha, A. M. Ajlouni and W. M. Al-Momani, J. Lumin., 2012, 132, 2832 CrossRef CAS.
- (a) K. Mohanan, R. Aswathy, L. P. Nitha, N. E. Mathews and B. S. Kumari, J. Rare Earths, 2014, 32, 379 CrossRef CAS; (b) M. Maciuca, A. C. Munteanu and V. Uivarosi, Molecules, 2020, 25, 1347 CrossRef; (c) G. R. Choppin and D. R. Peterman, Coord. Chem. Rev., 1998, 174, 283 Search PubMed; (d) J. Barańska, K. K. Szejn, M. Zabiszak, A. Grzeskiewicz, M. Skrobanska, M. Nowak, R. Jastrząb and M. T. Kaczmarek, Int. J. Mol. Sci., 2025, 26, 10379 CrossRef; (e) J.-C. G. Bünzli, Chem. Rev., 2010, 110, 2729 Search PubMed; (f) J.-C. G. Bünzli and C. Piguet, Chem. Rev., 2002, 102, 1897 CrossRef; (g) B. Shyni, T. S. Sikha, J. P. Remiya, Y. M. Thasneem and S. S. Beevy, J. Coord. Chem., 2023, 76, 878 CrossRef CAS; (h) O. Sun, T. Gao, J. Sun, G. Li, H. Li, H. Xu, C. Wang and P. Yan, CrystEngComm, 2014, 16, 10460 RSC.
- L. Zhang, D. Pornpattananangkul, C.-M. Hu and C.-M. Huang, Curr. Med. Chem., 2010, 17, 585 CrossRef CAS.
- B. Khameneh, R. Diab, K. Ghazvini and B. S. Bazzaz, Microb. Pathog., 2016, 95, 32 CrossRef CAS PubMed.
- C. Walsh, Nature, 2000, 406, 775 Search PubMed.
- M. S. Refat, W. F. El-Hawary and M. A. Mohamed, J. Mol. Struct., 2012, 1013, 45 Search PubMed.
- B. G. Tweedy, Phytopathology, 1964, 55, 910 Search PubMed.
- Y. Anjaneyula and R. P. Rao, Synth. React. Inorg. Met.-Org. Chem., 1986, 16, 257 Search PubMed.
- N. Nishat, S. Hasnain, S. Dhyani and J. Asma, Coord. Chem., 2010, 63, 3859 CrossRef CAS.
- A. E. Şabık, M. Karabork, G. Ceyhan, M. Tümer and M. Dıgrak, Int. J. Inorg. Chem., 2012, 2012, 1 Search PubMed.
- N. Dharmaraj, P. Viswanathamurthi and K. Natarajan, Transition Met. Chem., 2001, 26, 105 CrossRef CAS.
- Z. H. Chohan, M. Arif, A. Choukhtar and C. T. Supuran, Bioinorg. Chem. Appl., 2006, 2006, 1 Search PubMed.
- K. Siddappa, S. B. Mane and D. Manikprabhu, Spectrochim. Acta, Part A, 2014, 130, 634 CrossRef CAS.
- E. L. Gavey and M. Pilkington, Coord. Chem. Rev., 2015, 296, 125 CrossRef CAS.
- Z. Shen, D. Xu, N. Cheng, X. Zhou, X. Chen, Y. Xu and Q. He, J. Coord. Chem., 2011, 64, 2342 CrossRef CAS.
- A. Hameed, M. Al-Rashida, M. Uroos, S. A. Ali and K. M. Khan, Expert Opin. Ther. Pat., 2017, 27, 63 CrossRef CAS.
- M. S. More, P. G. Joshi, Y. K. Mishra and P. K. Khanna, Mater. Today Chem., 2019, 14, 100195 CrossRef CAS.
- X. Liu and J.-R. Hamon, Coord. Chem. Rev., 2019, 389, 94 CrossRef CAS.
- K. Prajapati, P. Prajapati, M. Brahmbhatt and J. Vora, Res. J. Life Sci., Bioinf., Pharm. Chem. Sci., 2018, 4, 803 CAS.
- A. Soroceanu and A. Bargan, Crystals, 2022, 12, 1436 CrossRef CAS.
- T. Rakshit, A. Haldar, S. Jana, S. Raha, R. Saha and D. Mandal, Results Chem., 2023, 6, 101224 CrossRef CAS.
- A. Nijs, R. Cartuyvels, A. Mewis, V. Peeters, J. L. Rummens and K. Magerman, J. Clin. Microbiol., 2003, 41, 3627 CrossRef CAS.
- B. A. Brown-Elliott, J. M. Brown, P. S. Conville and R. J. Wallace Jr, Clin. Microbiol. Rev., 2006, 19, 259 CrossRef CAS PubMed.
- K. Syal, R. Iriya, Y. Yang, H. Yu, S. Wang, S. E. Haydel and N. Tao, ACS Nano, 2016, 10, 845 CrossRef CAS PubMed.
- K. Das, R. K. Tiwari and D. K. Shrivastava, J. Med. Plants Res., 2010, 4, 104 Search PubMed.
- A. Taglietti, Y. A. Diaz Fernandez, E. Amato, L. Cucca, G. Dacarro, P. Grisoli and M. Patrini, Langmuir, 2012, 28, 8140 CrossRef CAS PubMed.
- L. Fguira, S. Fotso, R. B. Ameur-Mehdi, L. Mellouli and H. Laatsch, Res. Microbiol., 2005, 156, 341 CrossRef PubMed.
- K. Konate, J. F. Mavoungou, A. N. Lepengue, R. R. Aworet-Samseny, A. Hilou, A. Souza and B. M'Batchi, Ann. Clin. Microbiol. Antimicrob., 2012, 11, 18 CrossRef CAS PubMed.
- V. G. De Billerbeck, Phytother. Res., 2007, 5, 249 CrossRef CAS.
- N. S. Ncube, A. J. Afolayan and A. I. Okoh, Afr. J. Biotechnol., 2008, 7, 1797 CrossRef CAS.
- C. Valgas, S. M. Souza, E. Smania and A. Smania Jr, Braz. J. Microbiol., 2007, 38, 369 CrossRef.
- R. Ashraf and N. P. Shah, Int. Food Res. J., 2011, 18, 837 Search PubMed.
- Y. F. Qiao, L. Du, J. Zhou, Y. Hu, L. Li, B. Li and Q. Zhao, J. Coord. Chem., 2014, 67, 2615 CrossRef CAS.
- V. Kuete, T. K. Tabopda, B. Ngameni, F. Nana, T. E. Tshikalange and B. T. Ngadjui, S. Afr. J. Bot., 2010, 76, 125 CrossRef CAS.
- M. A. Pfaller, D. J. Sheehan and J. H. Rex, Clin. Microbiol. Rev., 2004, 17, 268 CrossRef CAS PubMed.
- P. Cos, A. J. Vlietinck, D. Vlietineck and L. Maes, J. Ethnopharmacol., 2006, 106, 290 CrossRef CAS PubMed.
- J. N. Eloff, P. Masoko and J. Picard, S. Afr. J. Bot., 2007, 73, 667 CrossRef CAS.
- D. Ma, S. Panda and J. D. Lin, EMBO J., 2011, 30, 4642 Search PubMed.
- T. G. McCloud, Molecules, 2010, 15, 4526 CrossRef CAS PubMed.
- D. Iacopetta, J. Ceramella, A. Catalano, A. Mariconda, F. Giuzio, C. Saturnino and M. S. Sinicropi, Inorganics, 2023, 11, 320 CrossRef CAS.
- I. Pospieszna-Markiewicz, M. A. Fik-Jaskółka, Z. Hnatejko, V. Patroniak and M. Kubicki, Molecules, 2022, 27, 8390 Search PubMed.
- L. Riccardi, V. Genna and M. De Vivo, Nat. Rev. Chem., 2018, 2, 100 CrossRef CAS.
- A. Omar and P. Nadworny, Adv. Drug Delivery Rev., 2017, 112, 61 CrossRef CAS.
- I. Kostova, Curr. Med. Chem., 2024, 31, 358 CrossRef CAS PubMed.
- A. Øwre, M. Vinum, M. Kern, J. Van Slageren, J. Bendix and M. Perfetti, Inorganics, 2018, 6, 72 CrossRef.
- S. Alfei, G. C. Schito, A. M. Schito and G. Zuccari, Int. J. Mol. Sci., 2024, 25, 7182 CrossRef CAS PubMed.
- S. Wankhede, A. Badule, S. Chaure, A. Damahe, M. Damahe and O. Porwal, J. Adv. Sci. Res., 2025, 16, 1 Search PubMed.
- S. B. Manjare, R. K. Mahadik, K. S. Manval, P. P. More and S. S. Dalvi, ACS Omega, 2022, 8, 473 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |