Open Access Article
Open Access ArticleSelf-assembling protein cages: from coiled-coil module to machine learning-driven de novo design of next-generation biomaterials
Arvind Kumar
Gupta
a,
Hana
Esih
a,
Helena
Gradišar
a and
Roman
Jerala
 *ab
*ab
aDepartment of Synthetic Biology and Immunology, National Institute of Chemistry, Ljubljana SI-1000, Slovenia
bEN-FIST Centre of Excellence, Ljubljana SI-1000, Slovenia. E-mail: Roman.Jerala@ki.si
First published on 23rd December 2025
Abstract
The rational design of self-assembling protein nanocages holds great promise for synthetic biology, biotechnology and biomedical applications. Protein nanocages are well-defined nanoparticles with an inner cavity formed by self-assembly of repetitive protein building blocks. These cavities can be tailored to encapsulate and protect cargo molecules such as drugs, enzymes, or imaging agents. The ability to design de novo protein cages has recently been revolutionized by new concepts of modular protein design, computational design of interacting surfaces and machine learning-based generative protein design. Protein cages can be designed in diverse architectures and sizes, and their assembly and disassembly can be regulated by chemical, biological, and physical signals. Here, we focus on the review of engineering strategies for the designed protein cages based on coiled coils or other modular protein domains, their functionalization and opportunities of customized engineered protein cages.
1. Introduction
The emergence of self-assembling protein nanostructures has opened new avenues in biomaterials, with applications spanning nanomedicine, drug delivery, vaccine development, enzymatic catalysis, and synthetic biology. Ranging from 10 to 200 nm in diameter, these nanocages possess internal cavities that could be used to encapsulate therapeutic molecules and reactive sites amenable to functionalization. Naturally occurring protein cages such as virus-like particles (VLPs), ferritin, heat-shock proteins, and chaperonins have been repurposed for biomedical and biotechnological applications, owing to their well-defined architectures and biocompatibility.1,2 These systems offer valuable insights into how symmetry, interface complementarity, and non-covalent interactions contribute to the stability of multimeric assemblies. Inspired by these natural systems, synthetic nanocages have been designed by precisely arranging protein subunits into diverse shapes and enabling the controlled encapsulation and targeted delivery of diverse molecular cargoes.3,4De novo design strategies allow the construction of user-defined protein nanocages with tailored geometries and functional surfaces. In recent years, approaches have relied on a combination of computational tools that operate at different stages of the design process. Rosetta provides a versatile framework for backbone remodeling and sequence optimization, with specialized protocols such as HBNet enabling the design of stabilizing hydrogen-bond networks at protein–protein interfaces.5,6 However, the most critical advances in nanocage design have come from the development of symmetry-aware rigid-body docking platforms, with RPXDock unifying and extending earlier methods such as tcDock, sicDock, and sicAxel.7–11 These algorithms systematically sample symmetric configurations of protein subunits, allowing the construction of highly ordered architectures with precise control over symmetry and assembly.7–11 In parallel, recent advances in machine learning-based generative design have streamlined and accelerated de novo protein design workflows, complementing established physics-based methods such as Rosetta and enabling more efficient exploration of diverse scaffold geometries.12–15
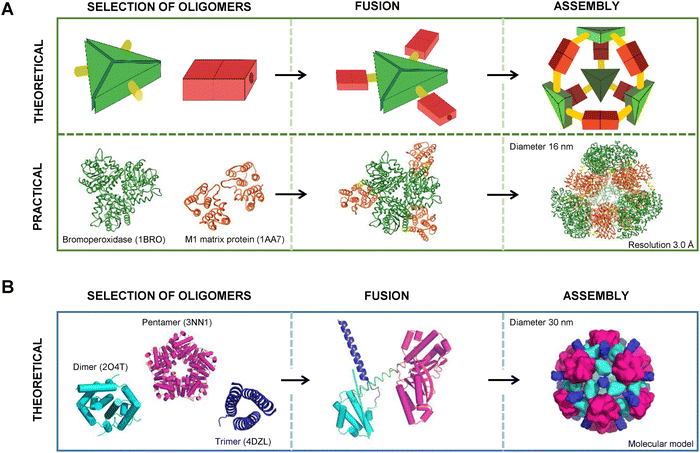
Depending on the design strategy, protein nanocages fall into four main categories: assemblies based on genetic fusion of natural oligomeric proteins; structures derived from computational interface design; geometries built using coiled-coil modules and machine learning-based generative protein cage design. In the genetic fusion approach, naturally oligomerizing protein domains are connected via rigid helical linkers to produce symmetric complexes.16–18 Computationally designed interfaces offer excellent control and precision, enabling the assembly of complex symmetrical cages from one or several different protein components.6,19–21 A transformative advance in this approach has been provided by machine learning/artificial intelligence (ML/AI). Tools such as ProteinMPNN enable rapid optimization of interface residues and large-scale sequence screening, supporting the design of megadalton-scale nanocages with improved assembly efficiency, stability, and solubility.13–15 While diffusion-based backbone generators are promising in other areas of protein design, their application to protein cage-level backbone generation remains limited and is under active development.12,22,23
Parallel to these developments, coiled-coil domains have emerged as a powerful modular platform for designing nanocages. The modularity and orthogonality of coiled-coil interactions enable precise control over nanocage geometry, size, and responsiveness.24,25 A highly useful tool is the CoCoPOD (coiled-coil protein origami design) platform, which uses orthogonal coiled-coil dimers and higher oligomeric modules arranged in a defined order to fold into single-chain polyhedral cages.26 Recent advances in computational modeling and protein engineering have expanded the capabilities of coiled-coil nanocages, enabling direct programming of folding pathways through stimuli-responsive modules. These modules are engineered structural elements that undergo conformational changes or trigger assembly and disassembly in response to specific environmental cues, such as pH, metal ions, redox conditions, temperature, or light. By incorporating such features, one can achieve dynamic and reversible control over nanocage formation, resulting in highly adaptable and customizable architectures suited for responsive biomedical applications.27,28
This review highlights recent advances in the rational and computational design of protein nanocages, particularly coiled coil-based modular architectures. We explore their applications in drug delivery, immunotherapy, and diagnostics, and discuss current challenges, including immune compatibility and stability. As design strategies continue to integrate synthetic biology and AI-guided optimization, protein nanocages are set to become central components in the future of biomolecular engineering and therapeutics.
2. Designed protein cages
Non-natural protein nanocages are at the forefront of nanomedicine, providing innovative solutions for targeted drug delivery, gene therapy, vaccine development, and biocatalysis.1,29–34 Computational tools for the design of protein nanocages have greatly enhanced their potential as molecular carriers capable of encapsulating a variety of therapeutic agents, including nucleic acids, and proteins.In the design of protein nanocages, symmetry defines the overall architecture, while the final dimensions also depend on the number and size of the constituent components. The overall stability of the nanocage, however, depends not only on the detailed molecular interactions at the interfaces but also on the degree of cooperativity imposed by the underlying symmetry. The seminal studies from the Yeates group16,35–37 provided both theoretical and experimental insights into the critical role of symmetry in building protein nanocages. Readers are encouraged to refer to these studies for a deeper understanding of symmetry principles in molecular design. However, in this review, we primarily focus on recently developed nanocages intended for therapeutic applications. We review two main groups of synthetic nanocages: (1) assembled from modular oligomeric protein domains, and (2) constructed using coiled-coil modules.
2.1 Nanocages based on modular oligomeric proteins
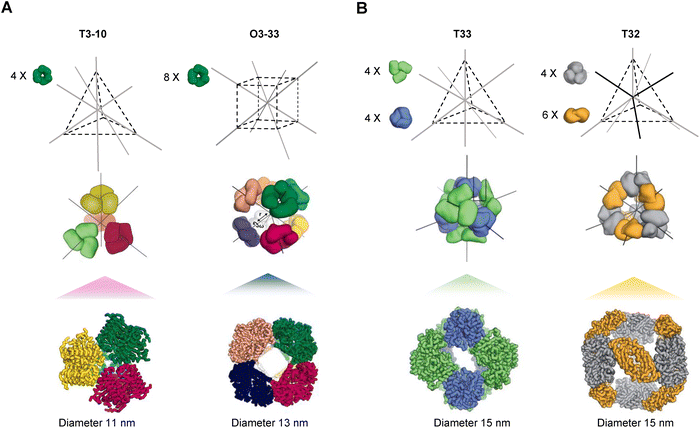
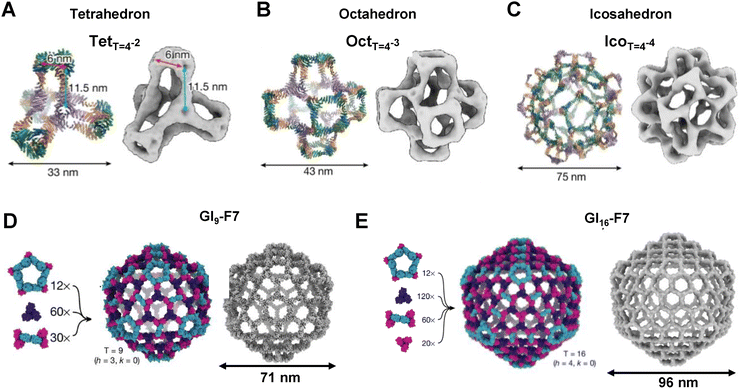
The structural and compositional features of protein nanocages assembled from modular oligomeric domains span a broad spectrum of architectures. Beginning with early symmetry-based designs from the Yeates group and later expanded by the Baker and King groups, the field has advanced toward more complex assemblies through programmed symmetry breaking. This strategy, related to concepts such as pseudo-symmetry and quasi-equivalence, introduces slight variations between subunits to enable deviations from perfect symmetry and supports the construction of larger and more versatile nanocages. These nanocages, typically ranging from 10 to 90 nm in size, are categorized here according to five dominant design methodologies: (1) genetic fusion of oligomerizing domain-based protein cages, (2) interface design-based de novo protein cages, (3) regulated cage assembly through interface interactions, (4) directed evolution-based redesign of protein nanocages, and (5) designs guided by ML/AI.Recent studies have significantly advanced protein nanocage design by introducing programmed symmetry breaking to create larger and more complex structures.32,47 The four-component nanocage strategy overcome the previous size and complexity limits by using pseudosymmetric heterotrimers to build tetrahedral (33 nm), octahedral (43 nm), and icosahedral (75 nm) architectures (Fig. 4A–C). However, through a hierarchical assembly strategy incorporating pseudosymmetry, designs were further scaled to produce icosahedral cages comprising up to 960 subunits and reaching diameters of 96 nm, representing the largest computationally designed protein structures to date (Fig. 4D and E).
Disulfide-mediated assembly leverages intra- and inter-subunit covalent linkages to induce structural transformations in a controlled manner.52,53 The study published by Zhao group presents a novel disulfide-mediated approach to create diverse protein nanocages using a single 8-mer bowl-like building block (NF-8).53 Through selective deletion of intra-subunit disulfide bonds and insertion of inter-subunit linkages, they successfully reprogrammed NF-8 into three distinct quaternary structures: a 24-mer ferritin-like cage, a 16-mer lenticular assembly, and a 48-mer hollow nanocage. Complementing these strategies, a chemically induced protein cage has been reported in which building blocks are covalently connected using DTME or BMH, forming stable cage-like architectures.54 This chemically driven assembly allows controlled cage opening and cargo release.
Hilvert's group used directed evolution to transform the non-viral enzyme lumazine synthase (AaLS) into virus-like nucleocapsids capable of packaging their own full-length RNA genomes.56,58 By fusing cationic peptides such as λN+ and applying iterative mutagenesis and selection, RNA encapsulation and stability were improved. Further work using error-prone PCR produced NC-4, a 240-subunit icosahedral capsid with significantly improved RNA packaging efficiency.56 Together, these studies show that iterative evolution can fine-tune features such as assembly yield, interface packing, cooperativity, and stability that are often difficult to optimize through rational design alone.60 With machine learning entering the field, there is an enormous potential to speed up directed evolution by predicting function-enhancing mutations and exploring sequence space more efficiently, although the tools still lack the sensitivity to point mutations and reliable ranking of most stable assemblies. While the fraction of successful designs is strongly improved, it is still required to screen tens of constructs.
Recent studies also highlight the transformative potential of ProteinMPNN in advancing protein nanocage engineering beyond traditional computational methods.14,15 De Haas et al. showed a fully automated workflow that eliminates the extensive manual optimization while reducing computational cost by orders of magnitude. Importantly, ProteinMPNN achieved an experimental success rate of ∼17% for two-component tetrahedral assemblies, comparable to the ∼18% reported for the original Rosetta-only designs, accomplished with much greater efficiency and without expert intervention.9,15 Their work further demonstrated that ProteinMPNN preferentially generates more polar interface residues than Rosetta, resulting in components with reduced aggregation propensity and improved in vitro assembly behavior.15 Parallel efforts using hybrid fragment-based and ML-guided protocols have similarly shown that integrating ProteinMPNN into cage design pipelines increases the diversity and quality of viable cage candidates compared with traditional fragment- or Rosetta-based sequence design alone.14
Additionally, hallucination-based approaches offer an alternative route for generating new protein structures by optimizing random amino acid sequences until deep neural networks predict well-defined distance and orientation maps. This method produces monomeric proteins with diverse all-α, all-β, and mixed α–β topologies, many of which fold as intended, as confirmed by X-ray crystallography and NMR.67 The strategy has also been extended to symmetric oligomers, where AlphaFold2-guided hallucination generates cyclic assemblies without predefined protomer structures, yielding oligomers with sequences and architectures distinct from natural proteins and validated by crystallography and cryo-EM.68
2.2 Designed nanocages based on modular coiled coils
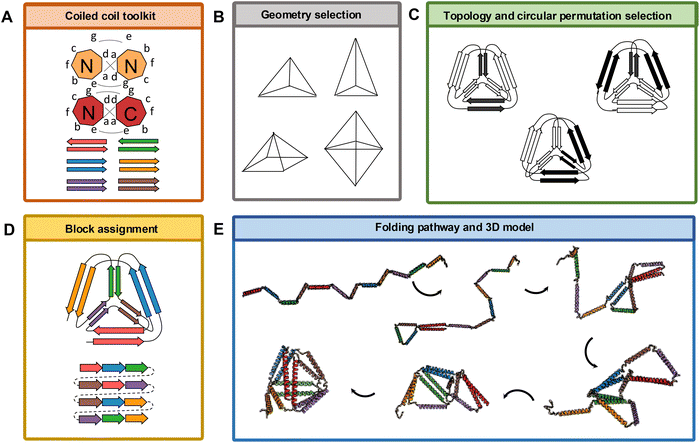
Coiled coils are a widespread protein structural motif that has been recognized as an excellent building module for the construction of de novo-designed protein nanostructures. The interactions that govern the specificity and orthogonality of coiled-coil dimers are well understood.24,69–72 The seven-amino acid periodicity provides the structural regularity to the coiled-coil dimer motif (heptad repeat), in which each residue is represented as a letter in the sequence abcdefg. The specificity of coiled-coil dimer pairing is primarily based on electrostatic interactions between residues at positions a and d, and e and g of the heptad repeat (Fig. 5A). The affinity, length, stability, and orthogonality of coiled-coil pairs can be precisely tuned through amino acid modifications and sequence design.24,69 Several sets of de novo designed, orthogonal coiled coils with different stabilities, sizes, affinities and orientations have been reported.24,73–81The Marsh group recognized the potential of the coiled coils for de novo protein design and introduced them as helical linkers fused to the natural oligomeric domains that self-assemble via coiled-coil dimerization.82–85 In a complementary approach, the Woolfson group demonstrated that short, designed coiled-coil forming peptides could assemble into bundles that form a hexagonal network with pores. These pores subsequently closed into larger spheres with diameters of approximately 100 nm.86
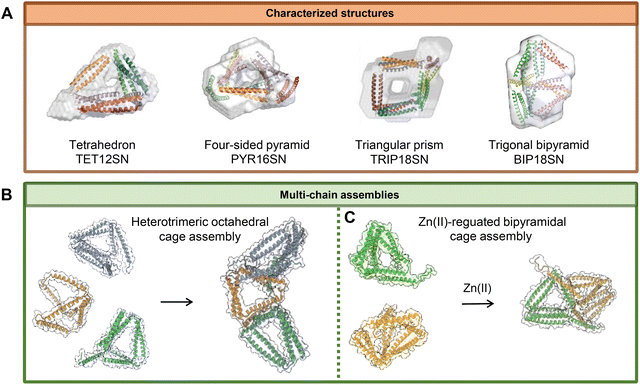
The proof of principle was first demonstrated experimentally using a tetrahedron, which is the simplest three-dimensional polyhedron. A tetrahedral nanocage (TET12), composed of 12 coiled-coil forming segments (each comprising four heptads) concatenated with flexible linkers, was successfully designed and characterized (Fig. 6A).87 Building on this work, the Jerala group expanded the CCPO platform to include more complex architectures, such as a four-sided pyramid (PYR16) and a triangular prism (TRIP18).26 The polypeptides for these second-generation structures had improved folding properties and were self-assembled in vivo under the physiological conditions in bacteria, mammalian cells and mice without causing inflammation. The structures of the tetrahedron, four-sided pyramid and triangular prism were validated using single-particle TEM reconstruction and SAXS analysis. These analyses revealed the maximum diameters of the structures to be in the range of 10–15 nm (Fig. 6A). Additionally, computational and experimental analyses revealed that CCPO folding proceeds through a sequential, stepwise pathway that is critically dependent on the precise arrangement of coiled-coil modules (Fig. 5E).91 Understanding this process enabled the control of the folding pathway and the construction of cages with multiple copies of the same module within a single chain while avoiding misfolding. Recently, tetrameric coiled-coil modules were introduced to CCPOs in addition to CC dimers. This expanded the range of topological solutions and allowed for the construction of cages from two identical polypeptide chains. This also facilitated structure determination by cryo-electron microscopy, as the tetrameric modules improved the stability at the air–water interface, which hindered previous attempts.89
Building on these advances, the Jerala group also described a strategy for assembling modular architectures based on structurally and covalently preorganized subunits. They employed the covalent cyclization of pre-organized subunits through the spontaneous self-splicing of split inteins and intramolecular connections. The cyclization and coiled-coil dimer-based interactions of the polypeptide chains provide the necessary structural constraints to facilitate the desired assembly (Fig. 6B).92 This strategy enables the self-assembly of higher-order nanostructures with improved folding fidelity and reduced conformational heterogeneity.
3. Application of designed protein nanocages
Nanostructures offer an exciting potential in cargo encapsulation and drug delivery, vaccine development, enzymatic catalysis and bioimaging (Fig. 7). The engineered protein nanocages provide precise control over size, shape, and surface characteristics, allowing for cargo loading through bioconjugation and non-covalent interactions. Their stable architectures protect therapeutic payloads from premature degradation, enhancing delivery accuracy and overall treatment efficacy. Moreover, the use of non-viral protein components in artificial nanocages can lower immunogenicity and decrease the risk of adverse immune responses upon repeated administration.30,93–95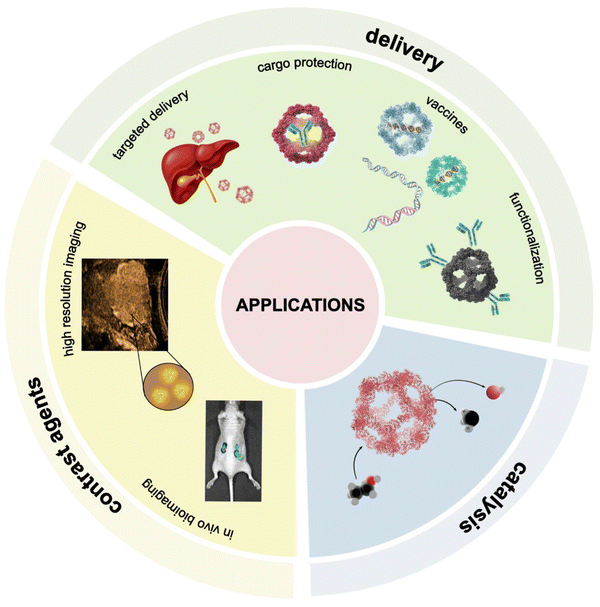 | ||
| Fig. 7 The schematic illustrations of protein nanocage applications, including cargo delivery, bioimaging, and catalysis. | ||
The Zhao group demonstrated the principle of therapeutic cargo encapsulation by engineering a 16-mer ferritin-derived nanocage capable of encapsulating curcumin, a yellow polyphenolic compound derived from the turmeric plant (Curcuma longa) known for its potent antioxidant, anti-inflammatory, anticancer, and antimicrobial properties.96 Despite these biological activities, curcumin suffers from extremely poor aqueous solubility, rapid degradation at physiological pH, and low bioavailability. The encapsulation significantly improved curcumin's chemical stability and bioavailability, thereby showcasing the nanocage's potential for delivering hydrophobic bioactives.97–99 Beyond encapsulation, nanocages can be functionalized with targeting modules such as tumor-homing peptides, cell-penetrating peptides, antibody-binding or receptor-specific ligands to achieve selective uptake and enhanced intracellular delivery of diverse cargoes.33,46,95,100–104 In addition to therapeutic delivery, protein nanocages can serve as confined environments that enhance enzymatic reactions. By restricting diffusion and stabilizing the enzyme structure, nanocages can increase catalytic efficiency while protecting sensitive proteins from degradation.105–107 Several natural and engineered systems including ferritin, lumazine synthase, encapsulins, and viral capsids have been used to encapsulate enzymes through electrostatic, affinity-based, or genetic fusion strategies. For example, engineered lumazine synthase variants (AaLS-neg, AaLS-13) efficiently encapsulate supercharged enzymes to enable controlled proteolysis or peroxidase reactions, while Thermotoga maritima ferritin (TmFtn) encapsulation enhances lysozyme activity through crowding effects.108 Encapsulins from Brevibacterium linens naturally load dye-decolorizing peroxidase (DyP), enabling the construction of cascade nanoreactors when paired with glucose oxidase.109
Engineered protein nanocages are being actively explored as platforms for the development of novel vaccines.31,110 Their symmetrical, virus-like architecture mimics natural pathogens and supports multivalent, ordered display of protein epitopes. This repetitive presentation is crucial for eliciting a strong humoral immune response. By presenting both B cell and T cell epitopes, protein nanocages can elicit robust IgG responses by enhancing antigen uptake and facilitating efficient B cell receptor cross-linking. This leads to potent B cell activation and subsequent differentiation into antibody-secreting plasma cells. Computationally designed scaffolds such as I3-01 and I53-50 have demonstrated that multivalent display of antigens, such as receptor-binding domain, significantly enhances the magnitude of neutralizing antibody titers compared to soluble antigen formulations alone.111–114 The well-defined geometry of protein nanocages allows for high-density, multivalent presentation of antigens with controlled spacing, improving immune receptor engagement. This strategy has been investigated in the development of vaccines against various infectious diseases, including hepatitis B, influenza, and respiratory syncytial virus.113,114 Several protein nanocage platforms have also progressed into clinical trials, including ferritin-based vaccines for SARS-CoV-2 and influenza, and, importantly, a designed icosahedral nanocage-based vaccine (I53-50) against SARS-CoV-2, which is marketed as SKYCovione™.115–120
Beyond humoral immunity, protein nanocages such as E2 and I3-01 can also promote robust cellular immune responses, including the activation of CD4+ helper T cells and CD8+ cytotoxic T lymphocytes.121–124 By incorporating specific T cell epitopes, these nanocages engage MHC class I and class II antigen presentation pathways. This dual engagement enables cytotoxic CD8+ T cells to directly target infected or malignant cells, while CD4+ helper T cells support sustained immune activity by promoting B cell maturation, CD8+ T cell memory formation, and coordinated cytokine signaling. The activation of a cell-mediated immune response can also be achieved by incorporating T cell epitopes at the interior side of the nanocage.
Building upon these advancements, protein nanocages have been systematically investigated as platforms for cancer immunotherapy and targeted drug delivery. Functionalizing nanocages with ligands that recognize specific cell surface receptors, such as transferrin or epidermal growth factor receptors, enhances cellular uptake and tumor targeting.125 Osiński et al. demonstrated that the introduction of histidine residues into metal-binding sites of protein cages allows for pH-sensitive disassembly, enabling selective fluorescent protein release under acidic conditions typical of tumor microenvironments.48 In a complementary approach, Yang et al. showed that pH-responsive trimeric building blocks can modulate the porosity of octahedral antibody nanoparticles, providing an additional mechanism for pH-dependent control of assembly and cargo accessibility.46 In the future, such cargo could be replaced with chemotherapeutic agents, immunostimulatory molecules or nucleic acids, which often face challenges in stability, solubility, or targeted delivery.
Nanocages based on natural proteins bring compelling advantages to biomedical imaging that distinguish them from established clinical agents such as small-molecule gadolinium (Gd) chelates or inorganic nanoparticles.126 These biologically derived structures like ferritin and heat shock proteins (Hsp16.5) are genetically programmable, uniform, and biocompatible, enabling the site-specific and high-density conjugation of imaging agents for magnetic resonance imaging (MRI), positron emission tomography (PET), fluorescence, and even ultrasound modalities.127–129 Beyond MRI, designed protein nanocages support multimodal imaging, such as: (1) tobacco mosaic virus-based nanoparticles co-loaded with Dy3+ and Cy7.5 fluorophores enabled dual MRI/near-infrared fluorescence imaging130 and (2) protein-based gas vesicles produced strong acoustic contrast in vivo for ultrasound applications.131
4. Challenges of designed protein cages
Despite major advances in protein nanocage engineering for catalysis, drug delivery, and bioimaging, several challenges remain to facilitate more effective clinical translation. Key issues include maintaining structural integrity, minimizing immunogenicity, and optimizing biological performance. While immunogenicity and biocompatibility remain important considerations for protein nanocages intended for therapeutic applications, these challenges are also shared by many other protein therapeutics and viral vector-based systems. Although nanocages are typically constructed from biocompatible parts, factors such as specific structural motifs or residual contaminants can still provoke immune response, potentially compromising therapeutic efficacy or increasing toxicity risks. Similar to other advanced delivery systems, ongoing efforts in protein engineering and purification strategies are critical for improving clinical translation.132 Another fundamental concern is the long-term stability and structural robustness of protein nanocages under the physiological conditions, which has been significantly improved by the hyperstability of ML-based designs. Many protein nanocages struggle to encapsulate sufficient quantities of drug molecules, especially hydrophobic or dual-mode therapeutics. Moreover, tunable and controlled release could be improved in sustained or stimuli-responsive delivery.103,133 Lastly, cellular permeability and tissue penetration remain inadequate in many formulations. Without surface modifications or targeting ligands, nanocages often demonstrate poor biodistribution and off-target accumulation. Together, these limitations highlight the need for continued innovation in both material design and production methodologies. On the other hand, recent advances are finding solutions to many of these issues. Single-chain designs significantly reduce heterogeneity, although the size of such cages is typically limited to about 100 kDa.5. Future directions
To enable broader clinical use, future work must address these challenges by integrating bioengineering strategies that improve structural stability, reduce immunogenicity, and enhance targeting and cargo release profiles. Innovative surface modifications such as the conjugation of tumor-targeting peptides, aptamers, and monoclonal antibodies, should be further investigated to enhance target specificity and therapeutic precision.134,135 Surface functionalization, particularly through polyethylene glycol (PEG) conjugation, offers several advantages such as prolonged circulation half-life, enhanced biocompatibility and protection from enzymatic degradation.136 Specifically for coiled-coil protein-based nanocages (CCPOs), systematic evaluation of diverse functionalization approaches remains necessary to optimize therapeutic delivery to specific targets. The modular nature of CCPOs provides a unique advantage, offering flexibility in size, shape, complexity, and functionalization, which can be tailored for both medical and biotechnological applications.Another emerging frontier in protein nanocage design is the development of high-throughput AI/ML pipelines to precisely predict and optimize self-assembly behavior. The field of de novo protein design has evolved dramatically over the past decades, transitioning from purely physics-based approaches to the integration of AI-driven methods.137,138 While advances such as AlphaFold have revolutionized structural prediction, they still fall short when applied to complex architectures like tetrahedral, octahedral, or icosahedral protein cages.139,140
In particular, AlphaFold fails to predict the global fold and 3D structures of de novo designed CCPOs, likely due to the lack of homologous templates, nonnatural topology and limitations in current training datasets. Recent advances in deep learning-based structure prediction are becoming an essential validation step for assessing the structure of complex multimeric assemblies.141 In addition to AlphaFold2 and AlphaFold3, which brought transformative accuracy to single-chain structure prediction, subsequent tools such as RoseTTAFold and AlphaFold-multimer have extended this capability to multimeric complexes, often using paired multiple sequence alignments (MSAs) to model inter-chain interactions.142–144 To expand capabilities further, Monte Carlo-based frameworks such as MoLPC were developed to simulate large symmetric assemblies through stepwise modeling, although they require prior stoichiometric knowledge of the system.145 Additionally, UniFold-symmetry demonstrated rapid and accurate modeling of high-order cyclic oligomers using symmetry-aware residue transformations.146 Although initially limited to cyclic assemblies, its extension to polyhedral nanocages appears promising. Meanwhile, the application of language-based deep learning models has introduced alternative routes for structural prediction without the need for explicit MSAs such as OmegaFold and ESMFold.147,148 While these models are not yet fully validated for highly complex protein assemblies, they represent scalable and rapid alternatives that may effectively complement traditional design methodologies. Incorporating such tools into synthetic protein design workflows could substantially enhance both predictive accuracy and development throughput.
On the other hand, ML-based designs are highly stable, can be engineered to be protease-resistant, and are also highly homogeneous. By incorporating diverse constraints, they can also avoid T-cell epitopes and immunogenic surfaces. Furthermore, the ability to regulate their assembly and disassembly, as well as cargo packaging, is being improved through the introduction of all-atom ML models.144,149
Author contributions
AKG: conceptualization, writing, visualization and original draft. HE: writing, visualization and review. HG: writing, visualization and review. RJ: conceptualization, review and supervision.Conflicts of interest
The authors declare that there are no conflicts of interest.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Acknowledgements
This work was funded by the Slovenian Research and Innovation Agency (projects N1-0377 and GC-0001, and program P4-0176), ERC AdG project 101141584 (PROFI) and European Innovation Council project LoopOfFun, Project 101070817.References
- T. G. W. Edwardson, M. D. Levasseur, S. Tetter, A. Steinauer, M. Hori and D. Hilvert, Protein Cages: From Fundamentals to Advanced Applications, Chem. Rev., 2022, 122, 9145–9197 CrossRef CAS PubMed.
- A. M. Demchuk and T. R. Patel, The biomedical and bioengineering potential of protein nanocompartments, Biotechnol. Adv., 2020, 41, 107547 CrossRef CAS PubMed.
- S. Panahandeh, S. Li, B. Dragnea and R. Zandi, Virus Assembly Pathways Inside a Host Cell, ACS Nano, 2022, 16, 317–327 Search PubMed.
- F. Fatehi and R. Twarock, An interaction network approach predicts protein cage architectures in bionanotechnology, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2303580120 CrossRef CAS PubMed.
- N. P. King, W. Sheffler, M. R. Sawaya, B. S. Vollmar, J. P. Sumida, I. André, T. Gonen, T. O. Yeates and D. Baker, Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy, Science, 2012, 336, 1171–1174 CrossRef CAS PubMed.
- J. B. Maguire, S. E. Boyken, D. Baker and B. Kuhlman, Rapid Sampling of Hydrogen Bond Networks for Computational Protein Design, J. Chem. Theory Comput., 2018, 14, 2751–2760 CrossRef CAS PubMed.
- W. Sheffler, E. C. Yang, Q. Dowling, Y. Hsia, C. N. Fries, J. Stanislaw, M. D. Langowski, M. Brandys, Z. Li, R. Skotheim, A. J. Borst, A. Khmelinskaia, N. P. King and D. Baker, Fast and versatile sequence-independent protein docking for nanomaterials design using RPXDock, PLoS Comput. Biol., 2023, 19, e1010680 CrossRef CAS PubMed.
- J. A. Fallas, G. Ueda, W. Sheffler, V. Nguyen, D. E. McNamara, B. Sankaran, J. H. Pereira, F. Parmeggiani, T. J. Brunette, D. Cascio, T. R. Yeates, P. Zwart and D. Baker, Computational design of self-assembling cyclic protein homo-oligomers, Nat. Chem., 2017, 9, 353–360 CrossRef CAS PubMed.
- N. P. King, J. B. Bale, W. Sheffler, D. E. McNamara, S. Gonen, T. Gonen, T. O. Yeates and D. Baker, Accurate design of co-assembling multi-component protein nanomaterials, Nature, 2014, 510, 103–108 CrossRef CAS PubMed.
- P. J. M. Brouwer, A. Antanasijevic, Z. Berndsen, A. Yasmeen, B. Fiala, T. P. L. Bijl, I. Bontjer, J. B. Bale, W. Sheffler, J. D. Allen, A. Schorcht, J. A. Burger, M. Camacho, D. Ellis, C. A. Cottrell, A.-J. Behrens, M. Catalano, I. Del Moral-Sánchez, T. J. Ketas, C. LaBranche, M. J. Van Gils, K. Sliepen, L. J. Stewart, M. Crispin, D. C. Montefiori, D. Baker, J. P. Moore, P. J. Klasse, A. B. Ward, N. P. King and R. W. Sanders, Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle, Nat. Commun., 2019, 10, 4272 Search PubMed.
- J. Marcandalli, B. Fiala, S. Ols, M. Perotti, W. De Van Der Schueren, J. Snijder, E. Hodge, M. Benhaim, R. Ravichandran, L. Carter, W. Sheffler, L. Brunner, M. Lawrenz, P. Dubois, A. Lanzavecchia, F. Sallusto, K. K. Lee, D. Veesler, C. E. Correnti, L. J. Stewart, D. Baker, K. Loré, L. Perez and N. P. King, Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus, Cell, 2019, 176, 1420–1431 Search PubMed.
- J. L. Watson, D. Juergens, N. R. Bennett, B. L. Trippe, J. Yim, H. E. Eisenach, W. Ahern, A. J. Borst, R. J. Ragotte, L. F. Milles, B. I. M. Wicky, N. Hanikel, S. J. Pellock, A. Courbet, W. Sheffler, J. Wang, P. Venkatesh, I. Sappington, S. V. Torres, A. Lauko, V. De Bortoli, E. Mathieu, S. Ovchinnikov, R. Barzilay, T. S. Jaakkola, F. DiMaio, M. Baek and D. Baker, De novo design of protein structure and function with RFdiffusion, Nature, 2023, 620, 1089–1100 Search PubMed.
- J. Dauparas, I. Anishchenko, N. Bennett, H. Bai, R. J. Ragotte, L. F. Milles, B. I. M. Wicky, A. Courbet, R. J. de Haas, N. Bethel, P. J. Y. Leung, T. F. Huddy, S. Pellock, D. Tischer, F. Chan, B. Koepnick, H. Nguyen, A. Kang, B. Sankaran, A. K. Bera, N. P. King and D. Baker, Robust deep learning–based protein sequence design using ProteinMPNN, Science, 2022, 378, 49–56 Search PubMed.
- K. Meador, R. Castells-Graells, R. Aguirre, M. R. Sawaya, M. A. Arbing, T. Sherman, C. Senarathne and T. O. Yeates, A suite of designed protein cages using machine learning and protein fragment-based protocols, Structure, 2024, 32, 751–765.e11 Search PubMed.
- R. J. De Haas, N. Brunette, A. Goodson, J. Dauparas, S. Y. Yi, E. C. Yang, Q. Dowling, H. Nguyen, A. Kang, A. K. Bera, B. Sankaran, R. De Vries, D. Baker and N. P. King, Rapid and automated design of two-component protein nanomaterials using ProteinMPNN, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2314646121 Search PubMed.
- J. E. Padilla, C. Colovos and T. O. Yeates, Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 2217–2221 Search PubMed.
- Y.-T. Lai, D. Cascio and T. O. Yeates, Structure of a 16-nm Cage Designed by Using Protein Oligomers, Science, 2012, 336, 1129 CrossRef CAS PubMed.
- K. A. Cannon, V. N. Nguyen, C. Morgan and T. O. Yeates, Design and Characterization of an Icosahedral Protein Cage Formed by a Double-Fusion Protein Containing Three Distinct Symmetry Elements, ACS Synth. Biol., 2020, 9, 517–524 Search PubMed.
- K. T. Simons, R. Bonneau, I. Ruczinski and D. Baker, Ab initio protein structure prediction of CASP III targets using ROSETTA, Proteins, 1999, 37, 171–176 CrossRef.
- A. Leaver-Fay, M. Tyka, S. M. Lewis, O. F. Lange, J. Thompson, R. Jacak, K. W. Kaufman, P. D. Renfrew, C. A. Smith, W. Sheffler, I. W. Davis, S. Cooper, A. Treuille, D. J. Mandell, F. Richter, Y.-E. A. Ban, S. J. Fleishman, J. E. Corn, D. E. Kim, S. Lyskov, M. Berrondo, S. Mentzer, Z. Popović, J. J. Havranek, J. Karanicolas, R. Das, J. Meiler, T. Kortemme, J. J. Gray, B. Kuhlman, D. Baker and P. Bradley, Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules, in Methods in Enzymology, ed. M. L. Johnson and L. Brand, Academic Press, 2011, vol. 487, pp. 545–574 Search PubMed.
- A. J. Wargacki, T. P. Wörner, M. Van De Waterbeemd, D. Ellis, A. J. R. Heck and N. P. King, Complete and cooperative in vitro assembly of computationally designed self-assembling protein nanomaterials, Nat. Commun., 2021, 12, 883 CrossRef CAS PubMed.
- J. Butcher, R. Krishna, R. Mitra, R. I. Brent, Y. Li, N. Corley, P. Kim, J. Funk, S. Mathis, S. Salike, A. Muraishi, H. Eisenach, E. Sehgal, B. Coventry, O. Zhang, B. Qiang, K. Didi, F. DiMaio and D. Baker, De novo Design of All-atom Biomolecular Interactions with RFdiffusion3, bioRxiv, 2025, preprint DOI:10.1101/2025.09.18.676967.
- W. Ahern, J. Yim, D. Tischer, S. Salike, S. M. Woodbury, D. Kim, I. Kalvet, Y. Kipnis, B. Coventry, H. R. Altae-Tran, M. Bauer, R. Barzilay, T. S. Jaakkola, R. Krishna and D. Baker, Atom level enzyme active site scaffolding using RFdiffusion2, bioRxiv, 2025, preprint DOI:10.1101/2025.04.09.648075.
- I. Drobnak, H. Gradišar, A. Ljubetič, E. Merljak and R. Jerala, Modulation of Coiled-Coil Dimer Stability through Surface Residues while Preserving Pairing Specificity, J. Am. Chem. Soc., 2017, 139, 8229–8236 CrossRef CAS PubMed.
- W. M. Park, Coiled-Coils: The Molecular Zippers that Self-Assemble Protein Nanostructures, Int. J. Mol. Sci., 2020, 21, 3584 CrossRef CAS PubMed.
- A. Ljubetič, F. Lapenta, H. Gradišar, I. Drobnak, J. Aupič, Ž. Strmšek, D. Lainšček, I. Hafner-Bratkovič, A. Majerle, N. Krivec, M. Benčina, T. Pisanski, T. Ć. Veličković, A. Round, J. M. Carazo, R. Melero and R. Jerala, Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo, Nat. Biotechnol., 2017, 35, 1094–1101 CrossRef PubMed.
- J. Aupič, F. Lapenta and R. Jerala, SwitCCh: Metal-Site Design for Controlling the Assembly of a Coiled-Coil Homodimer, ChemBioChem, 2018, 19, 2453–2457 CrossRef PubMed.
- J. Aupič, F. Lapenta, Ž. Strmšek, E. Merljak, T. Plaper and R. Jerala, Metal ion–regulated assembly of designed modular protein cages, Sci. Adv., 2022, 8, eabm8243 CrossRef PubMed.
- M. Rother, M. G. Nussbaumer, K. Renggli and N. Bruns, Protein cages and synthetic polymers: a fruitful symbiosis for drug delivery applications, bionanotechnology and materials science, Chem. Soc. Rev., 2016, 45, 6213–6249 RSC.
- E. Kianfar, Protein nanoparticles in drug delivery: animal protein, plant proteins and protein cages, albumin nanoparticles, J. Nanobiotechnol., 2021, 19, 159 CrossRef CAS PubMed.
- S. A. Kim, Y. Lee, Y. Ko, S. Kim, G. B. Kim, N. K. Lee, W. Ahn, N. Kim, G.-H. Nam, E. J. Lee and I.-S. Kim, Protein-based nanocages for vaccine development, J. Controlled Release, 2023, 353, 767–791 CrossRef CAS PubMed.
- Q. M. Dowling, Y.-J. Park, C. N. Fries, N. C. Gerstenmaier, S. Ols, E. C. Yang, A. J. Wargacki, A. Dosey, Y. Hsia, R. Ravichandran, C. D. Walkey, A. L. Burrell, D. Veesler, D. Baker and N. P. King, Hierarchical design of pseudosymmetric protein nanocages, Nature, 2025, 638, 553–561 Search PubMed.
- R. Divine, H. V. Dang, G. Ueda, J. A. Fallas, I. Vulovic, W. Sheffler, S. Saini, Y. T. Zhao, I. X. Raj, P. A. Morawski, M. F. Jennewein, L. J. Homad, Y.-H. Wan, M. R. Tooley, F. Seeger, A. Etemadi, M. L. Fahning, J. Lazarovits, A. Roederer, A. C. Walls, L. Stewart, M. Mazloomi, N. P. King, D. J. Campbell, A. T. McGuire, L. Stamatatos, H. Ruohola-Baker, J. Mathieu, D. Veesler and D. Baker, Designed proteins assemble antibodies into modular nanocages, Science, 2021, 372, eabd9994 CrossRef CAS PubMed.
- A. C. Walls, M. C. Miranda, A. Schäfer, M. N. Pham, A. Greaney, P. S. Arunachalam, M.-J. Navarro, M. A. Tortorici, K. Rogers, M. A. O’Connor, L. Shirreff, D. E. Ferrell, J. Bowen, N. Brunette, E. Kepl, S. K. Zepeda, T. Starr, C.-L. Hsieh, B. Fiala, S. Wrenn, D. Pettie, C. Sydeman, K. R. Sprouse, M. Johnson, A. Blackstone, R. Ravichandran, C. Ogohara, L. Carter, S. W. Tilles, R. Rappuoli, S. R. Leist, D. R. Martinez, M. Clark, R. Tisch, D. T. O’Hagan, R. Van Der Most, W. C. Van Voorhis, D. Corti, J. S. McLellan, H. Kleanthous, T. P. Sheahan, K. D. Smith, D. H. Fuller, F. Villinger, J. Bloom, B. Pulendran, R. S. Baric, N. P. King and D. Veesler, Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines, Cell, 2021, 184, 5432–5447.e16 Search PubMed.
- J. Laniado, K. A. Cannon, J. E. Miller, M. R. Sawaya, D. E. McNamara and T. O. Yeates, Geometric Lessons and Design Strategies for Nanoscale Protein Cages, ACS Nano, 2021, 15, 4277–4286 CrossRef CAS PubMed.
- J. Laniado and T. O. Yeates, A complete rule set for designing symmetry combination materials from protein molecules, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 31817–31823 CrossRef CAS PubMed.
- Y.-T. Lai, E. Reading, G. L. Hura, K.-L. Tsai, A. Laganowsky, F. J. Asturias, J. A. Tainer, C. V. Robinson and T. O. Yeates, Structure of a designed protein cage that self-assembles into a highly porous cube, Nat. Chem., 2014, 6, 1065–1071 CrossRef CAS PubMed.
- Y. Hsia, R. Mout, W. Sheffler, N. I. Edman, I. Vulovic, Y.-J. Park, R. L. Redler, M. J. Bick, A. K. Bera, A. Courbet, A. Kang, T. J. Brunette, U. Nattermann, E. Tsai, A. Saleem, C. M. Chow, D. Ekiert, G. Bhabha, D. Veesler and D. Baker, Design of multi-scale protein complexes by hierarchical building block fusion, Nat. Commun., 2021, 12, 2294 CrossRef CAS PubMed.
- T. F. Huddy, Y. Hsia, R. D. Kibler, J. Xu, N. Bethel, D. Nagarajan, R. Redler, P. J. Y. Leung, C. Weidle, A. Courbet, E. C. Yang, A. K. Bera, N. Coudray, S. J. Calise, F. A. Davila-Hernandez, H. L. Han, K. D. Carr, Z. Li, R. McHugh, G. Reggiano, A. Kang, B. Sankaran, M. S. Dickinson, B. Coventry, T. J. Brunette, Y. Liu, J. Dauparas, A. J. Borst, D. Ekiert, J. M. Kollman, G. Bhabha and D. Baker, Blueprinting extendable nanomaterials with standardized protein blocks, Nature, 2024, 627, 898–904 Search PubMed.
- I. Vulovic, Q. Yao, Y.-J. Park, A. Courbet, A. Norris, F. Busch, A. Sahasrabuddhe, H. Merten, D. D. Sahtoe, G. Ueda, J. A. Fallas, S. J. Weaver, Y. Hsia, R. A. Langan, A. Plückthun, V. H. Wysocki, D. Veesler, G. J. Jensen and D. Baker, Generation of ordered protein assemblies using rigid three-body fusion, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2015037118 CrossRef CAS PubMed.
- S. Wang, A. Favor, R. D. Kibler, J. M. Lubner, A. J. Borst, N. Coudray, R. Redler, H. T. Chiang, W. Sheffler, Y. Hsia, Z. Li, D. C. Ekiert, G. Bhabha, L. D. Pozzo and D. Baker, Bond-centric modular design of protein assemblies, Nat. Mater., 2025, 24, 1644–1652 CrossRef CAS PubMed.
- C. A. Rohl, C. E. M. Strauss, K. M. S. Misura and D. Baker, Methods in Enzymology, Elsevier, 2004, vol. 383, pp. 66–93 Search PubMed.
- K. A. Cannon, R. U. Park, S. E. Boyken, U. Nattermann, S. Yi, D. Baker, N. P. King and T. O. Yeates, Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering, Protein Sci., 2020, 29, 919–929 CrossRef CAS PubMed.
- J. B. Bale, S. Gonen, Y. Liu, W. Sheffler, D. Ellis, C. Thomas, D. Cascio, T. O. Yeates, T. Gonen, N. P. King and D. Baker, Accurate design of megadalton-scale two-component icosahedral protein complexes, Science, 2016, 353, 389–394 Search PubMed.
- Y. Hsia, J. B. Bale, S. Gonen, D. Shi, W. Sheffler, K. K. Fong, U. Nattermann, C. Xu, P.-S. Huang, R. Ravichandran, S. Yi, T. N. Davis, T. Gonen, N. P. King and D. Baker, Design of a hyperstable 60-subunit protein icosahedron, Nature, 2016, 535, 136–139 CrossRef CAS PubMed.
- E. C. Yang, R. Divine, M. C. Miranda, A. J. Borst, W. Sheffler, J. Z. Zhang, J. Decarreau, A. Saragovi, M. Abedi, N. Goldbach, M. Ahlrichs, C. Dobbins, A. Hand, S. Cheng, M. Lamb, P. M. Levine, S. Chan, R. Skotheim, J. Fallas, G. Ueda, J. Lubner, M. Somiya, A. Khmelinskaia, N. P. King and D. Baker, Computational design of non-porous pH-responsive antibody nanoparticles, Nat. Struct. Mol. Biol., 2024, 31, 1404–1412 Search PubMed.
- S. Lee, R. D. Kibler, G. Ahn, Y. Hsia, A. J. Borst, A. Philomin, M. A. Kennedy, B. Huang, B. Stoddard and D. Baker, Four-component protein nanocages designed by programmed symmetry breaking, Nature, 2025, 638, 546–552 CrossRef CAS PubMed.
- N. Osiński, K. Majsterkiewicz, Z. Pakosz-Stępień, Y. Azuma, A. P. Biela, S. Gaweł and J. G. Heddle, Designed, Programmable Protein Cages Utilizing Diverse Metal Coordination Geometries Show Reversible, pH-Dependent Assembly, Macromol. Rapid Commun., 2025, 46, 2400712 CrossRef PubMed.
- E. Golub, R. H. Subramanian, J. Esselborn, R. G. Alberstein, J. B. Bailey, J. A. Chiong, X. Yan, T. Booth, T. S. Baker and F. A. Tezcan, Constructing protein polyhedra via orthogonal chemical interactions, Nature, 2020, 578, 172–176 CrossRef CAS PubMed.
- D. J. E. Huard, K. M. Kane and F. A. Tezcan, Re-engineering protein interfaces yields copper-inducible ferritin cage assembly, Nat. Chem. Biol., 2013, 9, 169–176 CrossRef CAS PubMed.
- A. D. Malay, N. Miyazaki, A. Biela, S. Chakraborti, K. Majsterkiewicz, I. Stupka, C. S. Kaplan, A. Kowalczyk, B. M. A. G. Piette, G. K. A. Hochberg, D. Wu, T. P. Wrobel, A. Fineberg, M. S. Kushwah, M. Kelemen, P. Vavpetič, P. Pelicon, P. Kukura, J. L. P. Benesch, K. Iwasaki and J. G. Heddle, An ultra-stable gold-coordinated protein cage displaying reversible assembly, Nature, 2019, 569, 438–442 CrossRef CAS PubMed.
- C. Gu, Y. Mi, T. Zhang, G. Zhao and S. Wang, Construction of robust protein nanocage by designed disulfide bonds for active cargo molecules protection in the gastric environment, J. Colloid Interface Sci., 2025, 678, 637–647 CrossRef CAS PubMed.
- J. Zang, H. Chen, X. Zhang, C. Zhang, J. Guo, M. Du and G. Zhao, Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages, Nat. Commun., 2019, 10, 778 CrossRef CAS PubMed.
- I. Stupka, Y. Azuma, A. P. Biela, M. Imamura, S. Scheuring, E. Pyza, O. Woźnicka, D. P. Maskell and J. G. Heddle, Chemically induced protein cage assembly with programmable opening and cargo release, Sci. Adv., 2022, 8, eabj9424 CrossRef CAS PubMed.
- J. D. Bloom and F. H. Arnold, In the light of directed evolution: Pathways of adaptive protein evolution, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 9995–10000 CrossRef CAS PubMed.
- S. Tetter, N. Terasaka, A. Steinauer, R. J. Bingham, S. Clark, A. J. P. Scott, N. Patel, M. Leibundgut, E. Wroblewski, N. Ban, P. G. Stockley, R. Twarock and D. Hilvert, Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein, Science, 2021, 372, 1220–1224 CrossRef CAS PubMed.
- G. L. Butterfield, M. J. Lajoie, H. H. Gustafson, D. L. Sellers, U. Nattermann, D. Ellis, J. B. Bale, S. Ke, G. H. Lenz, A. Yehdego, R. Ravichandran, S. H. Pun, N. P. King and D. Baker, Evolution of a designed protein assembly encapsulating its own RNA genome, Nature, 2017, 552, 415–420 CrossRef CAS PubMed.
- N. Terasaka, Y. Azuma and D. Hilvert, Laboratory evolution of virus-like nucleocapsids from nonviral protein cages, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5432–5437 CrossRef CAS PubMed.
- B. Wörsdörfer, K. J. Woycechowsky and D. Hilvert, Directed Evolution of a Protein Container, Science, 2011, 331, 589–592 CrossRef PubMed.
- C. Zeymer and D. Hilvert, Directed Evolution of Protein Catalysts, Annu. Rev. Biochem., 2018, 87, 131–157 CrossRef CAS PubMed.
- J. Dauparas, G. R. Lee, R. Pecoraro, L. An, I. Anishchenko, C. Glasscock and D. Baker, Atomic context-conditioned protein sequence design using LigandMPNN, Nat. Methods, 2025, 22, 717–723 CrossRef CAS PubMed.
- A. Kubaney, A. Favor, L. McHugh, R. Mitra, R. Pecoraro, J. Dauparas, C. Glasscock and D. Baker, RNA sequence design and protein–DNA specificity prediction with NA-MPNN, bioRxiv, 2025, preprint DOI:10.1101/2025.10.03.679414.
- H. Dieckhaus, M. Brocidiacono, N. Z. Randolph and B. Kuhlman, Transfer learning to leverage larger datasets for improved prediction of protein stability changes, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2314853121 CrossRef CAS PubMed.
- A. J. Li, M. Lu, I. Desta, V. Sundar, G. Grigoryan and A. E. Keating, Neural network-derived Potts models for structure-based protein design using backbone atomic coordinates and tertiary motifs, Protein Sci., 2023, 32, e4554 CrossRef CAS PubMed.
- C. Hsu, R. Verkuil, J. Liu, Z. Lin, B. Hie, T. Sercu, A. Lerer and A. Rives, Learning inverse folding from millions of predicted structures, bioRxiv, 2022, preprint DOI:10.1101/2022.04.10.487779.
- D. Akpinaroglu, K. Seki, A. Guo, E. Zhu, M. J. S. Kelly and T. Kortemme, Structure-conditioned masked language models for protein sequence design generalize beyond the native sequence space, bioRxiv, 2023, preprint DOI:10.1101/2023.12.15.571823.
- I. Anishchenko, S. J. Pellock, T. M. Chidyausiku, T. A. Ramelot, S. Ovchinnikov, J. Hao, K. Bafna, C. Norn, A. Kang, A. K. Bera, F. DiMaio, L. Carter, C. M. Chow, G. T. Montelione and D. Baker, De novo protein design by deep network hallucination, Nature, 2021, 600, 547–552 CrossRef CAS PubMed.
- B. I. M. Wicky, L. F. Milles, A. Courbet, R. J. Ragotte, J. Dauparas, E. Kinfu, S. Tipps, R. D. Kibler, M. Baek, F. DiMaio, X. Li, L. Carter, A. Kang, H. Nguyen, A. K. Bera and D. Baker, Hallucinating symmetric protein assemblies, Science, 2022, 378, 56–61 CrossRef CAS PubMed.
- D. N. Woolfson, The Design of Coiled-Coil Structures and Assemblies, Advances in Protein Chemistry, Academic Press, 2005, vol. 70, pp. 79–112 Search PubMed.
- A. R. Thomson, C. W. Wood, A. J. Burton, G. J. Bartlett, R. B. Sessions, R. L. Brady and D. N. Woolfson, Computational design of water-soluble α-helical barrels, Science, 2014, 346, 485–488 CrossRef CAS PubMed.
- W. M. Dawson, E. J. M. Lang, G. G. Rhys, K. L. Shelley, C. Williams, R. L. Brady, M. P. Crump, A. J. Mulholland and D. N. Woolfson, Structural resolution of switchable states of a de novo peptide assembly, Nat. Commun., 2021, 12, 1530 CrossRef CAS PubMed.
- J. Zhu, N. Avakyan, A. Kakkis, A. M. Hoffnagle, K. Han, Y. Li, Z. Zhang, T. S. Choi, Y. Na, C.-J. Yu and F. A. Tezcan, Protein Assembly by Design, Chem. Rev., 2021, 121, 13701–13796 CrossRef CAS PubMed.
- E. H. C. Bromley, R. B. Sessions, A. R. Thomson and D. N. Woolfson, Designed α-Helical Tectons for Constructing Multicomponent Synthetic Biological Systems, J. Am. Chem. Soc., 2009, 131, 928–930 Search PubMed.
- A. W. Reinke, R. A. Grant and A. E. Keating, A Synthetic Coiled-Coil Interactome Provides Heterospecific Modules for Molecular Engineering, J. Am. Chem. Soc., 2010, 132, 6025–6031 Search PubMed.
- H. Gradišar and R. Jerala, De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers, J. Pept. Sci., 2011, 17, 100–106 CrossRef PubMed.
- J. M. Fletcher, A. L. Boyle, M. Bruning, G. J. Bartlett, T. L. Vincent, N. R. Zaccai, C. T. Armstrong, E. H. C. Bromley, P. J. Booth, R. L. Brady, A. R. Thomson and D. N. Woolfson, A Basis Set of de Novo Coiled-Coil Peptide Oligomers for Rational Protein Design and Synthetic Biology, ACS Synth. Biol., 2012, 1, 240–250 CrossRef CAS PubMed.
- K. E. Thompson, C. J. Bashor, W. A. Lim and A. E. Keating, SYNZIP Protein Interaction Toolbox: in Vitro and in Vivo Specifications of Heterospecific Coiled-Coil Interaction Domains, ACS Synth. Biol., 2012, 1, 118–129 CrossRef CAS PubMed.
- C. Negron and A. E. Keating, A Set of Computationally Designed Orthogonal Antiparallel Homodimers that Expands the Synthetic Coiled-Coil Toolkit, J. Am. Chem. Soc., 2014, 136, 16544–16556 CrossRef CAS PubMed.
- R. O. Crooks, A. Lathbridge, A. S. Panek and J. M. Mason, Computational Prediction and Design for Creating Iteratively Larger Heterospecific Coiled Coil Sets, Biochemistry, 2017, 56, 1573–1584 CrossRef CAS PubMed.
- T. Plaper, J. Aupič, P. Dekleva, F. Lapenta, M. M. Keber, R. Jerala and M. Benčina, Coiled-coil heterodimers with increased stability for cellular regulation and sensing SARS-CoV-2 spike protein-mediated cell fusion, Sci. Rep., 2021, 11, 9136 CrossRef CAS PubMed.
- T. Lebar, D. Lainšček, E. Merljak, J. Aupič and R. Jerala, A tunable orthogonal coiled-coil interaction toolbox for engineering mammalian cells, Nat. Chem. Biol., 2020, 16, 513–519 CrossRef CAS PubMed.
- D. P. Patterson, A. M. Desai, M. M. Banaszak Holl and E. N. G. Marsh, Evaluation of a symmetry-based strategy for assembling protein complexes, RSC Adv., 2011, 1, 1004–1012 Search PubMed.
- A. Sciore, M. Su, P. Koldewey, J. D. Eschweiler, K. A. Diffley, B. M. Linhares, B. T. Ruotolo, J. C. A. Bardwell, G. Skiniotis and E. N. G. Marsh, Flexible, symmetry-directed approach to assembling protein cages, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 8681–8686 CrossRef CAS PubMed.
- S. Badieyan, A. Sciore, J. D. Eschweiler, P. Koldewey, A. S. Cristie-David, B. T. Ruotolo, J. C. A. Bardwell, M. Su and E. N. G. Marsh, Symmetry-Directed Self-Assembly of a Tetrahedral Protein Cage Mediated by de Novo-Designed Coiled Coils, ChemBioChem, 2017, 18, 1888–1892 CrossRef CAS PubMed.
- A. S. Cristie-David, J. Chen, D. B. Nowak, A. L. Bondy, K. Sun, S. I. Park, M. M. Banaszak Holl, M. Su and E. N. G. Marsh, Coiled-Coil-Mediated Assembly of an Icosahedral Protein Cage with Extremely High Thermal and Chemical Stability, J. Am. Chem. Soc., 2019, 141, 9207–9216 CrossRef CAS PubMed.
- J. M. Fletcher, R. L. Harniman, F. R. H. Barnes, A. L. Boyle, A. Collins, J. Mantell, T. H. Sharp, M. Antognozzi, P. J. Booth, N. Linden, M. J. Miles, R. B. Sessions, P. Verkade and D. N. Woolfson, Self-Assembling Cages from Coiled-Coil Peptide Modules, Science, 2013, 340, 595–599 CrossRef CAS PubMed.
- H. Gradišar, S. Božič, T. Doles, D. Vengust, I. Hafner-Bratkovič, A. Mertelj, B. Webb, A. Šali, S. Klavžar and R. Jerala, Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments, Nat. Chem. Biol., 2013, 9, 362–366 Search PubMed.
- F. Lapenta, J. Aupič, M. Vezzoli, Ž. Strmšek, S. Da Vela, D. I. Svergun, J. M. Carazo, R. Melero and R. Jerala, Self-assembly and regulation of protein cages from pre-organised coiled-coil modules, Nat. Commun., 2021, 12, 939 CrossRef CAS PubMed.
- S. Vidmar, T. Šmidlehner, J. Aupič, Ž. Strmšek, A. Ljubetič, F. Xiao, G. Hu, C. Liu, F. Beck, P. S. Erdmann and R. Jerala, Beyond Dimerization: Harnessing Tetrameric Coiled-Coils for Nanostructure Assembly, Angew. Chem., Int. Ed., 2025, 64, e202422075 CrossRef CAS PubMed.
- T. Satler, S. Hadži and R. Jerala, Crystal Structure of de Novo Designed Coiled-Coil Protein Origami Triangle, J. Am. Chem. Soc., 2023, 145, 16995–17000 Search PubMed.
- J. Aupič, Ž. Strmšek, F. Lapenta, D. Pahovnik, T. Pisanski, I. Drobnak, A. Ljubetič and R. Jerala, Designed folding pathway of modular coiled-coil-based proteins, Nat. Commun., 2021, 12, 940 Search PubMed.
- J. Snoj, F. Lapenta and R. Jerala, Preorganized cyclic modules facilitate the self-assembly of protein nanostructures, Chem. Sci., 2024, 15, 3673–3686 Search PubMed.
- S. Bhaskar and S. Lim, Engineering protein nanocages as carriers for biomedical applications, NPG Asia Mater., 2017, 9, e371 Search PubMed.
- J. G. Heddle, S. Chakraborti and K. Iwasaki, Natural and artificial protein cages: design, structure and therapeutic applications, Curr. Opin. Struct. Biol., 2017, 43, 148–155 CrossRef CAS PubMed.
- N. F. Steinmetz, S. Lim and F. Sainsbury, Protein cages and virus-like particles: from fundamental insight to biomimetic therapeutics, Biomater. Sci., 2020, 8, 2771–2777 RSC.
- S. Zhang, J. Zang, W. Wang, H. Chen, X. Zhang, F. Wang, H. Wang and G. Zhao, Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues, Angew. Chem., Int. Ed., 2016, 55, 16064–16070 CrossRef CAS PubMed.
- T. Jiang, W. Liao and C. Charcosset, Recent advances in encapsulation of curcumin in nanoemulsions: A review of encapsulation technologies, bioaccessibility and applications, Food Res. Int., 2020, 132, 109035 CrossRef CAS PubMed.
- Y. Chen, Y. Lu, R. J. Lee and G. Xiang, Nano Encapsulated Curcumin: And Its Potential for Biomedical Applications, Int. J. Nanosci., 2020, 15, 3099–3120 CAS.
- Y. Hu, C. Qiu, D. Julian McClements, Y. Qin, J. Long, A. Jiao, X. Li, J. Wang and Z. Jin, Encapsulation, protection, and delivery of curcumin using succinylated-cyclodextrin systems with strong resistance to environmental and physiological stimuli, Food Chem., 2022, 376, 131869 CrossRef CAS PubMed.
- E. O. Bobylev, Y. Zeng, K. Weijgertse, E. Koelman, E. M. Meijer, B. De Bruin, A. Kros and J. N. H. Reek, The application of M12L24 nanocages as cell-specific siRNA delivery agents in vitro, Chem, 2023, 9, 1578–1593 CAS.
- H. Wang, N. Liu, F. Yang, N. Hu, M. Wang, M. Cui, N. Bruns and X. Guan, Bioengineered Protein Nanocage by Small Heat Shock Proteins Delivering mTERT siRNA for Enhanced Colorectal Cancer Suppression, ACS Appl. Bio Mater., 2022, 5, 1330–1340 CrossRef CAS PubMed.
- M. D. Levasseur, S. Mantri, T. Hayashi, M. Reichenbach, S. Hehn, Y. Waeckerle-Men, P. Johansen and D. Hilvert, Cell-Specific Delivery Using an Engineered Protein Nanocage, ACS Chem. Biol., 2021, 16, 838–843 CrossRef CAS PubMed.
- Y. Wang and T. Douglas, Protein nanocage architectures for the delivery of therapeutic proteins, Curr. Opin. Colloid Interface Sci., 2021, 51, 101395 CrossRef CAS.
- C. Rennie, C. Sives, I. Boyton, D. Diaz, C. Gorrie, O. Vittorio, L. Collins-Praino and A. Care, In Vivo Behavior of Systemically Administered Encapsulin Protein Nanocages and Implications for their use in Targeted Drug Delivery, Adv. Ther., 2024, 7, 2300360 CrossRef CAS.
- D. Ensign, M. Young and T. Douglas, Photocatalytic Synthesis of Copper Colloids from Cu(II) by the Ferrihydrite Core of Ferritin, Inorg. Chem., 2004, 43, 3441–3446 CrossRef CAS PubMed.
- Z. Varpness, J. W. Peters, M. Young and T. Douglas, Biomimetic Synthesis of a H2 Catalyst Using a Protein Cage Architecture, Nano Lett., 2005, 5, 2306–2309 CrossRef CAS PubMed.
- H. Li, G. Zheng and S. Zhu, Construction of an organelle-like nanodevice via supramolecular self-assembly for robust biocatalysts, Microb. Cell Fact., 2018, 17, 26 CrossRef PubMed.
- S. Chakraborti, T.-Y. Lin, S. Glatt and J. G. Heddle, Enzyme encapsulation by protein cages, RSC Adv., 2020, 10, 13293–13301 RSC.
- R. M. Putri, C. Allende-Ballestero, D. Luque, R. Klem, K.-A. Rousou, A. Liu, C. H.-H. Traulsen, W. F. Rurup, M. S. T. Koay, J. R. Castón and J. J. L. M. Cornelissen, Structural Characterization of Native and Modified Encapsulins as Nanoplatforms for in Vitro Catalysis and Cellular Uptake, ACS Nano, 2017, 11, 12796–12804 CrossRef CAS PubMed.
- R. Rappuoli and D. Serruto, Self-Assembling Nanoparticles Usher in a New Era of Vaccine Design, Cell, 2019, 176, 1245–1247 CrossRef CAS PubMed.
- M. N. Vu, H. G. Kelly, S. J. Kent and A. K. Wheatley, Current and future nanoparticle vaccines for COVID-19, EBioMedicine, 2021, 74, 103699 CrossRef CAS PubMed.
- L. A. Angalene, P. Pa, C. Y. Seng, J. H. Rhee and S. E. Lee, Protein nanocages: A new frontier in mucosal vaccine delivery and immune activation, Hum. Vaccines Immunother., 2025, 21, 2492906 CrossRef PubMed.
- S. A. Nelson, K. A. Richards, M. A. Glover, F. A. Chaves, M. C. Crank, B. S. Graham, M. Kanekiyo and A. J. Sant, CD4 T cell epitope abundance in ferritin core potentiates responses to hemagglutinin nanoparticle vaccines, npj Vaccines, 2022, 7, 1–13 CrossRef PubMed.
- D. Ellis, A. Dosey, S. Boyoglu-Barnum, Y.-J. Park, R. Gillespie, H. Syeda, G. B. Hutchinson, Y. Tsybovsky, M. Murphy, D. Pettie, N. Matheson, S. Chan, G. Ueda, J. A. Fallas, L. Carter, B. S. Graham, D. Veesler, M. Kanekiyo and N. P. King, Antigen spacing on protein nanoparticles influences antibody responses to vaccination, Cell Rep., 2023, 42, 113552 CrossRef CAS PubMed.
- B. L. O. Shepherd, P. T. Scott, J. N. Hutter, C. Lee, M. D. McCauley, I. Guzman, C. Bryant, S. McGuire, J. Kennedy, W.-H. Chen, A. Hajduczki, T. Mdluli, A. Valencia-Ruiz, M. F. Amare, G. R. Matyas, M. Rao, M. Rolland, J. R. Mascola, S. C. D. Rosa, M. J. McElrath, D. C. Montefiori, L. Serebryannyy, A. B. McDermott, S. A. Peel, N. D. Collins, M. G. Joyce, M. L. Robb, N. L. Michael, S. Vasan, K. Modjarrad, B. Gebrehana, M. E. Greenleaf, M. J. Hamer, N. K. Jansen, X. Jing, J. Kagai, K. Kourbanova, M. A. Koren, M. L. Martin, K. M. Wuertz, J. A. Regules, A. D. Sanborn, D. Wallace, L. Zhu, G. D. Gromowski, C. Corbitt, J. M. Darden, V. Dussupt, E. S. Golub, J. A. Headley, U. M. Jarral, J. King, S. J. Krebs, J. Lay, R. Lilly, J. Lynch, E. J. Martinez, S. V. Mayer, S. McGeehon, H. Lee, S. Schech, M. Tadesse, P. V. Thomas, Y. Romem, E. Zografos, B. C. Lin, S. R. Narpala, L. Wang, N. A. Doria-Rose, R. E. Carroll, A. Eaton, E. D. Badraslioglu, J. M. Koontz, U. E. Nwaeze, P. Dawson, A. J. Noll, C. M. Orndahl, A. Bray, R. Carrion, J. Patterson, V. Kulkarni, C. Hallam, O. Gonzalez and M. Gazi, SARS-CoV-2 recombinant spike ferritin nanoparticle vaccine adjuvanted with Army Liposome Formulation containing monophosphoryl lipid A and QS-21: a phase 1, randomised, double-blind, placebo-controlled, first-in-human clinical trial, Lancet Microbe, 2024, 5, e581–e593 CrossRef CAS PubMed.
- J. Y. Song, W. S. Choi, J. Y. Heo, E. J. Kim, J. S. Lee, D. S. Jung, S.-W. Kim, K.-H. Park, J. S. Eom, S. J. Jeong, J. Lee, K. T. Kwon, H. J. Choi, J. W. Sohn, Y. K. Kim, B. W. Yoo, I.-J. Jang, M. Z. Capeding, F. Roman, T. Breuer, P. Wysocki, L. Carter, S. Sahastrabuddhe, M. Song, N. D’Cor, H. Kim, J. H. Ryu, S. J. Lee, Y. W. Park and H. J. Cheong, Immunogenicity and safety of SARS-CoV-2 recombinant protein nanoparticle vaccine GBP510 adjuvanted with AS03: interim results of a randomised, active-controlled, observer-blinded, phase 3 trial, eClinicalMedicine, 2023, 64, 102140 CrossRef PubMed.
- K. V. Houser, G. L. Chen, C. Carter, M. C. Crank, T. A. Nguyen, M. C. Burgos Florez, N. M. Berkowitz, F. Mendoza, C. S. Hendel, I. J. Gordon, E. E. Coates, S. Vazquez, J. Stein, C. L. Case, H. Lawlor, K. Carlton, M. R. Gaudinski, L. Strom, A. R. Hofstetter, C. J. Liang, S. Narpala, C. Hatcher, R. A. Gillespie, A. Creanga, M. Kanekiyo, J. E. Raab, S. F. Andrews, Y. Zhang, E. S. Yang, L. Wang, K. Leung, W.-P. Kong, A. W. Freyn, R. Nachbagauer, P. Palese, R. T. Bailer, A. B. McDermott, R. A. Koup, J. G. Gall, F. Arnold, J. R. Mascola, B. S. Graham and J. E. Ledgerwood, Safety and immunogenicity of a ferritin nanoparticle H2 influenza vaccine in healthy adults: a phase 1 trial, Nat. Med., 2022, 28, 383–391 CrossRef CAS PubMed.
- P. Tang, E. Cui, J. Cheng, B. Li, J. Tao, Y. Shi, J. Jiao, E. Du, J. Wang and H. Liu, A ferritin nanoparticle vaccine based on the hemagglutinin extracellular domain of swine influenza A (H1N1) virus elicits protective immune responses in mice and pigs, Front. Immunol., 2024, 15, 1361323 CrossRef CAS PubMed.
- M. C. Miranda, E. Kepl, M. J. Navarro, C. Chen, M. Johnson, K. R. Sprouse, C. Stewart, A. Palser, A. Valdez, D. Pettie, C. Sydeman, C. Ogohara, J. C. Kraft, M. Pham, M. Murphy, S. Wrenn, B. Fiala, R. Ravichandran, D. Ellis, L. Carter, D. Corti, P. Kellam, K. Lee, A. C. Walls, D. Veesler and N. P. King, Potent neutralization of SARS-CoV-2 variants by RBD nanoparticle and prefusion-stabilized spike immunogens, npj Vaccines, 2024, 9, 1–14 CrossRef PubMed.
- P. A.-B. Weidenbacher, M. Sanyal, N. Friedland, S. Tang, P. S. Arunachalam, M. Hu, O. S. Kumru, M. K. Morris, J. Fontenot, L. Shirreff, J. Do, Y.-C. Cheng, G. Vasudevan, M. B. Feinberg, F. J. Villinger, C. Hanson, S. B. Joshi, D. B. Volkin, B. Pulendran and P. S. Kim, A ferritin-based COVID-19 nanoparticle vaccine that elicits robust, durable, broad-spectrum neutralizing antisera in non-human primates, Nat. Commun., 2023, 14, 2149 CrossRef CAS PubMed.
- U. K. Kar, J. Jiang, C. I. Champion, S. Salehi, M. Srivastava, S. Sharma, S. Rabizadeh, K. Niazi, V. Kickhoefer, L. H. Rome and K. A. Kelly, Vault Nanocapsules as Adjuvants Favor Cell-Mediated over Antibody-Mediated Immune Responses following Immunization of Mice, PLoS One, 2012, 7, e38553 CrossRef CAS PubMed.
- N. M. Molino, A. K. L. Anderson, E. L. Nelson and S.-W. Wang, Biomimetic Protein Nanoparticles Facilitate Enhanced Dendritic Cell Activation and Cross-Presentation, ACS Nano, 2013, 7, 9743–9752 CrossRef CAS PubMed.
- J.-A. Han, Y. J. Kang, C. Shin, J.-S. Ra, H.-H. Shin, S. Y. Hong, Y. Do and S. Kang, Ferritin protein cage nanoparticles as versatile antigen delivery nanoplatforms for dendritic cell (DC)-based vaccine development, Nanomedicine, 2014, 10, 561–569 CrossRef CAS PubMed.
- J.-S. Ra, H.-H. Shin, S. Kang and Y. Do, Lumazine synthase protein cage nanoparticles as antigen delivery nanoplatforms for dendritic cell-based vaccine development, Clin. Exp. Vaccine Res., 2014, 3, 227–234 CrossRef CAS PubMed.
- A. Naskalska, K. Borzęcka-Solarz, J. Różycki, I. Stupka, M. Bochenek, E. Pyza and J. G. Heddle, Artificial Protein Cage Delivers Active Protein Cargos to the Cell Interior, Biomacromolecules, 2021, 22, 4146–4154 CrossRef CAS PubMed.
- F. Sandra, N. U. Khaliq, A. Sunna and A. Care, Developing Protein-Based Nanoparticles as Versatile Delivery Systems for Cancer Therapy and Imaging, Nanomaterials, 2019, 9, 1329 CrossRef CAS PubMed.
- T. Kawano, M. Murata, J.-H. Kang, J. S. Piao, S. Narahara, F. Hyodo, N. Hamano, J. Guo, S. Oguri, K. Ohuchida and M. Hashizume, Ultrasensitive MRI detection of spontaneous pancreatic tumors with nanocage-based targeted contrast agent, Biomaterials, 2018, 152, 37–46 CrossRef CAS PubMed.
- K. Fan, C. Cao, Y. Pan, D. Lu, D. Yang, J. Feng, L. Song, M. Liang and X. Yan, Magnetoferritin nanoparticles for targeting and visualizing tumour tissues, Nat. Nanotechnol., 2012, 7, 459–464 CrossRef CAS PubMed.
- Z. Wang, P. Huang, O. Jacobson, Z. Wang, Y. Liu, L. Lin, J. Lin, N. Lu, H. Zhang, R. Tian, G. Niu, G. Liu and X. Chen, Biomineralization-Inspired Synthesis of Copper Sulfide–Ferritin Nanocages as Cancer Theranostics, ACS Nano, 2016, 10, 3453–3460 CrossRef CAS PubMed.
- H. Hu, Y. Zhang, S. Shukla, Y. Gu, X. Yu and N. F. Steinmetz, Dysprosium-Modified Tobacco Mosaic Virus Nanoparticles for Ultra-High-Field Magnetic Resonance and Near-Infrared Fluorescence Imaging of Prostate Cancer, ACS Nano, 2017, 11, 9249–9258 CrossRef CAS PubMed.
- M. G. Shapiro, P. W. Goodwill, A. Neogy, M. Yin, F. S. Foster, D. V. Schaffer and S. M. Conolly, Biogenic gas nanostructures as ultrasonic molecular reporters, Nat. Nanotechnol., 2014, 9, 311–316 CrossRef CAS PubMed.
- F. Silva, L. Sitia, R. Allevi, A. Bonizzi, M. Sevieri, C. Morasso, M. Truffi, F. Corsi and S. Mazzucchelli, Combined Method to Remove Endotoxins from Protein Nanocages for Drug Delivery Applications: The Case of Human Ferritin, Pharmaceutics, 2021, 13, 229 CrossRef CAS PubMed.
- Y. Li and J. A. Champion, Self-assembling nanocarriers from engineered proteins: Design, functionalization, and application for drug delivery, Adv. Drug Delivery Rev., 2022, 189, 114462 CrossRef CAS PubMed.
- C. Yin, P. Hu, L. Qin, Z. Wang and H. Zhao, The Current Status and Future Directions on Nanoparticles for Tumor Molecular Imaging, Int. J. Nanosci., 2024, 19, 9549–9574 CAS.
- F. Mainini, A. Bonizzi, M. Sevieri, L. Sitia, M. Truffi, F. Corsi and S. Mazzucchelli, Protein-Based Nanoparticles for the Imaging and Treatment of Solid Tumors: The Case of Ferritin Nanocages, a Narrative Review, Pharmaceutics, 2021, 13, 2000 CrossRef CAS PubMed.
- K. Shi, Y. Wang, X. Zhou, H. Gui, N. Xu, S. Wu, C. He and Z. Zhao, Tumor microenvironment targeting with dual stimuli-responsive nanoparticles based on small heat shock proteins for antitumor drug delivery, Acta Biomater., 2020, 114, 369–383 CrossRef CAS PubMed.
- X. Pan and T. Kortemme, Recent advances in de novo protein design: Principles, methods, and applications, J. Biol. Chem., 2021, 296, 100558 CrossRef CAS PubMed.
- T. Kortemme, De novo protein design—From new structures to programmable functions, Cell, 2024, 187, 526–544 CrossRef CAS PubMed.
- C. Mirabello, B. Wallner, B. Nystedt, S. Azinas and M. Carroni, Unmasking AlphaFold to integrate experiments and predictions in multimeric complexes, Nat. Commun., 2024, 15, 8724 CrossRef CAS PubMed.
- B. Shor and D. Schneidman-Duhovny, CombFold: predicting structures of large protein assemblies using a combinatorial assembly algorithm and AlphaFold2, Nat. Methods, 2024, 21, 477–487 CrossRef CAS PubMed.
- B. B. Mallik, J. Stanislaw, T. M. Alawathurage and A. Khmelinskaia, De Novo Design of Polyhedral Protein Assemblies: Before and After the AI Revolution, ChemBioChem, 2023, 24, e202300117 CrossRef CAS PubMed.
- M. Baek, F. DiMaio, I. Anishchenko, J. Dauparas, S. Ovchinnikov, G. R. Lee, J. Wang, Q. Cong, L. N. Kinch, R. D. Schaeffer, C. Millán, H. Park, C. Adams, C. R. Glassman, A. DeGiovanni, J. H. Pereira, A. V. Rodrigues, A. A. van Dijk, A. C. Ebrecht, D. J. Opperman, T. Sagmeister, C. Buhlheller, T. Pavkov-Keller, M. K. Rathinaswamy, U. Dalwadi, C. K. Yip, J. E. Burke, K. C. Garcia, N. V. Grishin, P. D. Adams, R. J. Read and D. Baker, Accurate prediction of protein structures and interactions using a three-track neural network, Science, 2021, 373, 871–876 CrossRef CAS PubMed.
- R. Evans, M. O’Neill, A. Pritzel, N. Antropova, A. Senior, T. Green, A. Žídek, R. Bates, S. Blackwell, J. Yim, O. Ronneberger, S. Bodenstein, M. Zielinski, A. Bridgland, A. Potapenko, A. Cowie, K. Tunyasuvunakool, R. Jain, E. Clancy, P. Kohli, J. Jumper and D. Hassabis, Protein complex prediction with AlphaFold-Multimer, bioRxiv, 2021, preprint DOI:10.1101/2021.10.04.463034.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard, J. Bambrick, S. W. Bodenstein, D. A. Evans, C.-C. Hung, M. O’Neill, D. Reiman, K. Tunyasuvunakool, Z. Wu, A. Žemgulytė, E. Arvaniti, C. Beattie, O. Bertolli, A. Bridgland, A. Cherepanov, M. Congreve, A. I. Cowen-Rivers, A. Cowie, M. Figurnov, F. B. Fuchs, H. Gladman, R. Jain, Y. A. Khan, C. M. R. Low, K. Perlin, A. Potapenko, P. Savy, S. Singh, A. Stecula, A. Thillaisundaram, C. Tong, S. Yakneen, E. D. Zhong, M. Zielinski, A. Žídek, V. Bapst, P. Kohli, M. Jaderberg, D. Hassabis and J. M. Jumper, Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature, 2024, 630, 493–500 CrossRef CAS PubMed.
- P. Bryant, G. Pozzati, W. Zhu, A. Shenoy, P. Kundrotas and A. Elofsson, Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search, Nat. Commun., 2022, 13, 6028 CrossRef CAS PubMed.
- Z. Li, S. Yang, X. Liu, W. Chen, H. Wen, F. Shen, G. Ke and L. Zhang, Uni-Fold Symmetry: Harnessing Symmetry in Folding Large Protein Complexes, bioRxiv, 2022, preprint DOI:10.1101/2022.08.30.505833.
- R. Wu, F. Ding, R. Wang, R. Shen, X. Zhang, S. Luo, C. Su, Z. Wu, Q. Xie, B. Berger, J. Ma and J. Peng, High-resolution de novo structure prediction from primary sequence, bioRxiv, 2022, preprint DOI:10.1101/2022.07.21.500999.
- Z. Lin, H. Akin, R. Rao, B. Hie, Z. Zhu, W. Lu, N. Smetanin, R. Verkuil, O. Kabeli, Y. Shmueli, A. dos Santos Costa, M. Fazel-Zarandi, T. Sercu, S. Candido and A. Rives, Evolutionary-scale prediction of atomic-level protein structure with a language model, Science, 2023, 379, 1123–1130 CrossRef CAS PubMed.
- R. Krishna, J. Wang, W. Ahern, P. Sturmfels, P. Venkatesh, I. Kalvet, G. R. Lee, F. S. Morey-Burrows, I. Anishchenko, I. R. Humphreys, R. McHugh, D. Vafeados, X. Li, G. A. Sutherland, A. Hitchcock, C. N. Hunter, A. Kang, E. Brackenbrough, A. K. Bera, M. Baek, F. DiMaio and D. Baker, Generalized biomolecular modeling and design with RoseTTAFold All-Atom, Science, 2024, 384, eadl2528 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |