Open Access Article
Open Access ArticleFLARE: a label-free fluorescence-assisted method for RNA engineering of three-way junctions
A. Murali
Krishna
a,
Nida
Fathima
a,
Jothi
Basu
b and
Ashwani
Sharma
 *ab
*ab
aDepartment of Chemistry, Indian Institute of Science Education and Research (IISER), Tirupati 517619, India. E-mail: a.sharma@iisertirupati.ac.in
bDepartment of Biology, Indian Institute of Science Education and Research (IISER), Tirupati 517619, India
First published on 26th November 2025
Abstract
RNA three-way junction (3WJ) motifs are important structural elements found in the majority of RNAs, and the thermal stability of these structural motifs is crucial in determining RNA's biological roles and functions. Engineering these RNA motifs for better stability would enhance the native folding of RNA and is believed to enhance RNA's functionality. However, there are no high-throughput methods reported in the literature to engineer RNAs. Here, we report a label-free, FLuorescence-Assisted method for RNA Engineering (FLARE) utilizing the light-up aptamer Baby Spinach and the highly stable pRNA 3WJ derived from the Phi29 bacteriophage. The generated FLARE scaffold could be used to engineer almost any RNA three-way junction, which is ubiquitously found in natural RNAs. Using FLARE, we engineered a 5S 3WJ of ribosomal RNA to enhance its thermal stability significantly, even more than the highly stable pRNA 3WJ. The engineered 5S 3WJ was further introduced into single guide RNA (sgRNA) of the CRISPR–Cas9 system to enhance its cleavage efficiency significantly and was also shown to form an RNA nanotriangle for therapeutic applications. The FLARE design is general in nature and could be used to engineer 3WJs found in natural or unnatural functional RNAs such as ribozymes, riboswitches or aptamers, and even would be helpful in engineering 3WJs found in various newly discovered RNAs, the functions of which are currently unknown, to understand their folding and function.
Introduction
Ribonucleic acids (RNAs) fold into complex secondary structures involving various motifs that are structural subunits of RNA and are critical determinants of RNA's folding, interaction with other molecules and function. These motifs include loops, loop junctions such as three-way junctions or four-way junctions, hairpins, bulges and knots that occur ubiquitously in many RNAs.1Among these motifs, RNA three-way junctions (3WJs), which result from three separate RNA helices meeting at a central unpaired junction, are thus far the most abundant loop junction structures found naturally in many RNAs including ribozymes, riboswitches, ribosomal RNAs and viral RNAs.2 Stability of these 3WJ structural motifs is critical for the function of many RNAs.3,4 For instance, in the hammerhead ribozyme, the 3WJ forms the catalytic core of the ribozyme.5 Bacteriophage Phi29 utilizes a 3WJ in its packaging RNA (pRNA) to facilitate efficient packaging of its genome.6 Purine nucleobases bind to a 3WJ central core in purine riboswitches.7 In ribosomes, the 3WJ in the large ribosomal subunit is the key motif in the organization of multiple junctions around the catalytic core where the peptidylation reaction occurs.2 Apart from their role in cellular systems, naturally occurring RNA 3WJs have also been utilized for applications in RNA nanotechnology,8,9 biosensing10 and RNA imaging.11,12 Guo and co-workers utilized the 3WJ motif present in the packaging RNA (pRNA) of the Phi29 bacteriophage to construct highly stable two-dimensional (2D) and three-dimensional (3D) RNA nanostructures.13 These RNA nanostructures have further been used in small-molecule drug delivery14,15 as well as siRNA16,17 delivery. The Phi29 3WJ, when appended with multiple functional RNAs such as the Spinach aptamer or a ribozyme on its arms, preserves the native folding of the attached functional modules.18 The Jaffrey lab used a slightly modified Phi29 3WJ that was named F30 and fused it to the Broccoli light-up aptamer to enable stable aptamer expression for imaging RNAs in live cells.12,19 Additionally, 3WJs have been utilized in designing fluorescence-based biosensors for the detection of various small-molecules20,21 as well as biomacromolecules.22,23
We envisioned that in a functional RNA containing a 3WJ motif, engineering the 3WJ motif for better thermal stability might lead to enhanced native folding of the RNA and in turn its enhanced functionality. This would be helpful in engineering functional RNAs with tailored properties for applications ranging from gene therapy24 to synthetic biology circuits,25 diagnostics26 and imaging.27 Additionally, 3WJ engineering would also be useful to understand the structure or function of naturally existing RNAs including newly discovered RNAs in cells for which the structure or function is unknown,28 assuming these RNAs have a 3WJ motif, which is abundantly present in many RNAs. We searched for existing methods to engineer RNAs in a high-throughput manner. Unfortunately, our search could not yield any high-throughput or fluorescence-based methods in the literature to engineer RNAs or RNA 3WJs. Although, advances in RNA structural analysis and machine learning have significantly enhanced the ability to predict RNA folding.29 However, understanding the stability of a particular RNA motif, especially a loop junction such as a 3WJ containing a few unpaired nucleotides, still remains a challenge. Current RNA folding algorithms predominantly compute energy-minimized secondary structures from base-pairing interactions. As a result, they are limited in accurately predicting the stabilization of 3WJs that depend on unpaired nucleotides. The reported methods to engineer different RNAs or to understand the stability of a particular motif involve exchanging the nucleotides in an RNA sequence one by one30–32 and checking for their stability or functionality. This has been performed for engineering single guide RNAs in the CRISPR system for gene-editing applications,30 for engineering ribozymes,31 for engineering aptamers32 and even for engineering 3WJ RNA motifs, where the stability of each individual motif was tested using UV optical melting.33 A different approach is to use a combinatorial approach by systematic evolution (SELEX)34,35 using a library of nucleic acids with multiple random nucleotides and selecting the sequences with the required functionality from the pool, which is a very time-consuming and tedious process.
We present here a FLuorescence-Assisted RNA Engineering (FLARE) method, a label-free tool to engineer an RNA 3WJ motif in order to tune its thermal stability. Using FLARE, we engineered a 5S 3WJ of ribosomal RNA to achieve exceptional thermal stability and show that introducing this highly stable engineered 5S 3WJ into a functional RNA such as sgRNA in the CRISPR–Cas9 system enhances the cleavage efficiency of a target DNA significantly. As 3WJs have been widely used in RNA nanotechnology to construct RNA-based nanostructures and apply them in targeted drug delivery, the engineered 5S 3WJ has also been used to construct an RNA nanotriangle for applications in RNA nanotechnology.13 In general, FLARE could be used to modulate the stability of any RNA 3WJ motif present in natural or synthetic RNAs, and we envision that reintroducing the engineered 3WJ back into the parent RNA should modulate the RNA's functionality.
Results and discussion
Designing of FLARE
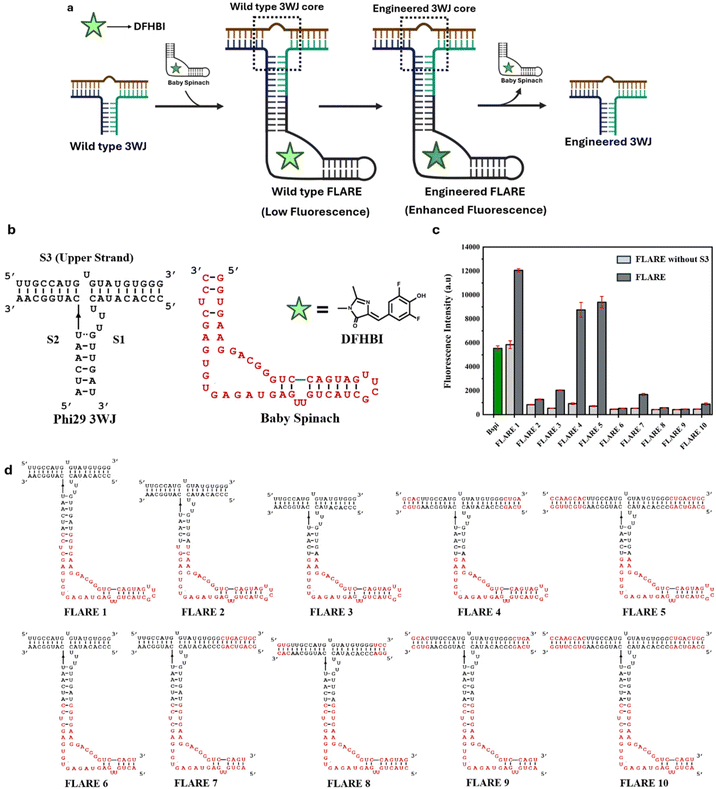
To design FLARE, a label-free fluorescence-assisted method to engineer RNA 3WJs, we hypothesized that the stability of an RNA 3WJ fused to a fluorescence-based RNA light-up aptamer (such as Spinach, a well-known light-up aptamer)36 should correlate with the fluorescence output of the light-up aptamer (Fig. 1a). Thus, any modification or engineering in the 3WJ sequence especially at the 3WJ core would alter the stability of the 3WJ, which in turn would affect the fluorescence output of the light-up aptamer fused to the 3WJ. The hypothesis is based on the previous work published by our group on the Baby Spinach-based minimal modified sensor (BSMS), in which the 3′ end of Baby Spinach was extended to make BSMS non-fluorescent.37 In the presence of DNA/RNA analytes complementary to the extended BSMS sequence, the resulting duplex stability after analyte binding led to an enhanced fluorescence output that was nearly 2.5 times more than the parent Baby Spinach aptamer. We anticipated that FLARE would behave in a similar fashion and would respond to even a single nucleotide modification especially at the core junction of the fused 3WJ. This would be useful to engineer the stability of any 3WJ RNA core introduced into the FLARE scaffold for applications in diagnostics,26 imaging,27 and nanotechnology.9,13,38 For our hypothesis to work, we utilized a highly stable RNA 3WJ motif derived from the pRNA of the Phi29 bacteriophage,9,13,17,39 which has been used widely in RNA nanotechnology (Fig. 1b).9,38 We constructed FLARE by fusing the Phi29 3WJ to a miniature variant of the well-known RNA light-up aptamer Spinach, named ‘Baby Spinach’, a 51-nucleotide RNA aptamer,40 which is known to bind and enhance the fluorescence of the nearly non-fluorescent small molecule DFHBI (3,5-difluoro-4-hydroxybenzylidene imidazolidinone) by several fold. This fusion provided a basic FLARE scaffold that could be used to engineer the 3WJ core, where the stability of the engineered 3WJ would relate to the fluorescence enhancement of FLARE.FLARE optimization
Initially, for the construction of FLARE 1 (Fig. 1d), the Phi29 3WJ was directly fused to the Baby Spinach aptamer. To achieve this, both the 5′ and 3′ ends of the Baby Spinach were flanked with two of the RNA strands S1 and S2 of the 3WJ, respectively (Fig. 1b). The third upper strand (S3) was further annealed to construct FLARE 1 (Fig. 1d). To check the fluorescence output of FLARE 1, we carried out fluorescence studies using a micro-plate reader. To confirm that the fluorescence output of FLARE 1 is mainly because of the complete formation of the 3WJ, we used FLARE 1 without the upper strand S3 of the 3WJ as a negative control. Baby Spinach was used as a positive control to compare the maximum fluorescence output of FLARE 1. The fluorescence data show that the increase of fluorescence of FLARE 1 (Fig. 1c) compared to FLARE 1 without S3 was only around 2.1-fold. In addition, the background fluorescence due to FLARE 1 without S3 (shown in light grey) was high and comparable to the fluorescence of Baby Spinach (shown in green).We speculated that to observe a significant fluorescence change caused by a single nucleotide alteration in the core of the fused 3WJ, the fluorescence fold ratio of FLARE compared to its negative control (FLARE without the upper strand S3) should be high. Thus, to improve the fluorescence fold ratio, we optimized FLARE by designing different FLARE constructs, keeping the fused Baby Spinach aptamer as full length (FLAREs 1–5) or split (FLAREs 6–10) as shown in Fig. 1d. The fluorescence data of all ten designed FLARE constructs (Fig. 1c) show a low fold-ratio for FLARE constructs containing a split Baby Spinach (FLAREs 6–10) compared to FLARE constructs containing a full-length Baby Spinach (FLAREs 1–5). FLARE constructs involving split Baby Spinach (FLARES 6–10) might possibly disrupt the overall FLARE folding, resulting in a very low fluorescence fold ratio. From the data, FLARE 4 and FLARE 5 showed excellent fold ratios of 9.6 and 13.2, respectively. However, we found in preliminary studies that FLARE 4 responded better to modification in the upper strand of the 3WJ (Fig. S1), and FLARE 4 was chosen for further studies.
Evaluation of the functionality of the FLARE system
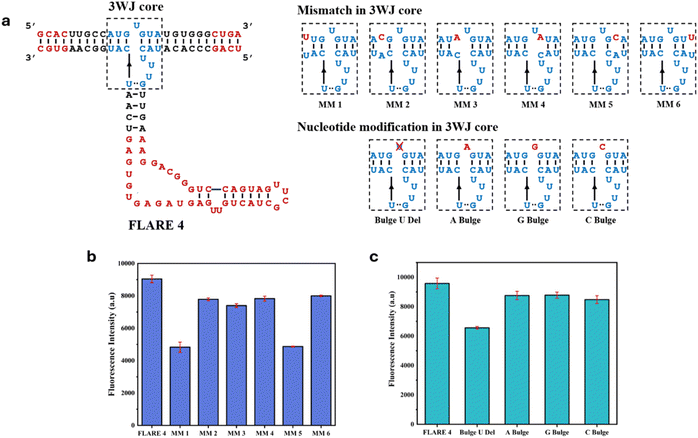
To further test our hypothesis that the stability of the 3WJ influences the fluorescence output of FLARE, we deliberately introduced a single-nucleotide bulge by introducing mismatches (MM1–MM6) into the 3WJ core of FLARE 4, as shown in Fig. 2a. The mismatches were expected to slightly destabilize the 3WJ core, which would diminish the fluorescence output of FLARE 4 to a small extent. Fig. 2b shows the fluorescence data for all the mismatch constructs. As expected, the fluorescence output for all mismatch constructs (MM1–MM6) was diminished to varying degrees compared to FLARE 4, depending on the position of the mismatches. This shows that FLARE 4 can respond well to a single-nucleotide modification in the 3WJ core and can be used to further engineer the Phi29 3WJ core.Engineering of the Phi29 3WJ using the FLARE system
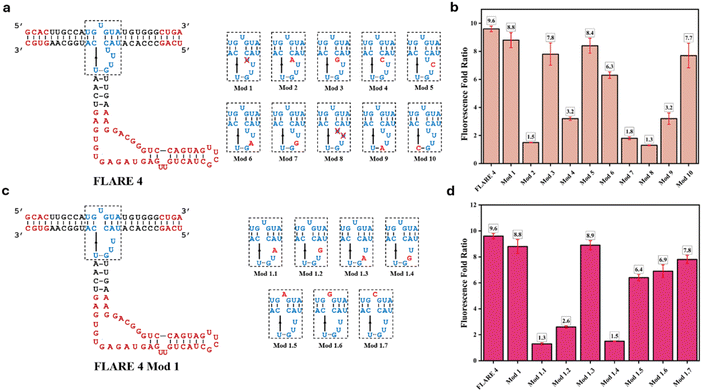
Furthermore, to engineer the 3WJ core in FLARE 4, we started first with engineering the upper strand of the Phi29 3WJ core. Deletion of the U bulge in the Phi29 3WJ destabilizes the 3WJ.17,33 We sought to reinvestigate the importance of the U bulge in the upper strand of the original Phi29 3WJ in maintaining the stability of the 3WJ core. For this, we deleted the U bulge in the upper strand of the 3WJ core (Fig. 2a, shown as ‘Bulge U Del’). In addition, the effect of nucleotide modifications through replacing U with A (A Bulge), G (G Bulge) or C (C Bulge) was also tested. The fluorescence data (Fig. 2c) show that deletion of the U bulge decreases the fluorescence output substantially, while replacing the nucleotide U with A, G or C had a minimal effect on fluorescence output. This shows that the bulge in the Phi29 3WJ core is required for the stability of the Phi29 3WJ and that the bulge with the U nucleotide has slightly higher 3WJ core stability than bulges with any other RNA nucleotides (A, G or C).Phi29 3WJ used in FLARE is already known to be a highly stable 3WJ as reported in the literature. We further aimed to investigate the significance of different nucleotides in the 3WJ that contribute to the stability of the 3WJ core. For this, we performed different modifications (Mod 1–Mod 10) in the core region of the 3WJ, as shown in Fig. 3a. Mainly, the unpaired nucleotides in the 3WJ core were either deleted or replaced by other nucleotides to make ten different constructs. Rational engineering of the 3WJ was not feasible here as the aim was to obtain an RNA 3WJ that is stable without any interactions with other small molecules or proteins. The unpaired nucleotides in the 3WJ core were altered on a trial-and-error approach. Fig. 3b shows the fluorescence fold ratio for all the FLARE constructs (Mod 1–Mod 10) with respect to the FLARE construct without the 3WJ upper strand.
The data show that deletion of one U (Mod 1) in the UUU bulge of the 3WJ core decreases the fluorescence output of FLARE 4 only slightly, from 9.6-fold to 8.8-fold. However, deleting two U's (Mod 8) in the UUU bulge reduces the fluorescence output by more than 90%, showing the least fluorescence fold among all constructs designed. This indicates that at least two U's are required in the UUU bulge of the Phi29 3WJ to maintain its stability. Replacing any of the U nucleotide in the UUU bulge with an A, G or C nucleotide overall decreases the fluorescence output of FLARE 4 to different extents depending on the position of the nucleotide in the UUU bulge and the nature of the nucleotide replaced (Mod 2–Mod 7). To understand the role of the U–G wobble base pair (represented in the 3WJ core as U⋯G), we replaced this wobble base pair with either a U–A (Mod 9) or C–G (Mod 10) base pair. Both modifications caused a significant drop in the fluorescence output compared to the U⋯G wobble base pair; however, the C–G base pair is better tolerated than U–A. This shows that the U–G wobble base pair is important for Phi29 3WJ stability and cannot be replaced with a Watson–Crick base pair such as C–G or U–A.
Based on the fluorescence data, we chose Mod 1 with one U deleted from the UUU bulge for further engineering in the quest to obtain a more stable 3WJ that would show a higher fluorescence fold ratio than FLARE 4. We synthesized seven new constructs by replacing one of the U's in the UU bulge of FLARE 4 Mod 1 (Fig. 3c) with A/G (Mod 1.1–Mod 1.4) or by replacing a single U bulge in the 3WJ upper strand with A/G/C (Mod 1.4–Mod 1.7), and checked the fluorescence output of all seven constructs. The fluorescence data revealed that Mod 1.3 showed a higher fluorescence fold ratio of 8.9 compared to the 8.8 for Mod 1; however, it was still slightly lower than or comparable to that of FLARE 4. Overall, these modifications suggest the importance of different nucleotides in the stability of Phi29 3WJ. However, we could not get an engineered 3WJ with better thermal stability compared to the Phi29 3WJ, which is already known to be one of the most stable 3WJs in the literature. Since Mod 1.3 showed a fluorescence fold ratio comparable to that of FLARE 4, we further performed UV-Tm studies on the isolated 3WJ of FLARE 4 and Mod 1.3, which consists of three RNA strands obtained by scission of the fused Baby Spinach from FLARE 4 and Mod 1.3 (see Fig. 1a). The UV-Tm data show a melting temperature of 57.5° C for the Phi 29 3WJ of FLARE 4 and 56.2° C for the engineered 3WJ of Mod 1.3 (Fig. S2 and S3a), which clearly match with their fluorescence results, emphasizing that the FLARE system could be used to engineer the stability of a 3WJ.
Although our results showed that the thermal stability of the Phi29 3WJ used in FLARE 4 could not be further improved using the set of nucleotide modifications shown above, FLARE 4 provides a basic FLARE scaffold to engineer any other 3WJs, which are abundantly present in many RNAs (Fig. 4a). We anticipated that any 3WJ core introduced into the empty FLARE scaffold should give a fluorescence fold ratio by virtue of its stability. A fluorescence fold ratio higher than that of FLARE 4 (9.6 fold) would make a 3WJ thermally more stable than the highly stable Phi29 3WJ. Thus, the FLARE system could also be used to compare the stabilities of 3WJ cores present in different RNAs.
Engineering of a 5S ribosomal RNA (rRNA) 3WJ using the FLARE system
As discussed, FLARE could be used to engineer almost any RNA 3WJ. To test this hypothesis, we used FLARE to engineer a 5S ribosomal RNA (rRNA) 3WJ that has been reported to be less stable than the Phi29 3WJ. It should be noted that accurate comparison of the thermal stabilities of two distinct 3WJs is challenging because variations in base-pairing number and composition across their arms can markedly affect the overall stability. However, FLARE would be able to measure the stability of a 3WJ core more accurately, as only the core junction is introduced into FLARE without changing any base pairing in the 3WJ arms. Thus, the thermal stability because of base pairing in the 3WJ arms while comparing two different 3WJs will be the same here.To engineer the 5S rRNA 3WJ, we introduced the 5S 3WJ core into the FLARE scaffold (obtained from FLARE 4) to get 5S-FLARE as shown in Fig. 4a. The fluorescence data revealed that 5S-FLARE showed a fluorescence fold ratio of only 1.8 (Fig. 4c). This shows that the 5S 3WJ core is not as stable as the Phi29 3WJ core (as in FLARE 4), which showed a much higher fluorescence fold ratio of 9.6. The 5S 3WJ had been shown in the literature to have lower thermal stability compared to the Phi29 3WJ in UV-Tm experiments.17 To engineer the 5S 3WJ core, we performed 13 modifications in the core junction of 5S-FLARE (5S-Mod 1 to 5S-Mod 13), as shown in Fig. 4b. First, in 5S-Mod 1, a CC mismatch was replaced with a GC base pair, and a U nucleotide was introduced to convert bulge A into an AU base pair (both modifications shown in blue in 5S-Mod 1).
However, this modification changed the fluorescence fold ratio to only 2.2 from 1.8 in 5S-FLARE (Fig. 4c). Keeping these modifications intact, we designed 12 more modifications (5S-Mod 2 to 5S-Mod 13) in the core region of the 5S 3WJ. All the constructs with nucleotides deleted or replaced in all three strands of the 5S 3WJ core are shown in Fig. 4b. The fluorescence data obtained from all the constructs (Fig. 4c) show that 5S-Mod 2 showed the highest fluorescence fold ratio of 10.1 compared to only 1.8 for 5S-FLARE. It is to be noted that the fluorescence fold ratio of 5S-Mod 2 (10.1-fold) was even higher than the fluorescence fold ratio of FLARE 4 (9.6-fold) having a highly stable Phi29 3WJ core. The results suggest that the engineered 5S 3WJ as in 5S-Mod 2 is substantially more stable than its wildtype (5S-FLARE). Additionally, the data also suggested that the engineered 5S 3WJ as in 5S-Mod 2 is even more stable than the already known highly stable Phi29 3WJ used in FLARE 4. To confirm this, we carried out UV-Tm experiments by using the isolated 3WJ cores of 5S-FLARE and 5S-Mod 2 and compared the results with the Phi29 3WJ core used in FLARE 4. The UV-Tm data and the derivative curves (Fig. 4d and e) showed that the wild-type 5S rRNA 3WJ used in 5S-FLARE had a Tm of 50.1° C, while the engineered 5S-Mod 2 3WJ used in the 5S-Mod 2 FLARE showed a Tm of 60.8° C, showing a stabilization by 10.7° C. Additionally, the Phi29 3WJ used in FLARE 4 showed a Tm of 57.5° C, showing that the engineered 5S-Mod 2 3WJ was even more stable than the known highly stable Phi29 3WJ by a Tm difference of 3.3° C. The UV-Tm results obtained were further validated using temperature-dependent circular dichroism (CD) spectroscopy (Fig. S4). Additionally, a head-to-head comparison of fluorescence and UV-Tm measurements was carried out for selected subtle FLARE constructs and their corresponding isolated 3WJs (Fig. S5 & Table S1). Thermal stabilities were also compared using denaturing polyacrylamide gel electrophoresis on an 8% gel containing 8 M or 10 M urea (Fig. S3b and S6). Thus, the fluorescence data obtained using the FLARE system align with the UV-Tm data, showing the fidelity of the FLARE system in engineering 3WJs.
Applications of the engineered 5S rRNA 3WJ in CRISPR-mediated gene editing and RNA nanotechnology
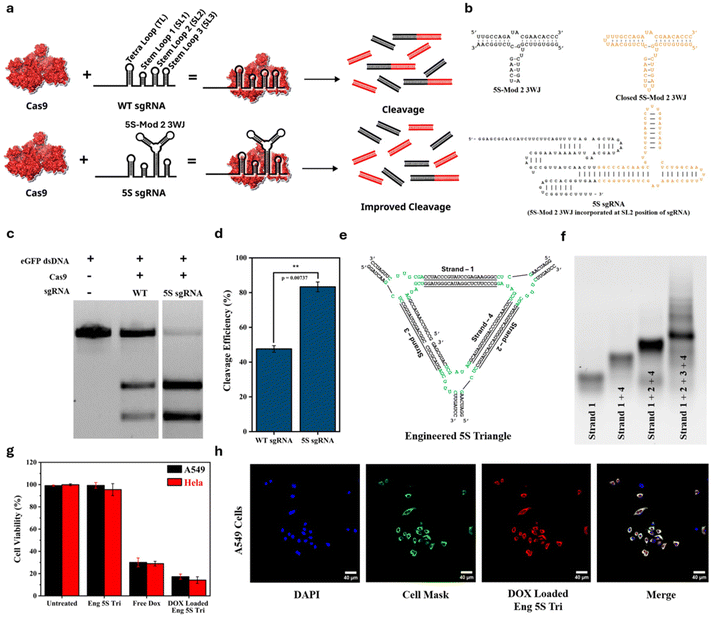
Next, we hypothesized that introducing a highly stable RNA 3WJ motif into a functional RNA should provide a nucleation site for RNA folding, stabilizing the native folding of the RNA and enhancing its function. To test this, we utilized the CRISPR–Cas9 system that has been used extensively for gene editing applications. We envisioned that the introduction of a highly stable 3WJ into single guide RNA (sgRNA) should stabilize its native folding and would enhance its cleavage efficiency compared to the WT sgRNA. Several regions in sgRNA including stem loop 2 (SL2) are amenable to engineering or modification without compromising the functioning of the CRISPR–Cas9 system.41 Thus, we introduced the engineered 5S-Mod 2 3WJ, after sealing its two ends with a UUU loop, into the SL2 region of single guide RNA (sgRNA) as shown in Fig. 5a and b to generate 5S sgRNA. Secondary structure prediction and thermodynamic parameters of 5S sgRNA, determined using NUPACK, are provided in Fig. S7. The in vitro cleavage efficiency of the 5S sgRNA was compared with that of the wild-type sgRNA using commercially available Cas9 protein and eGFP DNA as the target dsDNA. The in vitro cleavage assay gel and the corresponding gel band quantification (Fig. 5c and d) show that the 5S sgRNA has much better cleavage efficiency (83%) compared to the WT sgRNA (48%). Further studies on introducing the 5S-Mod 2 3WJ at different positions in sgRNA are currently in progress in our laboratory.Furthermore, we utilized the 3WJ core of 5S-Mod 2 to construct a nanotriangle following strategies used in the literature, and used it to deliver the anticancer drug doxorubicin to cancer cells. The engineered 5S triangle was constructed using four 2′-fluoro-modified RNA strands, as shown in Fig. 5e. The agarose gel (Fig. 5f) showed the stepwise assembly of the engineered 5S triangle. The hydrodynamic size of the engineered 5S triangle was determined using dynamic light scattering (DLS) (Fig. S8), revealing an average diameter of 14.5 ± 1.8 nm. The nanotriangle was further loaded with the anticancer drug doxorubicin, with a loading efficiency of approximately 80% (Fig. S9). The cytotoxicity of the doxorubicin-loaded nanotriangle was evaluated using an MTT assay against two different cancer cell lines, A549 (human lung adenocarcinoma) and HeLa (human cervical adenocarcinoma), representing distinct tissue origins to assess broader applicability. Untreated cells, cells treated with the engineered 5S triangle (Eng 5S Tri) and free doxorubicin (Dox) were used as controls. As shown in Fig. 5g, the Eng 5S Tri showed almost no cytotoxicity towards both A549 and HeLa cells, while the Dox-loaded Eng 5S Tri showed high cytotoxicity that was even higher compared to the free doxorubicin. This shows better cytotoxicity of doxorubicin towards cancer cell lines A549 and HeLa when doxorubicin is loaded onto the Eng 5S Tri compared to only free doxorubicin. The uptake of the Dox-loaded Eng 5S Tri was also confirmed using confocal microscopy where the intrinsic fluorescence of doxorubicin (DOX) was used for confocal imaging of A549 cells (Fig. 5h).
Conclusions
In this study, we developed FLARE, a label-free fluorescence-assisted method for RNA engineering, which was used to engineer RNA 3WJ motifs that are found ubiquitously in many RNAs. Utilizing FLARE, we engineered a weakly stable 5S ribosomal RNA 3WJ into a highly stable engineered 3WJ (5S-Mod 2), which exhibited a 10.7° C increase in thermal stability as evaluated using optical UV-melting. Additionally, the engineered 3WJ (5S-Mod 2) showed 3.3° C more thermal stabilization compared to the highly stable Phi29 3WJ. In many functional RNAs, the stability of 3WJ motifs is critical for the native folding of RNA and its function.4 Thus, the engineered 3WJ when introduced into the SL2 loop of sgRNA in the CRISPR–Cas9 system stabilized the native folding of sgRNA, resulting in an enhanced cleavage efficiency of 83% compared to the 48% for the wild-type sgRNA. The engineered 3WJ motif was also used to construct a nanotriangle that was loaded with the anticancer drug doxorubicin, and its cytotoxicity was checked against different cancer cell lines. In conclusion, FLARE is general in nature and could be used to engineer almost any 3WJ. Being a fluorescence-based method using a multimode plate reader, FLARE outperforms existing methods like UV-Tm in terms of throughput, sensitivity or speed. Stabilizing the 3WJ core motif of many functional RNAs such as ribozymes, riboswitches or aptamers would be helpful to enhance their functionality, which would be useful for a variety of biotechnological and therapeutic applications.9,38 This further would help us to understand the roles of various newly discovered RNAs, the functions of many of which are currently unknown.Author contributions
A. S. conceived and supervised the project. A. M. K. performed the experiments and contributed to the data analysis. N. F. participated in the experiments. J. B. performed CRISPR/Cas9 cleavage experiments and analyzed the data. A. S. wrote the manuscript, and A. M. K. revised the manuscript. All authors reviewed and approved the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article are included in the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ob01756d.Acknowledgements
This work was supported by the Indian Institute of Science Education and Research (IISER) Tirupati and the Anusandhan National Research Foundation (ANRF, grant CRG/2022/008274), India. J. B. was supported by a research fellowship from the Department of Biotechnology, India.References
- R. Schroeder, A. Barta and K. Semrad, Strategies for RNA folding and assembly, Nat. Rev. Mol. Cell Biol., 2004, 5(11), 908–919 CrossRef CAS PubMed.
- A. Lescoute and E. Westhof, Topology of three-way junctions in folded RNAs, RNA, 2006, 12(1), 83–93 CrossRef CAS.
- L. R. Ganser, M. L. Kelly, D. Herschlag and H. M. Al-Hashimi, The roles of structural dynamics in the cellular functions of RNAs, Nat. Rev. Mol. Cell Biol., 2019, 20(8), 474–489 CrossRef CAS PubMed.
- M. de la Peña, D. Dufour and J. Gallego, Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold, RNA, 2009, 15(11), 1949–1964 CrossRef.
- X. Zhan, T. J. Wilson, Z. Li, J. Zhang, Y. Yang, D. M. J. Lilley and Y. Liu, The structure and catalytic mechanism of a pseudoknot-containing hammerhead ribozyme, Nat. Commun., 2024, 15(1), 6628 CrossRef CAS PubMed.
- R. J. Reid, F. Zhang, S. Benson and D. Anderson, Probing the structure of bacteriophage phi 29 prohead RNA with specific mutations, J. Biol. Chem., 1994, 269(28), 18656–18661 CrossRef CAS PubMed.
- S. D. Gilbert, F. E. Reyes, A. L. Edwards and R. T. Batey, Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs, Structure, 2009, 17(6), 857–868 CrossRef CAS PubMed.
- W. W. Grabow and L. Jaeger, RNA Self-Assembly and RNA Nanotechnology, Acc. Chem. Res., 2014, 47(6), 1871–1880 CrossRef CAS PubMed.
- P. Guo, The emerging field of RNA nanotechnology, Nat. Nanotechnol., 2010, 5(12), 833–842 CrossRef CAS PubMed.
- J. Kehrli, C. Husser and M. Ryckelynck, Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives, Biosensors, 2024, 14(8), 376 CrossRef CAS.
- X. Chen, D. Zhang, N. Su, B. Bao, X. Xie, F. Zuo, L. Yang, H. Wang, L. Jiang, Q. Lin, M. Fang, N. Li, X. Hua, Z. Chen, C. Bao, J. Xu, W. Du, L. Zhang, Y. Zhao, L. Zhu, J. Loscalzo and Y. Yang, Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs, Nat. Biotechnol., 2019, 37(11), 1287–1293 CrossRef CAS PubMed.
- X. Li, H. Kim, J. L. Litke, J. Wu and S. R. Jaffrey, Fluorophore–promoted RNA folding and photostability enables imaging of single broccoli–tagged mRNAs in live mammalian cells, Angew. Chem., Int. Ed., 2020, 59(11), 4511–4518 CrossRef CAS PubMed.
- D. Jasinski, F. Haque, D. W. Binzel and P. Guo, Advancement of the emerging field of RNA nanotechnology, ACS Nano, 2017, 11(2), 1142–1164 CrossRef CAS PubMed.
- S. Guo, M. Vieweger, K. Zhang, H. Yin, H. Wang, X. Li, S. Li, S. Hu, A. Sparreboom, B. M. Evers, Y. Dong, W. Chiu and P. Guo, Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy, Nat. Commun., 2020, 11(1), 972 CrossRef CAS PubMed.
- X. Li, A. S. Bhullar, D. W. Binzel and P. Guo, The dynamic, motile and deformative properties of RNA nanoparticles facilitate the third milestone of drug development, Adv. Drug Delivery Rev., 2022, 186, 114316 CrossRef CAS PubMed.
- L. Pang, H. Shah, H. Wang, D. Shu, S. Y. Qian and V. Sathish, EpCAM-Targeted 3WJ RNA Nanoparticle Harboring Delta-5-Desaturase siRNA Inhibited Lung Tumor Formation via DGLA Peroxidation, Mol. Ther.-Nucleic Acids, 2020, 22, 222–235 CrossRef CAS PubMed.
- D. Shu, Y. Shu, F. Haque, S. Abdelmawla and P. Guo, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat. Nanotechnol., 2011, 6(10), 658–667 CrossRef CAS.
- D. Shu, E. F. Khisamutdinov, L. Zhang and P. Guo, Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor, Nucleic Acids Res., 2014, 42(2), e10–e10 CrossRef CAS PubMed.
- G. S. Filonov, C. W. Kam, W. Song and S. R. Jaffrey, In-Gel Imaging of RNA Processing Using Broccoli Reveals Optimal Aptamer Expression Strategies, Chem. Biol., 2015, 22(5), 649–660 CrossRef CAS PubMed.
- J. D. Moon, J. Wu, S. K. Dey, J. L. Litke, X. Li, H. Kim and S. R. Jaffrey, Naturally occurring three-way junctions can be repurposed as genetically encoded RNA-based sensors, Cell Chem. Biol., 2021, 28(11), 1569–1580 CrossRef CAS PubMed.
- J. S. Paige, T. Nguyen-Duc, W. Song and S. R. Jaffrey, Fluorescence imaging of cellular metabolites with RNA, Science, 2012, 335(6073), 1194–1194 CrossRef CAS PubMed.
- K. Huang, F. Doyle, Z. E. Wurz, S. A. Tenenbaum, R. K. Hammond, J. L. Caplan and B. C. Meyers, FASTmiR: an RNA-based sensor for in vitro quantification and live-cell localization of small RNAs, Nucleic Acids Res., 2017, 45(14), e130 CrossRef CAS PubMed.
- D. M. Kolpashchikov and A. A. Spelkov, Binary (Split) Light–up Aptameric Sensors, Angew. Chem., 2021, 133(10), 5040–5051 CrossRef.
- T. Li, Y. Yang, H. Qi, W. Cui, L. Zhang, X. Fu, X. He, M. Liu, P.-f. Li and T. Yu, CRISPR/Cas9 therapeutics: progress and prospects, Signal Transduction Targeted Ther., 2023, 8(1), 36 CrossRef PubMed.
- E. A. Davidson and A. D. Ellington, Synthetic RNA circuits, Nat. Chem. Biol., 2007, 3(1), 23–28 CrossRef CAS PubMed.
- A. F. Saju, A. Mukundan, M. Divyashree, R. Chandrashekhar and A. Mahadev Rao, RNA diagnostics and therapeutics: a comprehensive review, RNA Biol., 2025, 22(1), 1–11 CrossRef CAS.
- P. Le, N. Ahmed and G. W. Yeo, Illuminating RNA biology through imaging, Nat. Cell Biol., 2022, 24(6), 815–824 CrossRef CAS PubMed.
- L. Lorenzi, H.-S. Chiu, F. Avila Cobos, S. Gross, P.-J. Volders, R. Cannoodt, J. Nuytens, K. Vanderheyden, J. Anckaert and S. Lefever, The RNA Atlas expands the catalog of human non-coding RNAs, Nat. Biotechnol., 2021, 39(11), 1453–1465 CrossRef CAS PubMed.
- T. Shen, Z. Hu, S. Sun, D. Liu, F. Wong, J. Wang, J. Chen, Y. Wang, L. Hong and J. Xiao, Accurate RNA 3D structure prediction using a language model-based deep learning approach, Nat. Methods, 2024, 1–12 CAS.
- J. Filippova, A. Matveeva, E. Zhuravlev and G. Stepanov, Guide RNA modification as a way to improve CRISPR/Cas9-based genome-editing systems, Biochimie, 2019, 167, 49–60 CrossRef CAS PubMed.
- A. J. Che and T. F. Knight Jr, Engineering a family of synthetic splicing ribozymes, Nucleic Acids Res., 2010, 38(8), 2748–2755 CrossRef CAS PubMed.
- R. Oliveira, E. Pinho, A. L. Sousa, J. J. DeStefano, N. F. Azevedo and C. Almeida, Improving aptamer performance with nucleic acid mimics: de novo and post-SELEX approaches, Trends Biotechnol., 2022, 40(5), 549–563 CrossRef CAS PubMed.
- A. C. Hill and S. J. Schroeder, Thermodynamic stabilities of three-way junction nanomotifs in prohead RNA, RNA, 2017, 23(4), 521–529 CrossRef CAS.
- Y. Yokobayashi, High-throughput analysis and engineering of ribozymes and deoxyribozymes by sequencing, Acc. Chem. Res., 2020, 53(12), 2903–2912 CrossRef CAS PubMed.
- X. Huang, Y. Zhao, Q. Pu, G. Liu, Y. Peng, F. Wang, G. Chen, M. Sun, F. Du and J. Dong, Intracellular selection of trans-cleaving hammerhead ribozymes, Nucleic Acids Res., 2019, 47(5), 2514–2522 CrossRef CAS PubMed.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, RNA mimics of green fluorescent protein, Science, 2011, 333(6042), 642–646 CrossRef CAS PubMed.
- R. Soni, D. Sharma, A. M. Krishna, J. Sathiri and A. Sharma, A highly efficient Baby Spinach-based minimal modified sensor (BSMS) for nucleic acid analysis, Org. Biomol. Chem., 2019, 17(30), 7222–7227 RSC.
- D. W. Binzel, X. Li, N. Burns, E. Khan, W.-J. Lee, L.-C. Chen, S. Ellipilli, W. Miles, Y. S. Ho and P. Guo, Thermostability, tunability, and tenacity of RNA as rubbery anionic polymeric materials in nanotechnology and nanomedicine—specific cancer targeting with undetectable toxicity, Chem. Rev., 2021, 121(13), 7398–7467 CrossRef CAS PubMed.
- M. Bohmer, D. W. Binzel, W. Zhang and P. Guo, Constructing an active chimeric pRNA ring with a stoichiometry of six and identifying 12 domains of the pRNA ring binding to the 12-subunit channel of phi29 DNA-packaging motor, RNA, 2025, 31(6), 836–849 CrossRef CAS PubMed.
- K. D. Warner, M. C. Chen, W. Song, R. L. Strack, A. Thorn, S. R. Jaffrey and A. R. Ferré-D'Amaré, Structural basis for activity of highly efficient RNA mimics of green fluorescent protein, Nat. Struct. Mol. Biol., 2014, 21(8), 658–663 CrossRef CAS PubMed.
- G. Mullally, K. van Aelst, M. M. Naqvi, F. M. Diffin, T. Karvelis, G. Gasiunas, V. Siksnys and M. D. Szczelkun, 5′ modifications to CRISPR–Cas9 gRNA can change the dynamics and size of R-loops and inhibit DNA cleavage, Nucleic Acids Res., 2020, 48(12), 6811–6823 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |