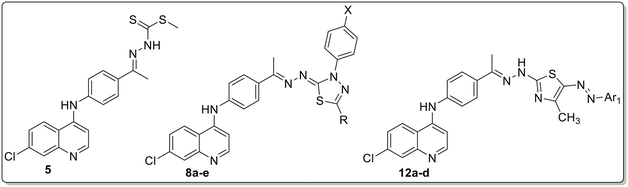
An exploration of potent antileishmanial agents derived from quinoline–thiazole and thiadiazole hybrids, targeting DHFR-TS and PTR1: design, synthesis, and computational analyses
Huda R. M.
Rashdan
 a,
Adnan A.
Bekhit
bc,
Veronika
Furlan
d,
Kikuko
Amagase
e,
Abdelsamed I.
Elshamy
a,
Adnan A.
Bekhit
bc,
Veronika
Furlan
d,
Kikuko
Amagase
e,
Abdelsamed I.
Elshamy
 f,
Nourhan
Elfar
g,
Mohamed. R.
Abdo
hi,
Tamer M.
Ibrahim
f,
Nourhan
Elfar
g,
Mohamed. R.
Abdo
hi,
Tamer M.
Ibrahim
 j,
Urban
Bren
dkl,
Wagdy M.
Eldehna
j,
Urban
Bren
dkl,
Wagdy M.
Eldehna
 *j and
Ahmed
Sabt
*j and
Ahmed
Sabt
 *f
*f
aChemistry of Natural and Microbial Products Department, Pharmaceutical and Drug Industries Research Institute, National Research Centre, Dokki, Cairo, 12622, Egypt
bDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Alexandria University, Alexandria, 21521, Egypt
cPharmacy Program, Allied Health Department, College of Health and Sport Sciences, University of Bahrain, P.O. Box 32038, Bahrain
dFaculty of Chemistry and Chemical Engineering, University of Maribor, Smetanova 17, SI-2000 Maribor, Slovenia
eLaboratory of Pharmacology & Pharmacotherapeutics, College of Pharmaceutical Sciences, Ritsumeikan University, Kusatsu, Shiga, Japan
fChemistry of Natural Compounds Department, Pharmaceutical and Drug Industries Research Institute, National Research Centre, Dokki, Cairo 12622, Egypt. E-mail: sabt.nrc@gmail.com
gDepartment of Biochemistry, School of Health and Social Work, University of Hertfordshire, hosted by Global Academic Foundation, New Administrative Capital, Cairo, 11578, Egypt
hMicrobiology and immunology department, faculty of pharmacy, Zagazig University, Zagazig City, Egypt
iDepartment of laboratory science, college of Pharmacy, University of Kut, Wasit, 52001, Iraq
jDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Kafrelsheikh University, Kafrelsheikh, 33516, Egypt. E-mail: wagdy2000@gmail.com
kFaculty of Mathematics, Natural Sciences and Information Technologies, University of Primorska, Glagoljaška 8, SI-6000 Koper, Slovenia
lInstitute of Environmental Protection and Sensors, Beloruska 7, SI-2000 Maribor, Slovenia
First published on 19th November 2025
Abstract
Neglected tropical diseases (NTDs) encompass a broad spectrum of infectious diseases predominantly found in tropical and subtropical regions. The limitations of current therapies underscore the critical demand for novel antileishmanial agents. In this investigation, we designed, synthesized, and evaluated ten hybrid compounds (5, 8a–e, and 12a–d) integrating a 7-chloroquinoline scaffold with thiadiazole and thiazole moieties, assessing their in vitro efficacy against Leishmania major. These hybrids exhibited potent activity against the promastigote stage, displaying IC50 values between 0.52 and 3.97 μM, outperforming miltefosine (IC50 = 7.83 μM). Additionally, they demonstrated strong inhibition of the intracellular amastigote form, with IC50 values ranging from 0.76 to 5.62 μM, compared to miltefosine's 8.07 μM. Notably, compound 5 emerged as a highly effective antileishmanial agent against both parasitic stages, while maintaining a favorable safety profile. Mechanistic studies revealed that compound 5 acts via an antifolate mechanism, selectively inhibiting key enzymes in the folate pathway: pteridine reductase 1 (PTR1) and dihydrofolate reductase-thymidylate synthase (DHFR-TS). Molecular docking and 100 ns molecular dynamics (MD) simulations demonstrated that the quinoline core occupies a hydrophobic pocket formed by residues Phe113, Leu188, Leu226, and Leu229, engaging in stable hydrophobic interactions and π–π stacking with Phe113. Furthermore, the quinoline scaffold and hydrazinecarbodithioate moiety formed hydrogen bonds with Tyr194, Gly225, and His241, reinforcing binding stability. Our findings introduce a promising new class of antileishmanial agents that disrupt the folate biosynthesis pathway, offering significant therapeutic potential for combating leishmaniasis.
1. Introduction
Neglected tropical diseases (NTDs) represent a heterogeneous group of infectious diseases that are attributable to protozoa, helminths, bacteria, viruses, and fungi, among other pathogens. These diseases are predominantly located in tropical and subtropical areas, characterized by high levels of poverty; however, instances of NTDs have also been documented in non-endemic areas, including developed nations.1 Globally, NTDs affect over 1.5 billion individuals and are responsible for more than 530![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities annually.2,3 In light of this situation, there has been a significant rise in studies concentrating on NTDs, driven by the imperative to develop efficient monitoring systems and improve current treatment programs, with the aim of reducing the effects of these diseases.4–6
000 fatalities annually.2,3 In light of this situation, there has been a significant rise in studies concentrating on NTDs, driven by the imperative to develop efficient monitoring systems and improve current treatment programs, with the aim of reducing the effects of these diseases.4–6
Leishmaniasis, a neglected tropical disease (NTD) caused by parasites of the Leishmania genus, affects approximately 12 million individuals annually and is responsible for an estimated 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 30
000 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities worldwide.7,8 A review of Leishmania's life cycle is intricate, characterized by the presence of two distinct morphological forms: the intracellular, flagellated amastigote, which resides within mammalian hosts such as humans, dogs, lizards, and rodents, and the motile promastigote, which is found in the insect vector.9,10 Currently, there are no vaccines available for human use, and the pharmacological treatments employed—namely pentavalent antimonials, amphotericin B, and miltefosine—often demonstrate suboptimal efficacy, adverse toxicological effects, and require lengthy treatment regimens. As a result, patients are less likely to adhere to their treatment and develop drug-resistant strains.11–14
000 fatalities worldwide.7,8 A review of Leishmania's life cycle is intricate, characterized by the presence of two distinct morphological forms: the intracellular, flagellated amastigote, which resides within mammalian hosts such as humans, dogs, lizards, and rodents, and the motile promastigote, which is found in the insect vector.9,10 Currently, there are no vaccines available for human use, and the pharmacological treatments employed—namely pentavalent antimonials, amphotericin B, and miltefosine—often demonstrate suboptimal efficacy, adverse toxicological effects, and require lengthy treatment regimens. As a result, patients are less likely to adhere to their treatment and develop drug-resistant strains.11–14
In this regard, organizations such as the Drugs for Neglected Diseases Initiative (DNDi) are crucial for promoting research and development aimed at discovering new therapeutic approaches.15,16 It has been reported that the inhibition of dihydrofolate reductase-thymidylate synthase (DHFR-TS) leads to the compensatory action of pteridine reductase 1 (PTR1), which supplies sufficient folate to ensure the survival of the parasite.17 This observation suggests that both DHFR-TS and PTR1 should be targeted in the development of effective leishmanicidal agents. However, this hypothesis remains contentious, as the knockout of the ptr1 gene has been shown to be lethal to the parasite, likely due to the essential role of reduction in pterin, a protein exclusively produced by PTR1, in parasite development and metacyclogenesis.18,19 Consequently, certain researchers consider PTR1 to be a validated target for pharmacological development and have engaged in active efforts to inhibit its function.20,21 Notwithstanding the advancements achieved in this field, the existing literature does not sufficiently address the potential emergence of resistant strains. As a result, medicinal chemistry has made significant strides in the systematic design and refinement of novel chemical scaffolds, with the objective of developing drug candidates that exhibit enhanced safety and efficacy characteristics.22,23
The natural quinoline structure represents a prevalent chemical framework found in numerous biologically active compounds,24–27 and it represents one of the most commonly utilized cores in a variety of antimalarial (antiparasitic) drugs.28–30 It is well known that the antimalarial agent chloroquine was initially developed as a derivative of the quinoline family of compounds.31–33 Furthermore, 8-aminoquinoline derivatives have demonstrated significant leishmanicidal efficacy, exemplified by the orally bioavailable structure sitamaquine, which exhibits antileishmanial activity against various Leishmania species. The effective dose for 50% inhibition (ED50) of amastigote forms was observed to range from 2.9 to 19.0 μM.34 In recent years, tafenoquine (I) has been thoroughly researched for its potential as an antileishmanial agent.35,36 Furthermore, derivatives of 4-aminoquinoline and 7-chloroquinoline II have been identified as effective antileishmanial agents.37–41 In previous research, our team discovered a bioactive hybrid molecule that contains isatin derivatives (structure III, Fig. 1) that exhibits significant antileishmanial activity.42
In addition to quinoline, various other chemical groups have been investigated for their antiprotozoal properties, with thiosemicarbazones emerging as particularly promising candidates.43,44 Furthermore, the cyclization of thiosemicarbazones into thiazole and carbodithioate derivatives within thiadiazole frameworks has been examined as a potential antiparasitic agent, especially for the inhibition of critical metabolic pathways in pathogenic protozoa.45,46 These compounds represent a significant class of heterocyclic compounds that exhibit a variety of biological functions, such as antitumor effects, antibacterial, and anti-inflammatory, and have recently shown notable efficacy against leishmaniasis.47–54 Despite the considerable potential of thiazole and thiadiazole pharmacophores as viable antileishmanial chemotypes, there are relatively few reports identifying these derivatives as candidates for antileishmanial activity (structures IV–VII, Fig. 1).55–58 This observation highlights the need to expand the chemical landscape for antileishmanial effects by exploring conjugations within the biologically active quinoline structure and thiazole and/or thiadiazole derivatives.
In pursuit of novel antileishmanial agents and building upon prior research focused on naturally derived bioactive candidates, the target compounds were rationally designed by employing FDA-approved quinoline-containing drugs as structural templates. For instance, tafenoquine, which has demonstrated promising antileishmanial activity, served as a model. Additionally, an isosteric replacement strategy was applied, substituting pteridine or quinazoline cores59,60 known potent antifolate agents and selective inhibitors of Leishmania DHFR/PTR1 commonly found in early lead compounds with a quinoline scaffold. This scaffold was further hybridized with bioactive thiadiazole and/or thiazole moieties, fragments previously reported to exhibit antileishmanial potential (Fig. 1). The hybrid design strategically preserved the essential pharmacophoric features, especially the 7-chloro substituent and 4-amino group of the quinoline core. This strategic combination resulted in the synthesis of compounds 8a–e and 12a–d (Fig. 1), which represent a novel chemical class designed to synergize the beneficial properties of each motif. The innovative scaffold was carefully engineered to evaluate its prospective efficacy against leishmaniasis, aiming to identify new lead molecules that may contribute to the development of more effective therapeutic interventions for this neglected tropical disease. Moreover, in silico analyses and molecular dynamics simulations were performed to elucidate the interactions between the lead compounds and the active binding site, thereby offering mechanistic insights into their potential antileishmanial activity.
2. Results and discussion
2.1. Chemistry
The synthetic methods employed for creating the target hybrids are detailed in Schemes 1 and 2. The key intermediate, 1-(4-((7-chloroquinolin-4-yl)amino)phenyl)ethan-1-one (3), was synthesized by refluxing 4,7-dichloroquinoline (1) and 4-aminoacetophenone (2) in absolute ethanol in the presence of hydrochloric acid as a catalyst. Compound 3 subsequently underwent condensation with methyl hydrazinecarbo-dithioate (4) in absolute ethanol, with the addition of hydrochloric acid at room temperature, resulting in the formation of methyl-2-(1-(4-((7-chloroquinolin-4-yl)amino)phenyl)ethylidene)hydrazine-1-carbodithioate (5). This compound served as a crucial intermediate and was reacted with hydrazonoyl halide derivatives (7a–e) in absolute ethanol, utilizing triethylamine (TEA) as a catalyst, which led to the synthesis of the corresponding thiadiazole derivatives (8a–e).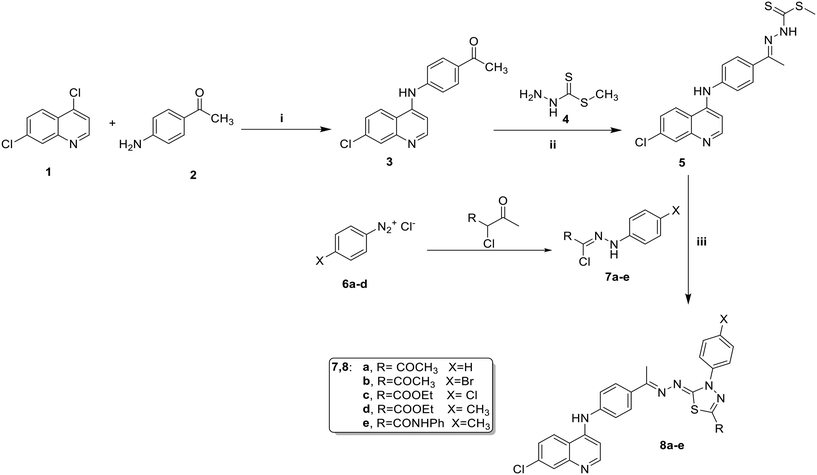 | ||
| Scheme 1 Synthesis of thidiazole derivatives 8a–e. i) EtOH, HCI, reflux, ii) EtOH, HCI, stirring, 3 h, rt, iii) EtOH, TEA, stirring, 4 h, rt. | ||
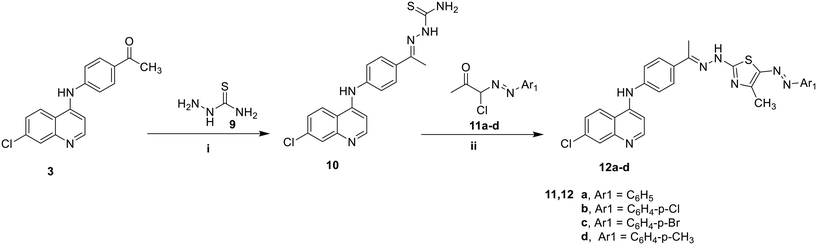 | ||
| Scheme 2 Synthesis of thiazole derivatives 12a–d, i) EtOH, AcOH, reflux, ii) Dioxane, TEA, reflux, 6 h. | ||
Additionally, thiosemicarbazone (10) was synthesized through the reaction of the key intermediate (3) with thiosemicarbazide in a mixture of ethanol and glacial acetic acid (Scheme 2). The heterocyclization of the synthesized thiosemicarbazone (10) with various hydrazonoyl chloride derivatives (11a–d) was conducted in refluxing dioxane containing a few drops of TEA, resulting in the formation of the corresponding thiazole compounds (12a–d) via an S-alkylation reaction followed by the elimination of a water or alcohol molecule.
The newly created hybrids were analyzed for their structures using various spectral techniques, such as proton nuclear magnetic resonance (1H-NMR), carbon-13 nuclear magnetic resonance (13C-NMR), and high-resolution mass spectrometry (HRMS).
2.2. Biological assessments
While antileishmanial efficacy was diminished by the presence of an acetyl group, as evidenced by compounds 8a and 8b, which exhibited IC50 values of 3.81 and 3.62 μM, respectively. These compounds demonstrated greater activity compared to the substituted anilide 8e, which showed an IC50 of 3.97 μM. Furthermore, the antileishmanial activity was also affected by the substituents on the phenyl diazo group at the C-5 position of the thiazole scaffold in derivatives 12a–d. These findings indicated that the unsubstituted phenyl diazo compound 12a exhibited the most substantial impediment against the promastigote at low micromolar levels, with an IC50 of 2.87 μM. In contrast, the activity was reduced upon substitution of the phenyl diazo group with halogen and/or methyl groups, as demonstrated by 4-chlorophenyl 12b (IC50 = 3.14 μM), 4-bromophenyl 12c (IC50 = 3.82 μM), and 4-methylphenyl 12d (IC50 = 3.82 μM), Table 1.
| Compound | X | R | Ar1 | IC50a (μM) | |
|---|---|---|---|---|---|
| Anti-promastigote | Anti-amastigote | ||||
| a IC50 values are the mean ± S.D. for three experiments. | |||||
| 5 | — | — | — | 0.52 ± 0.08 | 0.76 ± 0.06 |
| 8a | H | COCH3 | — | 3.81 ± 0.16 | 4.60 ± 0.14 |
| 8b | Br | COCH3 | — | 3.62 ± 0.18 | 5.26 ± 0.12 |
| 8c | Cl | COOC2H5 | — | 3.02 ± 0.22 | 3.88 ± 0.26 |
| 8d | CH3 | COOC2H5 | — | 3.38 ± 0.26 | 4.28 ± 0.24 |
| 8e | CH3 | CONHPh | — | 3.97 ± 0.36 | 5.62 ± 0.12 |
| 12a | — | — | C6H5 | 2.87 ± 0.26 | 4.34 ± 0.24 |
| 12b | — | — | C6H4-p-Cl | 3.14 ± 0.12 | 3.82 ± 0.14 |
| 12c | — | — | C6H4-p-Br | 3.82 ± 0.34 | 4.64 ± 0.22 |
| 12d | — | — | C6H4-p-CH3 | 3.82 ± 0.32 | 4.74 ± 0.18 |
| Miltefosine | — | — | — | 7.83 ± 0.34 | 8.07 ± 0.24 |
All compounds were then evaluated for their efficacy against the amastigote form of the parasite. Particularly, the majority of the compounds evaluated demonstrated superior antiamastigote activity compared to miltefosine, with IC50 values ranging from 0.76 μM to 5.62 μM, in contrast to miltefosine's IC50 of 8.07 μM. Consistent with the findings related to anti-promastigote activity, compound 5 showed the most pronounced inhibition of the amastigote form at low micromolar concentrations, achieving an IC50 value of 0.76 μM, which is over ten times more effective than miltefosine. This enhanced efficacy is likely attributable to the compounds' ability to optimally interact with the binding active site of PTR1. Similarly, the antiamastigote activity of the thiadiazole core in compounds 8a–e demonstrated an increase in potency in the following order: ester > acetyl > anilide, with 4-chlorophenyl derivative 8c (IC50 = 3.88 μM) being the most potent activity within the series. Furthermore, derivatives of the thiazole core (compounds 12a–d) also exhibited notable antiamastigote potency, particularly 4-chlorophenyl derivative 12b (IC50 = 3.82 μM), which was more active than the unsubstituted phenyl diazole compound 12a (IC50 = 4.34 μM) and the substituted derivatives 4-bromophenyl 12c and 4-methylphenyl 12d (IC50 = 4.64 and 4.74 μM, respectively), Table 1.
The results presented in Table 2 demonstrate the effectiveness of the antileishmanial treatment. Compound 5 was significantly reduced upon the introduction of folic acid into the experimental system, with parasite growth increasing to 81%. This observed reversal of inhibition strongly implies that the antileishmanial effects of this compound are primarily mediated through an anti-folate mechanism, likely involving the inhibition of the enzymes dihydrofolate reductase-thymidylate synthase (DHFR-TS) and pteridine reductase 1 (PTR1). To provide more support for this hypothesis, further experiments were carried out (data not shown) where extra folic acid was given to parasitic cells that had already been treated with the test compound. This approach was used to determine if the inhibition of DHFR and PTR1 could be reduced by high levels of folic acid.
| Compound | No competitor added | Folic acid | Folinic acid | ||
|---|---|---|---|---|---|
| 20 μM | 100 μM | 20 μM | 100 μM | ||
| a Percentage survival = 100 - % AP; where % AP is the percentage growth inhibition. | |||||
| 5 | 26% | 76% | 81% | 82% | 89% |
| Trimethoprim (100 μM) | 72% | — | 99% | — | — |
The selectivity indices (SI) presented in Table 3 highlighted that all the synthesized quinoline derivatives possess favorable safety profiles compared to the reference drug miltefosine. In particular, quinoline derivative 5 exhibited exceptional selectivity (SI = 410.22 for amastigotes), representing a 33-fold improvement over miltefosine (SI = 12.35). This remarkable selectivity, combined with its potent antileishmanial activity (IC50 = 0.52 μM against promastigotes and 0.76 μM against amastigotes), positions compound 5 as a highly promising lead candidate with an outstanding therapeutic window. The high SI indicates that this compound can effectively eliminate parasites at concentrations far below those causing mammalian cell toxicity.
| Compound | CC50a (μM) | SIb (amastigote) | SIc (promastigote) |
|---|---|---|---|
| a CC50 = concentration causing 50% cell death in VERO cells. b SI (amastigote) = CC50/IC50 (amastigote). c SI (promastigote) = CC50/IC50 (promastigote). | |||
| 5 | 311.77 | 410.22 | 599.56 |
| 8a | 147.55 | 31.95 | 38.72 |
| 8b | 126.89 | 24.13 | 35.02 |
| 8c | 175.44 | 45.21 | 58.09 |
| 8d | 163.11 | 38.10 | 48.25 |
| 8e | 119.34 | 21.23 | 30.06 |
| 12a | 154.53 | 35.60 | 53.84 |
| 12b | 186.98 | 48.94 | 59.55 |
| 12c | 133.66 | 28.80 | 34.98 |
| 12d | 130.56 | 27.54 | 34.18 |
| Miltefosine | 99.73 | 12.35 | 12.74 |
In addition, compounds 8c and 12b displayed the most favorable selectivity profiles with SI values of 45.21 and 48.94, respectively, representing approximately 4-fold improvements over miltefosine. All other derivatives (8a, 8b, 8d, 8e, 12a, 12c, and 12d) exhibited SI values ranging from 21.23 to 38.10, which remain 2- to 3-fold superior to miltefosine. It is worth mentioning that all the synthesized quinolines achieved SI values > 20, meeting the minimum threshold criteria for antileishmanial drug candidates and suggesting minimal off-target toxicity.
2.3. Molecular modeling
2.3.2.1. Analysis of RMSD and RMSF. To examine the conformational stability of the compound 5-LmPTR1 complex from molecular docking, a 100 ns MD simulation was performed. The root mean square deviation (RMSD) for both the ligand compound 5 and the protein LmPTR1 backbone atoms was then calculated. Fig. 2 shows the RMSD for ligand compound 5 and the LmPTR1 backbone atoms during the 100 ns MD simulation trajectory, respectively.
 | ||
| Fig. 2 RMSD curves of compound 5 (red) and LmPTR1 backbone atoms (blue) throughout the 100 ns MD simulation of the compound 5-LmPTR1 complex. | ||
Fig. 2 illustrates that the RMSD curves have stabilized, which indicates that the simulated compound 4 LmPTR1 complex has been well equilibrated. RMSD curve for ligand compound 5 stabilized at an average value of 1.53 ± 0.45 Å (Table 4). The rigidity of the quinoline core, as well as the electronegative functional groups in the hydrazine carbodithioate moiety, facilitated the stability of compound 5's atoms. The average RMSD value corresponds to the stable pose of compound 4 during a 100 ns MD simulation (below 2 Å).63
| Ligand | Average ligand RMSD (Å) | Average backbone RMSD (Å) | Average ligand RMSF (Å) | Average backbone RMSF (Å) |
|---|---|---|---|---|
| Compound 5 | 1.53 ± 0.45 | 1.99 ± 0.29 | 1.65 ± 0.31 | 1.66 ± 0.49 |
Moreover, from Fig. 2 and Table 4, it can be observed that the RMSD curves of the LmPTR1 backbone atoms converged below 2 Å, which indicates that equilibrium was established. The average RMSD backbone value for compound 5 in complex with LmPTR1 was 1.99 ± 0.29 Å during the 100 ns MD simulation. Moreover, the conformational stability of compound 5 is demonstrated by an average RMSF value of 1.65 ± 0.31 Å and is in agreement with the RMSD values presented in Table 4. From Table 4, it is also evident that average RMSF values remain below 2 Å, indicating that compound 5, as well as LmPTR1 amino acids, remain close to their initial conformations.
In Table 4, the average ligand compound 5 and LmPTR1 backbone RMSD values, as well as the average ligand and average backbone RMSF values of a 100 ns MD simulation, are provided.
To demonstrate the stability of the observed intermolecular interactions during the 100 ns MD simulation, the occupancies of the observed intermolecular interactions in the 25 ns intervals are presented in Table S2. Based on the binding mode illustrated in Fig. 3, it can be noted that compound 5, a derivative of quinoline, forms stable hydrophobic interactions with amino-acid residues Phe113 (98.30%), Leu188 (64.42%), Leu226 (67.40%), and Leu229 (78.03%) at distances of 3.32, 3.80, 3.46, and 3.57 Å, respectively. Compound 5 is further stabilized by the presence of hydrogen bonds with amino-acid residues Tyr194 (98%), Gly225 (54.32%), and His241 (88.45%) at distances of 4.07, 3.67, and 3.62 Å, respectively, as well as by a π–π stacking interaction with residue Phe113 (51.42%) at a distance of 3.95 Å.
As can be observed from Fig. 3 and Table S2, compound 5 formed beneficial interactions with the key amino acid residue Phe113 (occupancy 98.30%) and occupied the catalytic pocket facing the co-factor NADPH, forming favorable interactions with additional residues, as discussed above.23,42 Molecular docking and MD simulations, therefore, indicate the stable binding of compound 5 and support its potential to inhibit the catalytic activity of the Lm-PTR1 enzyme.
3. Conclusions
In summary, this study presents the design and synthesis of a novel series of 7-chloroquinoline/thiadiazole and thiazole hybrids, building upon our prior research aimed at developing new bioactive compounds with potential applications as antileishmanial agents. The antileishmanial activity assessment indicated that all ten synthesized compounds exhibited greater efficacy than miltefosine, with IC50 values ranging from 0.52 to 3.97 μM for promastigotes and from 0.76 to 5.62 μM for amastigotes, compared to 7.89 μM and 8.06 μM, respectively, for miltefosine. The most potent compound, designated as 5, demonstrated submicromolar activity, and its antileishmanial effects were negated when the parasites were exposed to varying concentrations of folic and folinic acids, with trimethoprim serving as a positive control, thereby confirming an antifolate mechanism of action. Furthermore, the safety profile of compound 5 was established, exhibiting a high selectivity index of 591.81 in the normal VERO cell line. Molecular docking and dynamics simulations confirmed that compound 5 has a strong and stable binding affinity for the L. major PTR1 enzyme. As a result, this class of compounds shows potential as a basis for further development and enhancement into more effective treatments for leishmaniasis.4. Experimental section
4.1. Chemistry
Melting points were measured using the Electrothermal IA 9000 apparatus, and no corrections were applied to these values. High-resolution mass spectrometry (HR-TOF-ESI-MS) data for all compounds were acquired utilizing the JEOL JMS-700 instrument, based in Tokyo, Japan. Nuclear magnetic resonance (NMR) analysis, encompassing both 1H and 13C-NMR spectra, was conducted with Bruker 500 NMR spectrometers located at the Faculty of Pharmaceutical Science, Tokushima Bunri University, Japan. Chemical shifts are reported in δ (ppm), while coupling constants are expressed in Hz. Thin-layer chromatography (TLC) was employed to monitor the reactions, utilizing silica gel on aluminum sheets (60 F254, Merck) with a chloroform/methanol (9.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 v/v) eluent, which was subsequently visualized using iodine–potassium spray. Compound 3 was synthesized according to previously established methods.65,66
0.2 v/v) eluent, which was subsequently visualized using iodine–potassium spray. Compound 3 was synthesized according to previously established methods.65,66
Methyl-2-(1-(4-((7-chloroquinolin-4-yl)amino)phenyl)ethylidene)hydrazine-1-carbodithioate (5). Yellow powder, m.p. > 300 °C, yield (167 mg, 4.18 mmol, 84%). HPLC: RT 5.77 min (purity: 99.40%); 1H NMR (500 MHz, DMSO-d6): δ = 2.42 (CH3), 2.52 (S–CH3), 6.93 (d, 1H, J = 7.0 Hz, H–Ar), 7.56 (d, 2H, J = 8.5 Hz, H–Ar), 7.80 (dd, 1H, J = 2.5 and 9.5 Hz, H–Ar), 7.99 (d, 2H, J = 8.5 Hz, H–Ar), 8.20 (d, 1H, J = 2.0 Hz, H–Ar), 8.53 (d, 1H, J = 7.0 Hz, H–Ar), 8.97 (d, 1H, J = 9.5 Hz, H–Ar), 11.41 (s, 1H, NH), 12.51 (s, 1H, NH–C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). 13C-NMR (126 MHz, DMSO) δ = 15.05 (CH3), 17.54 (CH3–S), 101.22, 116.67, 119.62, 125.30, 126.88, 127.82, 128.36, 136.27, 138.87, 139.03, 139.55, 143.85, 150.97, 154.78 (C
S). 13C-NMR (126 MHz, DMSO) δ = 15.05 (CH3), 17.54 (CH3–S), 101.22, 116.67, 119.62, 125.30, 126.88, 127.82, 128.36, 136.27, 138.87, 139.03, 139.55, 143.85, 150.97, 154.78 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 200.59 (C
N), 200.59 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). TOF-ESI-MS: [M + H]+: calcd for C19H17ClN4S2 401.0661; found 401.0662. Elemental analysis: calcd C, 56.92; H, 4.27; N, 13.97; found C, 57.11; H, 4.25; N, 14.05.
S). TOF-ESI-MS: [M + H]+: calcd for C19H17ClN4S2 401.0661; found 401.0662. Elemental analysis: calcd C, 56.92; H, 4.27; N, 13.97; found C, 57.11; H, 4.25; N, 14.05.
1-{-5-[(-1-{4-[(7-chloroquinolin-4-yl)amino]phenyl}ethylidene)hydrazineylidene]-4-phenyl-4,5-dihydro-1,3,4-thiadiazol-2-yl}ethan-1-one (8a). Yellow powder, m.p. 241–242 °C, yield (19 mg, 38 mmol, 77%). 1H NMR (500 MHz, DMSO-d6): δ = 2.44 (CH3), 2.60 (S–CH3), 7.14 (d, 1H, J = 5.5 Hz, H–Ar), 7.39–7.46 (m, 3H, H–Ar), 7.58–7.61 (m, 3H, H–Ar), 7.93 (d, 3H, J = 8.5 Hz, H–Ar), 8.07 (d, 2H, J = 7.5 Hz, H–Ar), 8.43 (d, 1H, J = 9.0 Hz, H–Ar), 8.53 (d, 1H, J = 5.5 Hz, H–Ar), 11.09 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 15.72 (CH3), 25.51 (COCH3), 103.69, 115.57, 119.23, 121.42, 122.51, 125.10, 125.71, 127.68, 128.08, 128.16, 129.63, 129.84, 132.48, 134.58, 139.28, 142.69, 147.55, 149.98, 151.13, 152.39, 160.38, 164.60 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 190.27 (C
N), 190.27 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). TOF-ESI-MS: [M + H]+: calcd for C27H22ClN6OS 513.1264; found 513.1265. Elemental analysis: calcd C, 63.21; H, 4.13; N, 16.38; found: C, 63.04; H, 4.16; N, 16.47.
O). TOF-ESI-MS: [M + H]+: calcd for C27H22ClN6OS 513.1264; found 513.1265. Elemental analysis: calcd C, 63.21; H, 4.13; N, 16.38; found: C, 63.04; H, 4.16; N, 16.47.
1-(4-(4-Bromophenyl)-5-((1-(4-((7-chloroquinolin-4-yl)amino)phenyl)ethylidene)hydrazono)-4,5-dihydro-1,3,4-thiadiazol-2-yl)ethan-1-one (8b). Yellow powder, m.p. 219–221 °C, yield (19 mg, 33 mmol, 67%). HPLC: RT 7.28 min (purity: 99.39%); 1H NMR (500 MHz, DMSO-d6): δ = 2.47 (s, 3H, CH3), 2.60 (s, 3H, S–CH3), 7.14 (brs, 1H, H–Ar), 7.44 (d, 2H, J = 7.0 Hz, H–Ar), 7.61 (d, 1H, J = 7.5 Hz, H–Ar), 7.78 (d, 2H, J = 7.5 Hz, H–Ar), 7.94 (brs, 3H, H–Ar), 8.07 (d, 2H, J = 7.5 Hz, H–Ar), 8.44 (d, 1H, J = 8.0 Hz, H–Ar), 8.54 (brs, 1H, H–Ar), 11.10 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 15.81 (CH3), 25.53 (COCH3), 103.61, 119.11, 119.82, 121.55, 124.10, 125.16, 125.83, 127.70, 128.23, 132.52, 134.80, 138.57, 142.62, 147.91, 151.52, 151.97, 160.72, 164.30 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 190.25 (C
N), 190.25 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). TOF-ESI-MS: [M + H]+: calcd for C27H20BrClN6OS 591.0369; found 591.0369. Elemental analysis: calcd: C, 54.79; H, 3.41; N, 14.20; found C, 54.98; H, 3.40; N, 14.11.
O). TOF-ESI-MS: [M + H]+: calcd for C27H20BrClN6OS 591.0369; found 591.0369. Elemental analysis: calcd: C, 54.79; H, 3.41; N, 14.20; found C, 54.98; H, 3.40; N, 14.11.
Ethyl-4-(4-chlorophenyl)-5-((1-(4-((7-chloroquinolin-4-yl)amino)phenyl)ethylidene)hydrazono)-4,5-dihydro-1,3,4-thiadiazole-2-carboxylate (8c). Yellow powder, m.p. 231–232 °C, yield (24 mg, 42 mmol, 86%). 1H NMR (500 MHz, DMSO-d6): δ = 1.33 (t, 3H, J = 7.0 Hz, CH3), 2.47 (s, 3H, CH3), 4.39 (q, 2H, J = 7.0 Hz, OCH2), 7.11 (d, 1H, J = 5.0 Hz, H–Ar), 7.48–7.49 (m, 2H, H–Ar), 7.59–7.70 (m, 3H, H–Ar), 7.97–7.99 (m, 3H, H–Ar), 8.06 (d, 2H, J = 8.5 Hz, H–Ar), 8.51–8.55 (m, 2H, H–Ar), 9.88 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 14.46 (CH3), 15.86 (CH3), 63.38 (CH2), 102.65, 118.00, 123.35, 123.45, 123.98, 125.71, 126.87, 128.35, 129.41, 129.62, 130.12, 131.55, 134.38, 138.06, 141.00, 143.76, 158.48, 160.44 (C2 of quinoline), 164.54 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 174.80 (C
N), 174.80 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). TOF-ESI-MS: [M + H]+: calcd for C28H23Cl2N6O2S 577.0980; found 577.0980. Elemental analysis: calcd: C, 58.24; H, 3.84; N, 14.55; found: C, 58.03; H, 3.87; N, 14.62.
O). TOF-ESI-MS: [M + H]+: calcd for C28H23Cl2N6O2S 577.0980; found 577.0980. Elemental analysis: calcd: C, 58.24; H, 3.84; N, 14.55; found: C, 58.03; H, 3.87; N, 14.62.
Ethyl 5-((1-(4-((7-chloroquinolin-4-yl)amino)phenyl)ethylidene)hydrazono)-4-(p-tolyl)-4,5-dihydro-1,3,4-thiadiazole-2-carboxylate (8d). Yellow powder, m.p. > 300 °C, yield (18 mg, 33 mmol, 68%). HPLC: RT 8.25 min (purity: 99.84%); 1H NMR (500 MHz, DMSO-d6): δ = 1.05 (t, 3H, J = 7.0 Hz, CH3), 2.44 (s, 3H, CH3), 2.53 (s, 3H, CH3), 4.36 (q, 2H, J = 7.0 Hz, OCH2), 6.95 (d, 1H, J = 7.0 Hz, H–Ar), 7.57–7.59 (m, 3H, H–Ar), 7.87 (dd, 2H, J = 3.0 and 10.0 Hz, H–Ar), 8.03–8.05 (m, 3H, H–Ar), 8.19 (d, 1H, J = 2.0 Hz, H–Ar), 8.55 (d, 2H, J = 7.5 Hz, H–Ar), 8.90 (d, 1H, J = 9.0 Hz, H–Ar), 11.28 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 15.06 (CH3), 17.52 (CH3), 60.96 (CH2), 101.30, 116.71, 119.80, 124.77, 125.38, 126.70, 127.95, 128.48, 130.12, 130.43, 136.41, 138.93, 139.05, 139.65, 144.07, 151.05, 154.88, 159.98, 160.82 (C2 of quinoline), 162.79 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 172.89 (C
N), 172.89 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). TOF-ESI-MS: [M + H]+: calcd for C29H26ClN6O2S 557.1526; found 557.2753. Elemental analysis: C, 62.53; H, 4.52; N, 15.09; found: C, 62.69; H, 4.50; N, 15.21.
O). TOF-ESI-MS: [M + H]+: calcd for C29H26ClN6O2S 557.1526; found 557.2753. Elemental analysis: C, 62.53; H, 4.52; N, 15.09; found: C, 62.69; H, 4.50; N, 15.21.
5-((1-(4-((7-Chloroquinolin-4-yl)amino)phenyl)ethylidene)hydrazono)-N-phenyl-4-(p-tolyl)-4,5-dihydro-1,3,4-thiadiazole-2-carboxamide (8e). Yellow powder, m.p 191–193 °C, yield (23 mg, 38 mmol, 77%). 1H NMR (500 MHz, DMSO-d6): δ = 2.39 (s, 3H, CH3), 2.45 (s, 3H, CH3), 7.11 (d, 1H, J = 5.5 Hz, H–Ar), 7.16 (t, 1H, J = 7.0 Hz, H–Ar), 7.38–7.41 (m, 4H, H–Ar), 7.45 (d, 2H, J = 9.0 Hz, H–Ar), 7.62 (dd, 1H, J = 2.5 and 9.0 Hz, H–Ar), 7.77 (d, 2H, J = 8.5 Hz, H–Ar), 7.94–7.95 (m, 3H, H–Ar), 8.10 (d, 2H, J = 8.0 Hz, H–Ar), 8.47 (d, 1H, J = 9.0 Hz, H–Ar), 8.53 (d, 1H, J = 5.5 Hz, H–Ar), 9.54 (s, 1H, NH). 10.55 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 15.68 (CH3), 21.08 (CH3), 103.32, 118.89, 121.45, 121.93, 122.20, 125.13, 125.26, 125.95, 127.05, 128.08, 129.25, 129.82, 130.11, 133.13, 135.10, 136.64, 137.23, 137.99, 142.07, 147.99, 148.46, 148.68, 151.35, 156.77 (C2 of quinoline), 159.53 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 164.92 (C
N), 164.92 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). TOF-ESI-MS: [M + H]+: calcd for C33H27ClN7OS 604.1686; found 604.1687. Elemental analysis: calcd: C, 65.61; H, 4.34; N, 16.23; found. C, 65.35; H, 4.37; N, 16.31.
O). TOF-ESI-MS: [M + H]+: calcd for C33H27ClN7OS 604.1686; found 604.1687. Elemental analysis: calcd: C, 65.61; H, 4.34; N, 16.23; found. C, 65.35; H, 4.37; N, 16.31.
7-Chloro-N-(4-(1-(2-(4-methyl-5-(phenyldiazenyl)thiazol-2-yl)hydrazono)ethyl)phenyl)quinolin-4-amine (12a). Red powder, m.p 252–253 °C, yield (23 mg, 0.45 mmol, 85%). HPLC: RT 10.63 min (purity: 99.57%); 1H NMR (500 MHz, DMSO-d6): δ = 2.52 (s, 3H, CH3), 2.59 (s, 3H, CH3), 6.98 (brs, 1H, H–Ar), 7.17 (d, 1H, J = 5.5 Hz, H–Ar), 7.31–7.37 (m, 4H, H–Ar), 7.46 (d, 2H, J = 8.5 Hz, H–Ar), 7.60–7.62 (m, 1H, H–Ar), 7.95 (d, 1H, J = 2.0 Hz, H–Ar), 8.02 (d, 2H, J = 8.0 Hz, H–Ar), 8.42 (d, 1H, J = 9.0 Hz, H–Ar), 8.55 (d, 1H, J = 5.0 Hz, H–Ar), 10.20 (s, 1H, NH). 10.57 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 14.25 (CH3), 19.02 (CH3), 103.33, 104.10, 114.67, 119.15, 119.37, 121.03, 121.44, 122.59, 125.07, 125.77, 128.25, 128.66, 129.75, 132.21, 133.04, 134.58, 138.63, 141.93, 143.58, 147.28, 147.92, 150.11, 152.51, 164.22 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 171.50 (C
N), 171.50 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C27H23ClN7S 512.1424; found 512.1425. Elemental analysis: calcd: C, 63.34; H, 4.33; N, 19.15; found: C, 63.47; H, 4.32; N, 19.08.
N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C27H23ClN7S 512.1424; found 512.1425. Elemental analysis: calcd: C, 63.34; H, 4.33; N, 19.15; found: C, 63.47; H, 4.32; N, 19.08.
7-Chloro-N-(4-(1-(2-(5-((4-chlorophenyl)diazenyl)-4-methylthiazol-2-yl)hydrazono)ethyl)phenyl)quinolin-4-amine (12b). Brown powder, m.p 242–243 °C, yield (20 mg, 0.37 mmol, 69%). 1H NMR (500 MHz, DMSO-d6): δ = 2.31 (s, 3H, CH3), 2.58 (s, 3H, CH3), 7.05 (d, 1H, J = 5.5 Hz, H–Ar), 7.35–7.38 (m, 3H, H–Ar), 7.46 (d, 1H, J = 8.5 Hz, H–Ar), 7.58–7.65 (m, 1H, H–Ar), 7.92–7.95 (m, 2H, H–Ar), 7.98 (d, 2H, J = 8.5 Hz, H–Ar), 8.02 (d, 1H, J = 8.5 Hz, H–Ar), 8.41–8.44 (m, 1H, H–Ar), 8.51 (d, 1H, J = 5.0 Hz, H–Ar), 10.20 (s, 1H, NH). 10.54 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 14.26 (CH3), 15.52 (CH3), 103.28, 104.10, 119.09, 119.34, 121.06, 121.52, 125.04, 125.67, 125.81, 127.94, 128.26, 128.69, 129.62, 130.49, 132.17, 133.12, 134.61, 134.64, 141.87, 147.84, 147.92, 149.81, 152.23, 164.50 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 171.26 (C
N), 171.26 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C27H22Cl2N7S 546.1034; found 546.1025. Elemental analysis: calcd: C, 59.34; H, 3.87; N, 17.94; found: C, 59.51; H, 3.85; N, 17.88.
N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C27H22Cl2N7S 546.1034; found 546.1025. Elemental analysis: calcd: C, 59.34; H, 3.87; N, 17.94; found: C, 59.51; H, 3.85; N, 17.88.
N-(4-(1-(2-(5-((4-Bromophenyl)diazenyl)-4-methylthiazol-2-yl)hydrazono)ethyl)phenyl)-7-chloroquinolin-4-amine (12c). Brown powder, m.p 237–238 °C, yield (26 mg, 0.45 mmol, 84%). HPLC: RT 14.86 min (purity: 98.11%); 1H NMR (500 MHz, DMSO-d6): δ = 2.31 (s, 3H, CH3), 2.59 (s, 3H, CH3), 7.06 (d, 1H, J = 5.5 Hz, H–Ar), 7.36 (d, 2H, J = 9.0 Hz, H–Ar), 7.46–7.50 (m, 2H, H–Ar), 7.59–7.63 (m, 1H, H–Ar), 7.92–7.95 (m, 2H, H–Ar), 7.98 (d, 2H, J = 8.5 Hz, H–Ar), 8.02 (d, 1H, J = 8.5 Hz, H–Ar), 8.41–8.44 (m, 1H, H–Ar), 8.51 (d, 1H, J = 5.5 Hz, H–Ar), 10.20 (s, 1H, NH), 10.44 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 14.25 (CH3), 19.01 (CH3), 103.28, 104.11, 116.57, 119.08, 119.34, 121.07, 121.54, 125.05, 125.09, 125.69, 125.83, 127.92, 128.27, 128.70, 132.49, 133.13, 134.62, 134.66, 141.86, 147.38, 147.86, 147.93, 149.79, 152.23, 164.54 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 171.24 (C
N), 171.24 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C27H22BrClN7S 590.0529; found 590.0522. Elemental analysis: calcd: C, 54.88; H, 3.58; N, 16.59; found: C, 55.04; H, 3.57; N, 16.45.
N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C27H22BrClN7S 590.0529; found 590.0522. Elemental analysis: calcd: C, 54.88; H, 3.58; N, 16.59; found: C, 55.04; H, 3.57; N, 16.45.
7-Chloro-N-(4-(1-(2-(4-methyl-5-(p-tolyldiazenyl)thiazol-2-yl)hydrazono)ethyl)phenyl)quinolin-4-amine (12d). Red powder, m.p 188–190 °C, yield (21 mg, 0.40 mmol, 75%). 1H NMR (500 MHz, DMSO-d6): δ = 2.25 (s, 3H, CH3), 2.52 (s, 3H, CH3), 2.58 (s, 3H, CH3), 7.13–7.18 (m, 3H, H–Ar), 7.25 (d, 2H, J = 8.0 Hz, H–Ar), 7.45 (d, 2H, J = 8.5 Hz, H–Ar), 7.61–7.63 (m, 1H, H–Ar), 7.98–8.02 (m, 2H, H–Ar), 8.42 (d, 2H, J = 9.0 Hz, H–Ar), 8.56 (d, 1H, J = 5.0 Hz, H–Ar), 9.37 (s, 1H, NH). 10.52 (s, 1H, NH). 13C NMR (126 MHz, DMSO) δ = 15.49 (CH3), 19.00 (CH3), 20.85 (CH3), 103.30, 104.06, 114.69, 119.32, 121.10, 121.52, 125.08, 125.81, 128.07, 128.27, 128.65, 130.20, 131.64, 132.31, 134.64, 138.02, 141.65, 143.51, 147.40, 149.96, 152.42, 164.04 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–), 171.63 (C
N–), 171.63 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C28H25ClN7S 526.1581; found 526.1572. Elemental analysis: calcd; C, 63.93; H, 4.60; N, 18.64; found: C, 64.08; H, 4.57; N, 18.54.
N-thiazole). TOF-ESI-MS: [M + H]+ calcd for C28H25ClN7S 526.1581; found 526.1572. Elemental analysis: calcd; C, 63.93; H, 4.60; N, 18.64; found: C, 64.08; H, 4.57; N, 18.54.
4.2. Biology
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells. The plates were subsequently incubated for 72 hours at 37 °C, under conditions of 95% humidity and 5% CO2. All experimental procedures adhered to established methodologies as documented in prior literature.62,69
000 cells. The plates were subsequently incubated for 72 hours at 37 °C, under conditions of 95% humidity and 5% CO2. All experimental procedures adhered to established methodologies as documented in prior literature.62,69
4.3. Molecular modeling
The docking employed DOCK.prm parameters, utilizing the rDOCK scoring function (SF3), which encompasses a sampling methodology that integrates a three-stage genetic algorithm search, low-temperature Monte Carlo simulations, and simplex minimization, executed over 100 iterations.73 The efficacy of rDOCK's sampling and scoring was validated against various target proteins73,74 and RNA,73,75 demonstrating its superior performance relative to other open-source software.76 The complex structure of compound 4 with Lm-PTR1, which yielded the lowest docking score, was subsequently selected for further molecular dynamics (MD) simulations.
To mitigate potential steric clashes and to refine the atomic coordinates of the protein-ligand complex, an initial optimization was performed involving 50 steps of steepest descent followed by 50 steps of adopted basis Newton–Raphson energy minimization on the combined coordinate files of the Lm-PTR1 protein and associated water molecules. Subsequently, a 0.125 ns equilibration molecular dynamics (MD) simulation was conducted within the NVT ensemble, employing the HOOVER thermostat and an integration timestep of 1 fs at a temperature of 310.15 K. Following this, a 100 ns production run was executed in the NPT ensemble under periodic boundary conditions conditions with harmonic constraints on the center of mass of ligand and the protein binding site aminoacids in the radius of 6 Å. During this phase, the HOOVER thermostat and barostat were maintained at 310.15 K and 1 bar, respectively, with a timestep of 2 fs. The van der Waals interactions were truncated using the force switch method (VFSWIt) at distances between 10 and 12 Å, while the electrostatic potential was similarly truncated at 12 Å using the force shifting method (FSHIft). Long-range electrostatic interactions were computed utilizing the particle mesh Ewald summation technique.84 Additionally, the SHAKE algorithm was employed to constrain the bonds involving hydrogen atoms.
The coordinate trajectory was visualized utilizing the VMD85 and Pymol86 software applications. An automated protein–ligand interaction profiler (PLIP) was employed to ascertain the most stable intermolecular interactions involving compound 4-Lm-PTR1.87,88 The root-mean-square deviations (RMSD) for both the protein backbone and the ligand were computed using the Python library MD Analysis.89 Furthermore, the root-mean-square fluctuations (RMSF) of the ligand and protein atoms throughout the molecular dynamics (MD) simulation were also determined.89 In addition, the MD Analysis library was utilized to evaluate the occupancies of intermolecular interactions for all atoms located within a 5.5 Å radius of the protein and ligand.
Author contributions
Huda R. M. Rashdan: conceptualization, investigation, validation, writing – review & editing; Adnan A. Bekhit: methodology, investigation, writing – review & editing; Veronika Furlan: formal analysis, investigation, writing – original draft; Kikuko Amagase: methodology, investigation, writing – review & editing; Abdelsamed I. Elshamy: formal analysis, investigation, writing – review & editing; Nourhan Elfar: methodology, data curation, visualization; Mohamed. R. Abdo: methodology, data curation, visualization; Tamer M. Ibrahim: validation, writing – review & editing; Urban Bren: methodology, investigation, writing – review & editing; Wagdy M. Eldehna: conceptualization, investigation, writing – original draft, writing – review & editing; Ahmed Sabt: conceptualization, investigation, validation, supervision, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors alone are responsible for the content and writing of the paper.Data availability
ALL required data inserted in manuscript and supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5md00709g.References
- T. K. Mackey, B. A. Liang, R. Cuomo, R. Hafen, K. C. Brouwer and D. E. Lee, Emerging and reemerging neglected tropical diseases: a review of key characteristics, risk factors, and the policy and innovation environment, Clin. Microbiol. Rev., 2014, 27(4), 949–979 CrossRef PubMed.
- World Health Organization, Global report on neglected tropical diseases 2023, World Health Organization, 2023 Search PubMed.
- B. Baral, K. Mamale, S. Gairola, C. Chauhan, A. Dey and R. K. Kaundal, Infectious diseases and its global epidemiology, in Nanostructured Drug Delivery Systems in Infectious Disease Treatment, Academic Press, 2024, pp. 1–24 Search PubMed.
- A. K. Ng'etich, K. Voyi and C. M. Mutero, Development and validation of a framework to improve neglected tropical diseases surveillance and response at sub-national levels in Kenya, PLoS Neglected Trop. Dis., 2021, 15(10), e0009920 CrossRef PubMed.
- C. M. Wolfe, A. Barry, A. Campos, B. Farham, D. Achu, E. Juma, A. Kalu and B. Impouma, Control, elimination, and eradication efforts for neglected tropical diseases in the World Health Organization African region over the last 30 years: A scoping review, Int. J. Infect. Dis., 2024, 141, 106943 CrossRef PubMed.
- M. P. Namratha, P. Mittal, A. Sharma and V. Mittal, Strategies to Overcome the Impact of Neglected Diseases on the World, in Emerging Approaches to Tackle Neglected Diseases: From Molecule to End Product, 2024, pp. 16–29 Search PubMed.
- S. S. Santos, R. V. de Araujo, J. Giarolla, O. El Seoud and E. I. Ferreira, Searching for drugs for Chagas disease, leishmaniasis and schistosomiasis: A review, Int. J. Antimicrob. Agents, 2020, 55(4), 105906 CrossRef CAS.
- World Health Organization (WHO), World health statistics 2018: monitoring health for the SDGs, sustainable development goals, World Health Organization (WHO), Geneva, 2018, Licence: CC BY-NC-SA 3.0 IGO. IGO Search PubMed.
- P. Albajar-Vinas and J. Jannin, The hidden Chagas disease burden in Europe, Eurosurveillance, 2011, 16(38), 19975 CrossRef PubMed.
- D. M. Walker, S. Oghumu, G. Gupta, B. S. McGwire, M. E. Drew and A. R. Satoskar, Mechanisms of cellular invasion by intracellular parasites, Cell. Mol. Life Sci., 2014, 71(7), 1245–1263 CrossRef CAS.
- K. Chanda, An overview on the therapeutics of neglected infectious diseases—Leishmaniasis and Chagas diseases, Front. Chem., 2021, 9, 622286 CrossRef.
- A. Greco, R. Karki, Y. Gadiya, C. Deecke, A. Zaliani and S. Gul, Pharmacological profiles of neglected tropical disease drugs, Artif. Intell. Life Sci., 2024, 6, 100116 CAS.
- F. T. Akinsolu, P. O. Nemieboka, D. W. Njuguna, M. N. Ahadji, D. Dezso and O. Varga, Emerging resistance of neglected tropical diseases: A scoping review of the literature, Int. J. Environ. Res. Public Health, 2019, 16, 1925 CrossRef PubMed.
- G. Joshi, S. S. Quadir and K. S. Yadav, Road map to the treatment of neglected tropical diseases: Nanocarriers interventions, J. Controlled Release, 2021, 339, 51–74 CrossRef CAS PubMed.
- K. K. Maheshwari and D. Bandyopadhyay, Heterocycles in the Treatment of Neglected Tropical Diseases, Curr. Med. Chem., 2021, 28, 472–495 CrossRef CAS.
- J. A. Shiran, B. Kaboudin, N. Panahi and N. Razzaghi-Asl, Privileged small molecules against neglected tropical diseases: A perspective from structure activity relationships, Eur. J. Med. Chem., 2024, 271, 116396 CrossRef.
- A. Bhattacharya, P. Leprohon and M. Ouellette, Combined gene deletion of dihydrofolate reductase-thymidylate synthase and pteridine reductase in Leishmania infantum, PLoS Neglected Trop. Dis., 2021, 15(4), e0009377 CrossRef.
- B. V. F. Teixeira, A. L. B. Teles, S. G. D. Silva, C. C. B. Brito, H. F. Freitas, A. B. L. Pires, T. Q. Froes and M. S. Castilho, Dual and selective inhibitors of pteridine reductase 1 (PTR1) and dihydrofolate reductase-thymidylate synthase (DHFR-TS) from Leishmania chagasi, J. Enzyme Inhib. Med. Chem., 2019, 34(1), 1439–1450 CrossRef CAS PubMed.
- M. Sezavar, I. Sharifi, P. Ghasemi Nejad Almani, B. Kazemi, N. Davoudi, S. Salari, E. Salarkia, A. Khosravi and M. Bamorovat, The potential therapeutic role of PTR1 gene in non-healing anthroponotic cutaneous leishmaniasis due to Leishmania tropica, J. Clin. Lab. Anal., 2021, 35(3), e23670 CrossRef CAS PubMed.
- S. Sundar and B. Singh, Emerging therapeutic targets for treatment of leishmaniasis, Expert Opin. Ther. Targets, 2018, 22(6), 467–486 CrossRef CAS.
- A. Shtaiwi, Thiadiazine-thiones as inhibitors of leishmania pteridine reductase (PTR1) target: Investigations and in silico approach, J. Biomol. Struct. Dyn., 2024, 42(16), 8588–8597 CrossRef CAS.
- L. S. Goupil and J. H. McKerrow, Introduction: Drug discovery and development for neglected diseases, Chem. Rev., 2014, 114, 11131–11137 CrossRef CAS.
- N. W. Hassan, A. Sabt, M. A. El-Attar, M. Ora, A. E. Bekhit, K. Amagase, A. A. Bekhit, A. S. Belal and P. A. Elzahhar, Modulating leishmanial pteridine metabolism machinery via some new coumarin-1, 2, 3-triazoles: Design, synthesis and computational studies, Eur. J. Med. Chem., 2023, 253, 115333 CrossRef CAS.
- A. Sabt, M. H. Abdulla, M. S. Ebaid, J. Pawełczyk, H. A. Abd El Salam, N. T. Son, N. X. Ha, M. A. Vaali Mohammed, T. Traiki, A. E. Elsawi and B. Dziadek, Identification of 2-(N-aryl-1, 2, 3-triazol-4-yl) quinoline derivatives as antitubercular agents endowed with InhA inhibitory activity. Frontiers, Chemistry, 2024, 12, 1424017 CAS.
- A. Sabt, E. F. Khaleel, M. A. Shaldam, M. S. Ebaid, R. M. Badi, A. K. Allayeh, W. M. Eldehna and J. Dziadek, Discovery of new quinoline derivatives bearing 1-aryl-1, 2, 3-triazole motif as influenza H1N1 virus neuraminidase inhibitors, Bioorg. Chem., 2024, 151, 107703 CrossRef CAS PubMed.
- A. Sabt, M. Abdelraof, M. F. Hamissa and M. A. Noamaan, Antibacterial activity of quinoline-based derivatives against methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa: design, synthesis, DFT and molecular dynamic simulations, Chem. Biodiversity, 2023, 20(11), e202300804 CrossRef CAS.
- B. Kumaraswamy, K. Hemalatha, R. Pal, G. S. Matada, K. R. Hosamani, I. Aayishamma and N. V. Aishwarya, An insight into sustainable and green chemistry approaches for the synthesis of quinoline derivatives as anticancer agents, Eur. J. Med. Chem., 2024, 116561 CrossRef CAS.
- R. Mishra, J. da Cunha Xavier, N. Kumar, G. Krishna, P. K. Dhakad, H. S. dos Santos, P. N. Bandeira, T. H. Soares Rodrigues, D. R. Gondim, W. H. Ribeiro and D. Sales da Silva, Exploring Quinoline Derivatives: Their Antimalarial Efficacy and Structural Features, Med. Chem., 2025, 21(2), 96–121 CrossRef CAS PubMed.
- C. Peron, R. S. Gonçalves and S. Moura, Advances in the Potential of Quinoline-Derived Metal Complexes as Antimalarial Agents: A Review, Appl. Organomet. Chem., 2025, 39(3), e70050 CrossRef CAS.
- S. B. Srinivasa, S. N. Ullal and B. S. Kalal, Quinoline conjugates for enhanced antimalarial activity: a review on synthesis by molecular hybridization and structure-activity relationship (SAR) investigation, Am. J. Transl. Res., 2025, 17(2), 1335 CrossRef.
- N. R. Rossi, S. N. Fialho, A. de Jesus Gouveia, A. S. Ferreira, M. A. da Silva, L. D. Martinez, W. D. do Nascimento, A. Gonzaga Jr., D. S. de Medeiros, N. B. de Barros and R. de Cássia Alves, Quinine and chloroquine: Potential preclinical candidates for the treatment of tegumentary leishmaniasis, Acta Trop., 2024, 252, 107143 CrossRef CAS.
- F. Malik, M. M. Hanif and G. Mustafa, Comparing the efficacy of oral chloroquine versus oral Tetracycline in the treatment of cutaneous leishmaniasis, J. Coll. Physicians Surg. Pak., 2019, 29, 403–405 CrossRef PubMed.
- L. Herrera, A. Llanes, J. Álvarez, K. Degracia, C. M. Restrepo, R. Rivera, D. E. Stephens, H. T. Dang, O. V. Larionov, R. Lleonart and P. L. Fernández, Antileishmanial activity of a new chloroquine analog in an animal model of Leishmania panamensis infection, Int. J. Parasitol.: Drugs Drug Resist., 2020, 14, 56–61 Search PubMed.
- T. Garnier, M. B. Brown, M. J. Lawrence and S. L. Croft, In-vitro and in-vivo studies on a topical formulation of sitamaquine dihydrochloride for cutaneous leishmaniasis, J. Pharm. Pharmacol., 2006, 58, 1043–1054 CrossRef CAS.
- V. Yardley, F. Gamarro and S. L. Croft, Antileishmanial and antitrypanosomal activities of the 8-aminoquinoline tafenoquine, Antimicrob. Agents Chemother., 2010, 54, 5356–5358 CrossRef CAS.
- S. I. Shah, F. Nasir, N. S. Malik, M. Alamzeb, M. Abbas, I. U. Rehman, F. Khuda, Y. Shah, K. W. Goh, A. Zeb and L. C. Ming, Efficacy evaluation of 10-hydroxy chondrofoline and tafenoquine against leishmania tropica (HTD7), Pharmaceuticals, 2022, 15, 1005 CrossRef CAS.
- M. Yousuf, D. Mukherjee, A. Pal, S. Dey, S. Mandal, C. Pal and S. Adhikari, Synthesis and biological evaluation of ferrocenylquinoline as a potential antileishmanial agent, ChemMedChem, 2015, 10, 546–554 CrossRef CAS PubMed.
- E. S. Coimbra, R. Carvalhaes, R. M. Grazul, P. A. Machado, M. V. De Souza and A. D. Da Silva, Synthesis, cytotoxicity and antileishmanial activity of some N-(2-(indol-3-yl) ethyl)-7-chloroquinolin-4-amines, Chem. Biol. Drug Des., 2010, 75, 628–631 CrossRef CAS PubMed.
- E. Valdivieso, F. Mejías, C. Torrealba, G. Benaim, V. V. Kouznetsov, F. Sojo, F. A. Rojas-Ruiz, F. Arvelo and F. Dagger, In vitro 4-Aryloxy-7-chloroquinoline derivatives are effective in mono- and combined therapy against Leishmania donovani and induce mitocondrial membrane potential disruption, Acta Trop., 2018, 183, 36–42 CrossRef CAS PubMed.
- A. H. Romero, N. Rodríguez, S. E. López and H. Oviedo, Identification of dehydroxy isoquine and isotebuquine as promising antileishmanial agents, Arch. Pharm., 2019, 352, e1800281 CrossRef.
- J. Konstantinović, M. Videnović, S. Orsini, K. Bogojević, S. D'Alessandro, D. Scaccabarozzi, N. Terzić Jovanović, L. Gradoni, N. Basilico and B. A. Šolaja, Novel aminoquinoline derivatives significantly reduce parasite Load in leishmania infantum infected mice, ACS Med. Chem. Lett., 2018, 9, 629–634 CrossRef.
- A. Sabt, W. M. Eldehna, T. M. Ibrahim, A. A. Bekhit and R. Z. Batran, New antileishmanial quinoline linked isatin derivatives targeting DHFR-TS and PTR1: Design, synthesis, and molecular modeling studies, Eur. J. Med. Chem., 2023, 246, 114959 CrossRef CAS PubMed.
- A. Ibáñez-Escribano, C. Fonseca-Berzal, M. Martínez-Montiel, M. Álvarez-Márquez, M. Gómez-Núñez, M. Lacueva-Arnedo, T. Espinosa-Buitrago, T. Martín-Pérez, J. A. Escario, P. Merino-Montiel and S. Montiel-Smith, Thio-and selenosemicarbazones as antiprotozoal agents against Trypanosoma cruzi and Trichomonas vaginalis, J. Enzyme Inhib. Med. Chem., 2022, 37(1), 781–791 CrossRef PubMed.
- T. M. de Aquino, P. H. França, É. E. Rodrigues, I. J. Nascimento, P. F. Santos-Júnior, P. G. Aquino, M. S. Santos, A. C. Queiroz, M. V. Araújo, M. S. Alexandre-Moreira and R. R. Rodrigues, Synthesis, antileishmanial activity and in silico studies of Aminoguanidine Hydrazones (AGH) and Thiosemicarbazones (TSC) against Leishmania chagasi amastigotes, Med. Chem., 2022, 18(2), 151–169 CrossRef CAS.
- P. S. Tada da Cunha, A. L. Rodriguez Gini, C. Man Chin, J. L. Dos Santos and C. B. Scarim, Recent Progress in Thiazole, Thiosemicarbazone, and Semicarbazone Derivatives as Antiparasitic Agents Against Trypanosomatids and Plasmodium spp, Molecules, 2025, 30(8), 1788 CrossRef PubMed.
- T. M. Almutairi, N. Rezki, M. R. Aouad, M. Hagar, B. A. Bakr, M. T. Hamed, M. K. Hassen, B. H. Elwakil and E. A. Moneer, Exploring the antiparasitic activity of tris-1, 3, 4-thiadiazoles against Toxoplasma gondii-infected mice, Molecules, 2022, 27(7), 2246 CrossRef CAS PubMed.
- M. S. Ebaid, H. A. Ibrahim, A. F. Kassem and A. Sabt, Recent studies on protein kinase signaling inhibitors based on thiazoles: review to date, RSC Adv., 2024, 14(50), 36989–37018 RSC.
- A. Petrou, M. Fesatidou and A. Geronikaki, Thiazole ring—A biologically active scaffold, Molecules, 2021, 26(11), 3166 CrossRef CAS PubMed.
- T. A. Farghaly, G. H. Alfaifi and S. M. Gomha, Recent literature on the synthesis of thiazole derivatives and their biological activities, Mini-Rev. Med. Chem., 2024, 24(2), 196–251 CrossRef CAS.
- C. M. Queiroz, G. B. de Oliveira Filho, J. W. Espíndola, A. V. do Nascimento, A. S. Aliança, V. M. de Lorena, A. P. Feitosa, P. R. da Silva, L. C. Alves, A. C. Leite and F. A. Brayner, Thiosemicarbazone and thiazole: In vitro evaluation of leishmanicidal and ultrastructural activity on Leishmania infantum, Med. Chem. Res., 2020, 29, 2050–2065 CrossRef.
- A. Tahghighi and F. Babalouei, Thiadiazoles: The appropriate pharmacological scaffolds with leishmanicidal and antimalarial activities: A review, Iran. J. Basic Med. Sci., 2017, 20(6), 613 Search PubMed.
- H. R. Rashdan, M. T. Abdelrahman, A. C. De Luca and M. Mangini, Towards a new generation of hormone therapies: Design, synthesis and biological evaluation of novel 1, 2, 3-triazoles as estrogen-positive breast cancer therapeutics and non-steroidal aromatase inhibitors, Pharmaceuticals, 2024, 17(1), 88 CrossRef CAS PubMed.
- (a) H. R. Rashdan and A. H. Abdelmonsef, Towards Covid-19 TMPRSS2 enzyme inhibitors and antimicrobial agents: Synthesis, antimicrobial potency, molecular docking, and drug-likeness prediction of thiadiazole-triazole hybrids, J. Mol. Struct., 2022, 15(1268), 133659 CrossRef; (b) H. R. Rashdan and A. H. Abdelmonsef, In silico study to identify novel potential thiadiazole-based molecules as anti-Covid-19 candidates by hierarchical virtual screening and molecular dynamics simulations, Struct. Chem., 2022, 33(5), 1727–1739 CrossRef CAS PubMed.
- H. R. Rashdan, A. H. Abdelmonsef, M. M. Abou-Krisha and T. A. Yousef, Synthesis, identification, computer-aided docking studies, and ADMET prediction of novel benzimidazo-1, 2, 3-triazole based molecules as potential antimicrobial agents, Molecules, 2021, 26(23), 7119 CrossRef CAS PubMed.
- A. Mijoba, N. Parra-Giménez, E. Fernandez-Moreira, H. Ramírez, X. Serrano, Z. Blanco, S. Espinosa and J. E. Charris, Synthesis of Hybrid Molecules with Imidazole-1,3,4-thiadiazole Core and Evaluation of Biological Activity on Trypanosoma cruzi and Leishmania donovani, Molecules, 2024, 29, 4125 CrossRef CAS PubMed.
- A. Tahghighi, F. R. Marznaki, F. Kobarfard, S. Dastmalchi, J. S. Mojarrad and S. Razmi, et al., Synthesis and antileishmanial activity of novel 5-(5-nitrofuran-2-y1)-1,3,4-thiadiazoles with piperazinyl-linked benzamidine substituents, Eur. J. Med. Chem., 2011, 46, 2602–2608 CrossRef CAS PubMed.
- V. V. G. de Oliveira, M. A. A. de Souza, R. R. M. Cavalcanti, M. V. de Oliveira Cardoso, A. C. L. Leite, V. A. da Silva Junior and R. C. B. Q. de Figueiredo, Study of in vitro biological activity of thiazoles on Leishmania (Leishmania)infantum, J. Global Antimicrob. Resist., 2020, 22, 414–421 CrossRef.
- M. Haroon, T. Akhtar, H. Mehmood, A. C. da Silva Santos, J. M. da Conceição, G. L. Brondani, R. D. Silva Tibúrcio, D. C. Galindo Bedor, J. W. Viturino da Silva, P. A. Sales Junior and V. R. Alves Pereira, Synthesis of hydrazinyl–thiazole ester derivatives, in vitro trypanocidal and leishmanicidal activities, Future Med. Chem., 2024, 16(3), 221–238 CrossRef CAS PubMed.
- A. H. Romero, N. Rodríguez and H. Oviedo, 2-Aryl-quinazolin-4(3H)-ones as an inhibitor of leishmania folate pathway: in vitro biological evaluation, mechanism studies and molecular docking, Bioorg. Chem., 2019, 83, 145–153 CrossRef CAS.
- K. S. Van Horn, X. Zhu, T. Pandharkar, S. Yang, B. Vesely, M. Vanaerschot, J. C. Dujardin, S. Rijal, D. E. Kyle, M. Z. Wang, K. A. Werbovetz and R. Manetsch, Antileishmanial activity of a series of N2 ,N4 -disubstituted quinazoline-2,4- diamines, J. Med. Chem., 2014, 57, 5141–5156 CrossRef CAS.
- C. Mendoza-Martínez, N. Galindo-Sevilla, J. Correa-Basurto, V. M. Ugalde-Saldivar, R. G. Rodríguez-Delgado, J. Hernández-Pineda, C. Padierna-Mota, M. Flores-Alamo and F. Hernández-Luis, Antileishmanial activity of quinazoline derivatives: synthesis, docking screens, molecular dynamic simulations and electrochemical studies, Eur. J. Med. Chem., 2015, 92, 314–331 CrossRef.
- M. Tonelli, E. Gabriele, F. Piazza, N. Basilico, S. Parapini, B. Tasso, R. Loddo, F. Sparatore and A. Sparatore, Benzimidazole derivatives endowed with potent antileishmanial activity, J. Enzyme Inhib. Med. Chem., 2018, 33, 210–226 CrossRef CAS.
- K. Liu and H. Kokubo, Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations: a cross-docking study, J. Chem. Inf. Model., 2017, 57, 2514–2522 CrossRef CAS.
- S. Salentin, S. Schreiber, V. J. Haupt, M. F. Adasme and M. Schroeder, PLIP: fully automated protein–ligand interaction profiler, Nucleic Acids Res., 2015, 43, W443–W447 CrossRef CAS PubMed.
- T. Al-Warhi, A. Sabt, M. Korycka-Machala, A. F. Kassem, M. A. Shaldam, H. A. Ibrahim, M. Kawka, B. Dziadek, M. Kuzioła, W. M. Eldehna and J. Dziadek, Benzenesulfonohydrazide-tethered non-fused and fused heterocycles as potential anti-mycobacterial agents targeting enoyl acyl carrier protein reductase (InhA) with antibiofilm activity, RSC Adv., 2024, 14(41), 30165–30179 RSC.
- R. Z. Batran, S. M. El-Daly, W. A. El-Kashak and E. Y. Ahmed, Design, synthesis, and molecular modeling of quinoline-based derivatives as anti-breast cancer agents targeting EGFR/AKT signaling pathway, Chem. Biol. Drug Des., 2022, 99(3), 470–482 CrossRef CAS.
- T. M. Ibrahim, G. Abada, M. Dammann, R. M. Maklad, W. M. Eldehna, R. Salem, M. M. Abdelaziz, R. A. El-Domany, A. A. Bekhit and F. M. Beockler, Tetrahydrobenzo [h] quinoline derivatives as a novel chemotype for dual antileishmanial-antimalarial activity graced with antitubercular activity: Design, synthesis and biological evaluation, Eur. J. Med. Chem., 2023, 257, 115534 CrossRef CAS PubMed.
- A. A. Bekhit, M. N. Saudi, A. M. Hassan, S. M. Fahmy, T. M. Ibrahim, D. Ghareeb, A. M. El-Seidy, S. M. Al-Qallaf, H. J. Habib and A. E. A. Bekhit, Synthesis, molecular modeling and biological screening of some pyrazole derivatives as antileishmanial agents, Future Med. Chem., 2018, 10, 2325–2344 CrossRef CAS.
- W. M. Eldehna, H. Almahli, T. M. Ibrahim, M. Fares, T. Al-Warhi, F. M. Boeckler, A. A. Bekhit and H. A. Abdel-Aziz, Synthesis, in vitro biological evaluation and in silico studies of certain arylnicotinic acids conjugated with aryl (thio) semicarbazides as a novel class of anti-leishmanial agents, Eur. J. Med. Chem., 2019, 179, 335–346 CrossRef CAS PubMed.
- M. C. Teixeira, R. de Jesus Santos, R. B. Sampaio, L. Pontes-de-Carvalho and W. L. dosSantos, A simple and reproducible method to obtain large numbers of axenic amastigotes of different Leishmania species, Parasitol. Res., 2002, 88, 963–968 CrossRef PubMed.
- L. Quiñonez-Díaz, J. Mancilla-Ramírez, M. Avila-García, J. Ortiz-Avalos, A. Berron, S. González, Y. Paredes and N. Galindo-Sevilla, Effect of ambient temperature on the clinical manifestations of experimental diffuse cutaneous leishmaniasis in a rodent model, Vector-Borne Zoonotic Dis., 2012, 12, 851–860 CrossRef.
- S. Lešnik, M. Jukič and U. Bren, Mechanistic insights of polyphenolic compounds from Rosemary bound to their protein targets obtained by molecular dynamics simulations and free-energy calculations, Foods, 2023, 12, 408 CrossRef.
- S. Ruiz-Carmona, D. Alvarez-Garcia, N. Foloppe, A. B. Garmendia-Doval, S. Juhos, P. Schmidtke, X. Barril, R. E. Hubbard and S. D. Morley, rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids, PLoS Comput. Biol., 2014, 10, e1003571 CrossRef PubMed.
- M. J. Hartshorn, M. L. Verdonk, G. Chessari, S. C. Brewerton, W. T. Mooij, P. N. Mortenson and C. W. Murray, Diverse, high-quality test set for the validation of protein− ligand docking performance, J. Med. Chem., 2007, 50, 726–741 CrossRef CAS PubMed.
- S. D. Morley and M. Afshar, Validation of an empirical RNA-ligand scoring function for fast flexible docking using RiboDock®, J. Comput.-Aided Mol. Des., 2004, 18, 189–208 CrossRef CAS PubMed.
- D. Soler, Y. Westermaier and R. Soliva, Extensive benchmark of rDock as a peptide-protein docking tool, J. Comput.-Aided Mol. Des., 2019, 33, 613–626 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, Scalable Molecular Dynamics with NAMD, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- J. C. Phillips, D. J. Hardy, J. D. C. Maia, J. E. Stone, J. V. Ribeiro, R. C. Bernardi, R. Buch, G. Fiorin, J. Hénin and W. Jiang, et al., Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD, J. Chem. Phys., 2020, 153, 044130 CrossRef CAS PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, CHARMM-GUI: a web-based graphical user interface for CHARMM, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- J. Huang and A. D. MacKerell Jr., CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data, J. Comput. Chem., 2013, 34, 2135–2145 CrossRef CAS PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. De Groot, H. Grubmüller and A. D. MacKerell Jr., CHARMM36m: an improved force field for folded and intrinsically disordered proteins, Nat. Methods, 2017, 14, 71–73 CrossRef CAS PubMed.
- H. M. Khan, A. D. MacKerell Jr. and N. Reuter, Cation-π interactions between methylated ammonium groups and tryptophan in the CHARMM36 additive force field, J. Chem. Theory Comput., 2018, 15, 7–12 CrossRef.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes and I. Vorobyov, CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields, J. Comput. Chem., 2010, 31, 671–690 CrossRef CAS.
- T. Darden, L. Perera, L. Li and L. Pedersen, New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations, Structure, 1999, 7, R55–R60 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, VMD: visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS.
- W. L. DeLano, Pymol: An open-source molecular graphics tool, CCP4 Newsletter on protein crystallography, 2002, 40, 82–92.
- M. F. Adasme, K. L. Linnemann, S. N. Bolz, F. Kaiser, S. Salentin, V. J. Haupt and M. Schroeder, PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA, Nucleic Acids Res., 2021, 49, W530–W534 CrossRef CAS PubMed.
- S. Salentin, S. Schreiber, V. J. Haupt, M. F. Adasme and M. Schroeder, PLIP: fully automated protein–ligand interaction profiler, Nucleic Acids Res., 2015, 43, W443–W447 CrossRef CAS PubMed.
- R. J. Gowers, M. Linke, J. Barnoud, T. J. E. Reddy, M. N. Melo, S. L. Seyler, J. Domanski, D. L. Dotson, S. Buchoux and I. M. Kenney, MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations, Los Alamos National Lab.(LANL), Los Alamos, NM (United States), 2019, pp. 2575–9752 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |