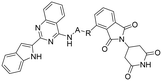
Design and evaluation of an HSP70-targeting PROTAC in synergy with an HSF1 inhibitor for enhanced antitumor activity
Wei-Hao
Huang
,
Teng-Yu
Mao
,
Guo-Yao
Dai
,
Jian-Mei
Ye
,
Jia-Bao
Li
,
Shuo-Bin
Chen
 ,
Jia-Heng
Tan
,
Jia-Heng
Tan
 ,
Zhi-Shu
Huang
and
Shi-Liang
Huang
,
Zhi-Shu
Huang
and
Shi-Liang
Huang
 *
*
School of Pharmaceutical Sciences, Guangdong Provincial Key Laboratory of New Drug Design and Evaluation, Sun Yat-sen University, Guangzhou 510006, China. E-mail: lsshsl@mail.sysu.edu.cn
First published on 18th November 2025
Abstract
Heat shock protein (HSP) 70 represents a clinically promising anti-tumor target, yet the development of effective inhibitors faces numerous technical challenges. To address this, we developed novel non-ATP site Proteolysis-targeting Chimeras (PROTACs) that selectively degrade HSP70 by engaging the E3 ubiquitin ligase CRBN. However, the PROTACs exhibited limited degradation activity, potentially due to heat shock response-mediated HSP70 upregulation. To circumvent this resistance mechanism, we explored combination therapy with the heat shock factor 1 (HSF1) inhibitor DTHIB to disrupt the heat shock feedback loop, markedly enhancing HSP70 degradation. The combination strategy showed synergistic and selective anti-tumor activity across a panel of cancer cell lines. This success relied on the distinct profile of C4, which preferentially targets cytosolic HSP70 and, unlike conventional inhibitors, effectively circumvents compensatory HSP70 upregulation.
Introduction
70 kDa heat shock proteins (HSP70s) are a class of highly conserved molecular chaperones that are crucial for maintaining cellular proteostasis, modulating cellular stress responses, and suppressing apoptosis.1 HSP70 overexpression in malignancies drives tumor initiation, progression, and metastasis by promoting cell survival, enhancing drug resistance, and inhibiting apoptosis.2–5 Consequently, HSP70 is considered as an important target for anti-tumor therapy. However, developing effective inhibitors remains challenging due to several major obstacles: first, the exceptionally high affinity of the HSP70 nucleotide-binding domain (NBD) for ATP, coupled with millimolar intracellular ATP concentrations, impedes effective competition for the ATP-binding site.6 Second, high conformational dynamics and incomplete structural characterization hinder rational design of allosteric modulators targeting allosteric sites.5,7 Third, conventional active-site inhibitors often induce compensatory HSP70 accumulation, triggering stress responses and fostering resistance via binding-site mutations.8–13 These constraints severely limit the advancement of HSP70 inhibitors. Notably, the high abundance of HSP70 in tumor cells and its multiple homologous isoforms (e.g., HSP70α, HSP70β, HSP76, BiP) present additional challenges for selective targeting, as these isoforms play distinct roles in tumor survival and drug resistance.Proteolysis-targeting Chimera (PROTAC) technology offers a promising alternative by recruiting E3 ubiquitin ligases to ubiquitinate and degrade target proteins.14–16 This strategy overcomes key limitations of occupancy-driven inhibitors: it requires only transient target engagement (not necessarily high-affinity binding to functional sites) and reduces mutation-driven resistance by eliminating the target protein.17,18 Therefore, developing HSP70 degraders based on the PROTAC strategy is considered as a promising anti-tumor approach. Nonetheless, HSP70-targeted PROTAC therapy still faces major technical difficulties: (1) extremely high HSP70 abundance, particularly within the tumor microenvironment, may saturate degrader capacity. (2) Chronic HSP70 inhibition activates HSF1-mediated feedback, upregulating chaperone expression and diminishing long-term efficacy. (3) Traditional HSP70 ATP-site binders might exhibit binding affinity for other ATP-dependent targets; PROTACs might effectively degrade even low-affinity off-targets, potentially compromising selectivity. Combining HSP70 degraders with HSF1 inhibitors offers a strategy to address these issues. HSF1 inhibition blocks the HSP70 negative feedback loop, potentially enhancing anti-tumor efficacy. Furthermore, identifying non-ATP site HSP70 binders is critical for improving selectivity.
In this study, compound R17 was identified through DSF screening and confirmed as a non-ATP site HSP70 binder. Its derivatives were used as warheads to design and synthesize a series of small-molecule PROTACs capable of degrading HSP70. To enhance the degradation effect, the HSF1 inhibitor DTHIB was employed to block the heat shock feedback loop, aiming to achieve more effective suppression of HSP70 and provide a novel therapeutic strategy for tumor treatment.
Results and discussion
Design of HSP70 small-molecule degraders
In our preliminary screening, compound R17, a bouchardatine analog, was identified from our in-house compound library via differential scanning fluorimetry (DSF) (Fig. 1a–d). R17 significantly reduced the peak fluorescence intensity of HSP70 but did not alter its melting temperature (Tm). This result indicated that R17 binds to HSP70, yet this binding does not affect the protein's thermal stability or the hydrophobic unfolding process. Further analysis revealed that R17 had no effect on HSP70-mediated ATP hydrolysis or the refolding of denatured luciferase protein (Fig. 1e and f), indicating that it binds to a non-ATP, non-allosteric site on HSP70. Notably, R17 contains an unstable aldehyde group. However, its derivative deCHOR-17, lacking the aldehyde, and the parent compound 3, with the dimethylamino side chain removed, both retained HSP70-binding activity in DSF assays. Therefore, 3 was selected as the warhead for the development of HSP70-targeting PROTACs, and the position bearing the chloro substituent was chosen as the linker attachment site for conjugation to an E3 ligase ligand.Although over 600 E3 ligases exist, only about six are commonly used in PROTACs. Among these, CRBN,19,20 VHL,21 MDM2,22 and cIAP23 are the most widely applied. Thalidomide, as a ligand for the E3 ligase CRBN, is widely used in the design of PROTAC molecules due to its advantages such as low molecular weight and easy synthesis. Based on the differences in the type of substituent groups introduced at position 1 of thalidomide, as well as variations in the length and properties of the linker, we designed three series comprising a total of ten PROTAC compounds (Table 1 and Scheme 3):
| Comp. | A | R | Degradation ratea (%) | Cytotoxicityb (inhibition rate, %) | Synergistic cytotoxicityc (inhibition rate, %) |
|---|---|---|---|---|---|
| a The degradation rates of HSP70 were measured by western blot assay. Values were the means ± SEM of three independent experiments. b The effects of individual compounds (8 μM) on RKO cell viability after 48 h treatment. The inhibition rates are shown as means ± SEM of three independent experiments. c The effects of compounds (8 μM) in combination with DTHIB (4 μM) on RKO cell viability after 48 h treatment. The inhibition rates are shown as means ± SEM of three independent experiments. d n.d.: no degradation. | |||||
| A1 |
|
|
n.d.d | 9 ± 1 | 38 ± 4 |
| A2 |
|
|
n.d. | 6 ± 1 | 55 ± 4 |
| A3 |
|
|
n.d. | 12 ± 3 | 53 ± 6 |
| B1 |
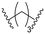
|
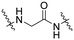
|
21 ± 4 | 30 ± 5 | 78 ± 2 |
| B2 |
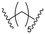
|
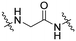
|
n.d. | 17 ± 3 | 57 ± 8 |
| B3 |
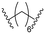
|
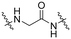
|
n.d. | 15 ± 1 | 56 ± 8 |
| C1 |
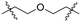
|
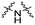
|
n.d. | 21 ± 3 | 66 ± 3 |
| C2 |
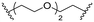
|
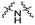
|
n.d. | 9 ± 2 | 50 ± 3 |
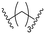
| C3 |
|
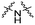
|
19 ± 1 | 5 ± 5 | 52 ± 1 |
| C4 |
|
|
56 ± 1 | 26 ± 4 | 78 ± 5 |
| DTHIB | — | — | 20 ± 3 | 44 ± 2 | — |
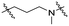
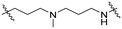
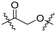
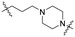
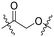
Series A: retained the R17N,N-dimethyl side chain and conjugated it to thalidomide via alkyloxy linkers using different diamines (N-(3-aminopropyl)-N-methylpropane-1,3-diamine, 3-(piperazin-1-yl) propan-1-amine, or N-methylpropane-1,3-diamine). This approach preserved the R17 structure while systematically investigating the effect of linker length on degradation activity.
Series B: utilized pomalidomide (a CRBN ligand) connected via amide linkers of varying alkyl chain lengths. This series aimed to assess the impact of removing the R17N,N-dimethyl side chain on HSP70 degradation activity.
Series C: employed alkyl or PEG chains as flexible linkers, replacing the amide bond with an alkylamine connection point, aiming to maximize linker flexibility, enhancing the opportunity for the protein of interest (POI) to engage the ubiquitin ligase complex.24
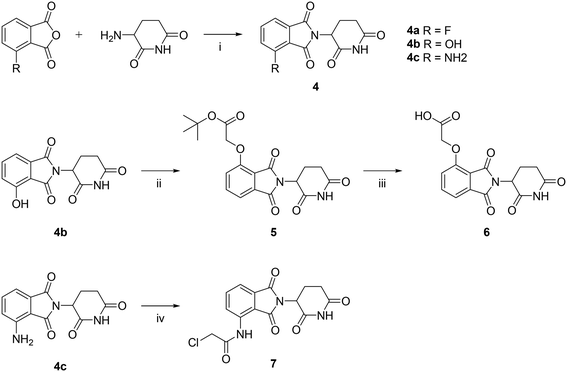
Synthesis of degrader compounds
The synthesis of the starting material deCHOR-17 derivative and the thalidomide derivative are shown in Schemes 1 and 2, through which compounds 3, 6, and 7 were obtained. | ||
| Scheme 1 The synthetic route for compound 3. (i) a. SOCl2, chloroform, 70 °C, 4 h. b. Pyridine, chloroform, 70 °C, 2 h; (ii) NaOH, EtOH, 70 °C, 2 h; (iii) POCl3, 120 °C, 6 h. | ||
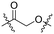
As shown in Scheme 3, the synthetic routes for the three series of target compounds, A, B, and C, all started from 3, which underwent nucleophilic substitution with the –NH2 group of different linkers (L1–L5). The resulting intermediates were then treated with TFA to remove the Boc protecting group at the terminal end of the linker. Subsequent nucleophilic substitution or amide coupling with intermediates 6, 7, or 4a afforded the final compounds of series A, B, and C, respectively.
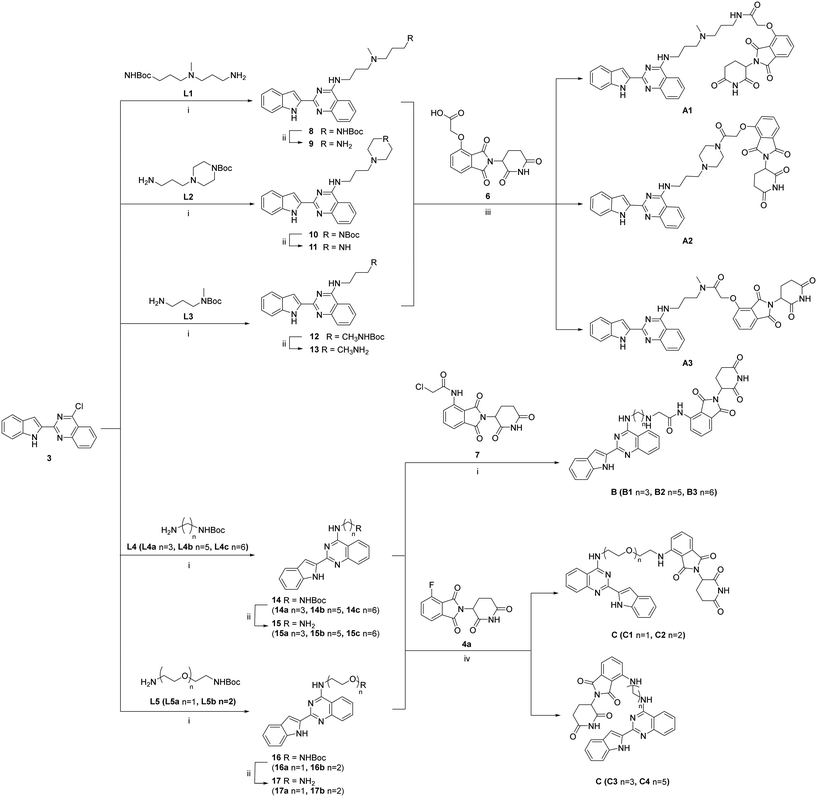 | ||
| Scheme 3 The synthetic route for compound series A, B and C. (i) K2CO3, DMF, r.t., 2 h; (ii) TFA, DCM, r.t., 2 h; (iii) HATU, DIPEA, DMF, r.t., 2 h. (iv) NMP, DIPEA, 60 °C, 30 min. | ||
The negative control compound C4-Me, was synthesized analogously from methylation of 4a (Scheme 4).
 | ||
| Scheme 4 The synthetic route for the negative control compound. (i) Iodomethane, K2CO3, DMF, r.t., overnight; (ii) NMP, DIPEA, 60 °C, 30 min. | ||
Evaluation of the activity and summary of structure–activity relationships of the synthesized compounds
We first performed western blot analysis to screen for cell lines with high expression of HSP70 (Fig. 2a and b). HSP70 protein levels were significantly elevated in RKO cells compared to those in LX-2, NCM460, H-358, and MIA-Paca-2 cells. Therefore, RKO cells were selected for subsequent experiments.The degradation efficiency of the synthesized PROTAC compounds on HSP70 and their cytotoxic activity against the RKO cell line were evaluated by western blot and CCK-8 assay, respectively. The results are shown in Fig. 2c and d and Table 1. The structure–activity relationships (SARs) of the synthesized HSP70-targeting PROTAC compounds revealed that the efficiency is highly dependent on the type of linker group attached to thalidomide and the properties and length of the linker. Compounds with alkylamine linkers (series C) exhibited the highest HSP70 degradation rates, with C4 reaching 56% degradation and exhibiting 26% cytotoxicity at 8 μM. Aminocarbonyl linkers (series B) showed weaker degradative activity (21% for B1) with a corresponding cytotoxicity of 21%. Alkoxy linkers (series A) exhibited almost no protein degradation and cytotoxicity. The optimal linker length appears to be 5–8 atoms, balancing flexibility and steric considerations, which is crucial for effective ternary complex formation and subsequent degradation. Additionally, the presence of heteroatoms or bulky groups in the linker can introduce geometric constraints or non-productive binding sites, potentially hindering complex formation and reducing both degradation efficiency and cytotoxicity. These findings have underscored the importance of linker composition and length in modulating the efficacy of PROTACs targeting HSP70.
Based on these results, compounds B1 and C4 were selected for further mechanistic investigation. Immunoblotting confirmed that R17 alone did not reduce intracellular HSP70 levels (Fig. 3a and b). The degradation efficiency of B1 and C4 against HSP70 was assessed at various concentrations after 48 h. B1 showed only weak activity (Fig. 3c and d), whereas C4 exhibited a concentration-dependent degradation effect (Fig. 3e and f). Time-course analysis revealed that treatment with 16 μM C4 led to noticeable HSP70 degradation after 48 h, with further reduction observed at 72 h (Fig. 3g and h). Importantly, the negative control C4-Me failed to induce HSP70 degradation at either 16 or 32 μM (Fig. 3i and j), supporting the conclusion that C4 mediates HSP70 degradation via recruitment of the E3 ubiquitin ligase through its thalidomide-derived moiety.
Mechanistic study and HSP70 isoform selectivity of the lead compound C4
To investigate the mechanism of C4-mediated HSP70 degradation, competition assays were conducted using either R17 (HSP70 binder) or thalidomide (CRBN ligand). Immunoblotting (Fig. 4a and b) showed that both compounds significantly blocked HSP70 degradation by C4, suggesting that they occupy the respective binding sites and prevent the formation of the ternary complex required for ubiquitination. Additionally, co-treatment with the proteasome inhibitor MG132 also inhibited degradation, confirming that C4 acts through the proteasomal pathway. These findings demonstrated that C4-dependent HSP70 degradation relies on simultaneous engagement of both HSP70 and CRBN, followed by proteasomal degradation. To assess ternary complex formation, a pull-down assay was performed using HSP70 antibody-conjugated magnetic beads. Western blot analysis (Fig. 4c and d) showed that C4 effectively recruited CRBN to HSP70. However, this interaction was blocked by co-treatment with R17 or thalidomide, indicating competitive binding and disruption of complex assembly. To provide definitive genetic evidence for CRBN dependence, we performed CRBN knockdown using siRNA. As shown in Fig. 4e and f, CRBN depletion substantially impaired C4-mediated HSP70 degradation, confirming the essential role of CRBN in this process. These results confirmed that C4 facilitates the formation of an HSP70–C4–CRBN ternary complex, consistent with the mechanistic conclusions drawn above.Co-expression of Flag-HSP70 and HA-Ub in RKO cells showed that C4 significantly increased HSP70 ubiquitination (Fig. 4g and h). Consistent with this, total ubiquitin levels also showed an upward trend (Fig. 4i and j), further supporting the induction of proteasome-dependent degradation. These data confirmed that C4 promotes ubiquitin-dependent degradation of HSP70 through a PROTAC-mediated mechanism.
Given the high sequence homology among HSP70 family members and their distinct roles in cellular processes, we evaluated the isoform selectivity of C4. As shown in Fig. 4k and l, C4 demonstrated degradation activity against cytosolic HSP70 isoforms HSP70α, HSP70β, HSP76 and BiP at 8 μM, respectively, but showed minimal effect on the endoplasmic reticulum (ER)-resident BiP. This selectivity correlates with the genetic homology: HSP70β shares >99% sequence identity with HSP70α, HSP76 shares ∼92%, while BiP shares only ∼81%. The observed differences likely reflect structural variations at the R17 binding site. Importantly, the preserved BiP function may help maintain a therapeutic window, as full disruption of ER proteostasis could cause significant toxicity in normal cells. Such isoform selectivity offers valuable guidance for optimizing HSP70-targeted therapies and supports the potential for isoform-specific targeting strategies.
Synergistic enhancement of HSP70 degradation and antitumor activity by C4 in combination with DTHIB
As a stress-induced and highly abundant chaperone protein, HSP70 is not only constitutively expressed at high levels in tumor cells, but also further upregulated in response to external stimuli.3 The relatively low degradation efficiency of our synthesized compounds may be attributed to the overwhelming intracellular abundance of HSP70 and its compensatory upregulation through negative feedback mechanisms. Previous studies suggested that co-targeting HSP70 and its upstream regulator HSF1 could effectively suppress this stress response and enhance PROTAC efficacy through synergistic inhibition.25Consistent with this hypothesis, RT-qPCR (Fig. 5a) showed that C4 treatment alone significantly upregulated HSP70 mRNA, reflecting activation of the heat shock feedback loop triggered by HSP70 degradation. This compensatory response substantially limits the efficacy of single-agent PROTAC therapy, as evidenced by the moderate degradation efficiency despite effective ternary complex formation. In contrast, RT-qPCR and immunoblotting (Fig. 5b and c) demonstrated that co-treatment with C4 and DTHIB markedly suppressed HSP70 transcription and enhanced its degradation relative to C4 alone, indicating that DTHIB effectively blocks HSF1-mediated feedback. We also directly compared C4 with representative ATP-competitive (VER-155008) and allosteric (MKT-077) HSP70 inhibitors. Both conventional inhibitors induced significant upregulation of HSP70 protein levels, consistent with the activation of the heat shock response pathway. Notably, DTHIB at 4 μM had minimal impact on HSP70 levels when administered alone, but in combination with C4, it strongly potentiated protein degradation. Similarly, as presented in Table 1, a synergistic cytotoxic effect was observed when the synthesized compounds were co-administered with DTHIB. This result indicates that the PROTAC-based strategy effectively circumvented the compensatory HSP70 upregulation that limits the efficacy of conventional inhibitors. This comparative analysis demonstrates that targeted protein degradation, particularly when combined with HSF1 inhibition, represents a superior strategy for overcoming the intrinsic resistance mechanisms associated with HSP70 targeting.
Cell growth inhibition assays using a high-content imaging system (Fig. 5d) further demonstrated that the combination of C4 and DTHIB strongly suppresses RKO cell proliferation in a concentration- and time-dependent manner. We expanded our evaluation to include two additional cancer cell lines (HL60 leukemia and HCT116 colon cancer) and one normal cell line (NCM460). Cell viability assays revealed a pronounced therapeutic window for the C4/DTHIB combination (Fig. 5e–g). Time-course analysis demonstrated that tumor cells exhibited accelerated cell death after 72 hours of combination treatment, consistent with proteostasis disruption, whereas normal cells showed minimal toxicity within the therapeutic window. Notably, DTHIB concentrations below 2 μM showed minimal synergistic activity across all cell lines, while the 4–8 μM range demonstrated a steep dose–response curve, corresponding to the concentration range required for effective disruption of the HSF1-mediated feedback loop.
To further evaluate the biological consequences of this combination treatment, we assessed apoptosis induction via immunoblotting and flow cytometry. Immunoblot analysis (Fig. 5h–k) showed that combined treatment led to a concentration-dependent decrease in full-length PARP and caspase-3, along with increased cleaved caspase-3, indicating enhanced apoptotic activity. Flow cytometry with PI/Annexin V staining (Fig. 5l–n) confirmed a significant increase in both early and late apoptotic cells following combination treatment, with an elevated PI signal suggesting an increase in mid-to-late-stage apoptosis. These findings collectively demonstrated that the combination of C4 and DTHIB synergistically enhance HSP70 degradation, suppress tumor cell proliferation, and effectively induce apoptosis.
Conclusion
HSP70 is a critical yet challenging target in cancer therapy due to its high ATP-binding affinity, conformational flexibility, and activation of compensatory stress responses, all of which hinder conventional inhibitor development. In this study, we introduce a novel strategy to overcome these limitations through PROTAC-mediated targeted protein degradation. We developed C4, the first PROTAC degrader that targets HSP70 via a non-active site binding mode, enabling selective and efficient degradation of this oncoprotein.Crucially, we show that combining C4 with the HSF1 inhibitor DTHIB significantly enhances its efficacy by disrupting the heat shock feedback loop, which otherwise limits HSP70 degradation. This synergistic combination not only improves degradation efficiency but also triggers robust antitumor effects in cancer cells.
Our results establish a new paradigm for targeting HSP70 and other feedback-regulated oncoproteins. Although C4 demonstrated moderate single-agent activity, our research provides a robust foundation for understanding the challenges and opportunities in HSP70-targeted therapy. Further optimization is needed to enhance C4's potency and pharmacological properties, particularly to improve its selectivity profile among HSP70 isoforms. Nonetheless, this work establishes a compelling proof-of-concept for a dual-targeting strategy that simultaneously degrades HSP70 and disrupts its compensatory feedback loop, offering a promising framework for overcoming tumor survival mechanisms and potentially applicable to other traditionally ‘undruggable’ targets with similar resistance mechanisms.
Author contributions
S. H. and W. H. conceived the study and designed the research. W. H., T. M., and G. D. performed the experiments. W. H., T. M., J. Y. and J. L. analyzed the data. S. H. and W. H. wrote the original draft of the paper. S. C., J. T., Z. H. and S. H. supervised the project. All authors critically reviewed and approved the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5md00726g.Acknowledgements
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2025A1515010764, 2024A1515011751) and the National Natural Science Foundation of China (22377156).Notes and references
- A. Tissieres, H. K. Mitchell and U. M. Tracy, J. Mol. Biol., 1974, 84, 389–398 CrossRef CAS PubMed.
- D. R. Ciocca and S. K. Calderwood, Cell Stress Chaperones, 2005, 10, 86–103 CrossRef CAS PubMed.
- D. R. Ciocca, G. M. Clark, A. K. Tandon, S. A. Fuqua, W. J. Welch and W. L. McGuire, J. Natl. Cancer Inst., 1993, 85, 570–574 CrossRef CAS PubMed.
- K. Hellman, A. A. Alaiya, K. Schedvins, W. Steinberg, A. C. Hellstrom and G. Auer, Br. J. Cancer, 2004, 91, 319–326 CrossRef CAS PubMed.
- M. E. Murphy, Carcinogenesis, 2013, 34, 1181–1188 CrossRef CAS PubMed.
- A. J. Massey, J. Med. Chem., 2010, 53, 7280–7286 CrossRef CAS.
- R. Rosenzweig, N. B. Nillegoda, M. P. Mayer and B. Bukau, Nat. Rev. Mol. Cell Biol., 2019, 20, 665–680 CrossRef CAS PubMed.
- X. Cao, Y. Zhou, H. Sun, M. Xu, X. Bi, Z. Zhao, B. Shen, F. Wan, Z. Hong, L. Lan, L. Luo, Z. Guo and Z. Yin, Cancer Lett., 2018, 424, 84–96 CrossRef CAS PubMed.
- I. D. Ferguson, Y. T. Lin, C. Lam, H. Shao, K. M. Tharp, M. Hale, C. Kasap, M. C. Mariano, A. Kishishita, B. Patino Escobar, K. Mandal, V. Steri, D. Wang, P. Phojanakong, S. T. Tuomivaara, B. Hann, C. Driessen, B. Van Ness, J. E. Gestwicki and A. P. Wiita, Cell Chem. Biol., 2022, 29(1288–1302), e1287 Search PubMed.
- Y. Li, J. Kang, J. Fu, H. Luo, Y. Liu, Y. Li and L. Sun, Int. J. Mol. Sci., 2021, 22, 2537 CrossRef CAS PubMed.
- F. Wang, C. Xu, G. Li, P. Lv and J. Gu, Exp. Cell Res., 2021, 409, 112910 CrossRef CAS PubMed.
- X. Yang, J. Wang, Y. Zhou, Y. Wang, S. Wang and W. Zhang, Cancer Lett., 2012, 321, 137–143 CrossRef PubMed.
- C. W. Yun, H. J. Kim, J. H. Lim and S. H. Lee, Cells, 2019, 9, 60 CrossRef PubMed.
- M. Bekes, D. R. Langley and C. M. Crews, Nat. Rev. Drug Discovery, 2022, 21, 181–200 CrossRef CAS PubMed.
- A. C. Lai and C. M. Crews, Nat. Rev. Drug Discovery, 2017, 16, 101–114 CrossRef CAS PubMed.
- Y. Zou, D. Ma and Y. Wang, Cell Biochem. Funct., 2019, 37, 21–30 CrossRef CAS.
- K. C. Carmony and K. B. Kim, Methods Mol. Biol., 2012, 832, 627–638 CrossRef CAS PubMed.
- C. Shi, H. Zhang, P. Wang, K. Wang, D. Xu, H. Wang, L. Yin, S. Zhang and Y. Zhang, Cell Death Dis., 2019, 10, 815 CrossRef PubMed.
- J. Lu, Y. Qian, M. Altieri, H. Dong, J. Wang, K. Raina, J. Hines, J. D. Winkler, A. P. Crew, K. Coleman and C. M. Crews, Chem. Biol., 2015, 22, 755–763 CrossRef CAS.
- G. E. Winter, D. L. Buckley, J. Paulk, J. M. Roberts, A. Souza, S. Dhe-Paganon and J. E. Bradner, Science, 2015, 348, 1376–1381 CrossRef CAS PubMed.
- C. Galdeano, M. S. Gadd, P. Soares, S. Scaffidi, I. Van Molle, I. Birced, S. Hewitt, D. M. Dias and A. Ciulli, J. Med. Chem., 2014, 57, 8657–8663 CrossRef CAS PubMed.
- C. Wang, Y. Zhang, Y. Wu and D. Xing, Eur. J. Med. Chem., 2021, 225, 113749 CrossRef CAS PubMed.
- N. Ohoka, K. Okuhira, M. Ito, K. Nagai, N. Shibata, T. Hattori, O. Ujikawa, K. Shimokawa, O. Sano, R. Koyama, H. Fujita, M. Teratani, H. Matsumoto, Y. Imaeda, H. Nara, N. Cho and M. Naito, J. Biol. Chem., 2017, 292, 4556–4570 CrossRef CAS PubMed.
- T. A. Bemis, J. J. La Clair and M. D. Burkart, J. Med. Chem., 2021, 64, 8042–8052 CrossRef CAS PubMed.
- S. M. Xu, X. Z. Liu, L. Wang, W. H. Huang, Y. T. Hu, S. B. Chen, Z. S. Huang and S. L. Huang, Biochim. Biophys. Acta, Mol. Basis Dis., 2025, 1871, 167630 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |