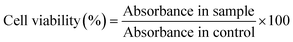Open Access Article
Open Access ArticleSolvent-driven sod-ZIF-8 ↔ ZIF-C phase transformation preserves nucleic acid functionality for gene delivery
Shakil Ahmed
Polash
ab,
Arpita
Poddar
c,
Francesco
Carraro
 d,
Gary
Bryant
d,
Gary
Bryant
 b,
Paolo
Falcaro
b,
Paolo
Falcaro
 *d and
Ravi
Shukla
*d and
Ravi
Shukla
 *abe
*abe
aSir Ian Potter NanoBiosensing Facility, NanoBiotechnology Research Laboratory, RMIT University, Melbourne, Victoria 3000, Australia. E-mail: ravi.shukla@rmit.edu.au
bSchool of Science, STEM College, RMIT University, Melbourne, Victoria 3000, Australia
cFiona Elsey Cancer Research Institute, Ballarat, Victoria 3350, Australia
dInstitute of Physical and Theoretical Chemistry, Graz University of Technology, Graz 8010, Austria. E-mail: paolo.falcaro@tugraz.at
eCentre for Advanced Materials & Industrial Chemistry, RMIT University, Melbourne, Victoria 3000, Australia
First published on 19th December 2025
Abstract
Metal–organic frameworks (MOFs) built from zinc ions and 2-methylimidazole (HmIM) are widely explored carriers for gene delivery. Although preliminary reports show phase-dependent properties, often studies label distinct crystalline phases as “ZIF-8” and overlook how routine washing steps influence the property-to-function relationship. Here, we examine in depth how solvent history tunes the crystal phase and biological performance in plasmid DNA (pDNA) encapsulated in Zn–mIM carriers. An aqueous biomimetic mineralization at 25 °C (HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn2+ = 4
Zn2+ = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) yields defect-rich sodalite (sod) ZIF-8 with an ∼80% encapsulation efficiency of the input plasmid and four times the loading obtained from an identical ethanolic synthesis. Post-synthetic solvent washes or aging govern phase transformations: media containing ≥70% water trigger a solution-mediated transformation into a carbonate–imidazolate framework (ZIF-C), whereas absolute ethanol stabilizes sod ZIF-8 topology. Despite the extensive recrystallization processes into chemically distinct solids, pDNA loss remains ≤12% for sod → ZIF-C and ≤20% for ZIF-C → sod transformation. The green fluorescent protein (GFP) assays confirm that the recovered pDNA retains full transcriptional activity. Comparative cytotoxicity tests on PC-3 cells show that water-aged ZIF-C sustains ≥85% viability and superior colloidal stability, while ethanol-stabilized sod-ZIF-8 possesses a higher pDNA loading and an on-demand burst release upon first contact with water. By examining how post-processing with ethanol/water yields pure sod, mixed sod/ZIF-C, and pure ZIF-C carriers, this work provides the first solvent-phase guide for MOF gene carriers and establishes simple washing protocols as tools to tune release kinetics, stability, and biocompatibility without altering the ZIF precursors or the reaction temperature. By enabling phase-engineered DNA@ZIF carriers with predictable gene-delivery performance, this study advances MOF-mediated nucleic acid therapies toward reproducible biomedical applications.
1) yields defect-rich sodalite (sod) ZIF-8 with an ∼80% encapsulation efficiency of the input plasmid and four times the loading obtained from an identical ethanolic synthesis. Post-synthetic solvent washes or aging govern phase transformations: media containing ≥70% water trigger a solution-mediated transformation into a carbonate–imidazolate framework (ZIF-C), whereas absolute ethanol stabilizes sod ZIF-8 topology. Despite the extensive recrystallization processes into chemically distinct solids, pDNA loss remains ≤12% for sod → ZIF-C and ≤20% for ZIF-C → sod transformation. The green fluorescent protein (GFP) assays confirm that the recovered pDNA retains full transcriptional activity. Comparative cytotoxicity tests on PC-3 cells show that water-aged ZIF-C sustains ≥85% viability and superior colloidal stability, while ethanol-stabilized sod-ZIF-8 possesses a higher pDNA loading and an on-demand burst release upon first contact with water. By examining how post-processing with ethanol/water yields pure sod, mixed sod/ZIF-C, and pure ZIF-C carriers, this work provides the first solvent-phase guide for MOF gene carriers and establishes simple washing protocols as tools to tune release kinetics, stability, and biocompatibility without altering the ZIF precursors or the reaction temperature. By enabling phase-engineered DNA@ZIF carriers with predictable gene-delivery performance, this study advances MOF-mediated nucleic acid therapies toward reproducible biomedical applications.
1. Introduction
An ideal nucleic acid delivery system must protect the biotherapeutic cargo from degradation before it reaches its intracellular targets. Engineered viral vectors meet this criterion and have been widely used for nucleic acid delivery; however, they are costly to manufacture and are burdened by immunogenicity concerns.1 These limitations have driven intensive research into non-viral delivery platforms.2,3 The literature reports different particle systems used for the delivery of biotherapeutics,4 and, among these, metal–organic frameworks (MOFs) show strong potential in biomedicine.5–7 MOFs are porous hybrid materials composed of metal ions (or clusters) coordinated to organic bridging ligands.8 Their high surface areas, tunable pore apertures, crystallinity, and chemical versatility enable diverse biological applications.8 Specifically, within the MOF family, zeolitic imidazolate frameworks (ZIFs) have emerged as materials of interest for biological and biomedical uses: Zn-based ZIFs can form in water under mild aqueous conditions around biomolecules, possess thermal and chemical stability, are generally biocompatible, and the release of the biotherapeutic cargo can be triggered on demand.9 However, identical Zn2+ and methylimidazole (HmIM) precursors can yield different Zn–mIM-based crystalline structures.Despite the use of identical building blocks, different Zn–mIM based crystalline structures possess distinct physicochemical properties, porosity, accessible surface area, specific volume, and chemical stability.10 Examples include ZIF-8 sodalite (sod), diamondoid (dia), and a carbonated derivative (ZIF-C) that, for biomedical applications, exhibit diverse encapsulation efficiencies, degradation kinetics, and release profiles, cytotoxicity, and cellular uptake.11,12 For example, the sod phase is an open, highly porous framework that can support high biomolecule loading but is comparatively less stable in aqueous or physiological media.13,14 In contrast, the denser ZIF-C phase that incorporates carbonate anions in the nonporous framework offers stability and cargo protection; these distinct properties are desirable in customized delivery systems.15,16 Therefore, for ZIF-based biomedical systems, fine-tuning of ZIF phases enables the precise control over crucial functional properties.
Despite growing awareness of ZIF polymorphism, the literature often labels distinct Zn–mIM crystalline materials as “ZIF-8”, overlooking fundamental structure–function relationships. In addition, post-synthesis washing steps-commonly performed with alcohols (e.g., ethanol) or water to remove unreacted precursors before microscopy, X-ray diffraction (XRD), or surface area measurements - can themselves induce crystal phase changes. When this goes unnoticed or unreported, the characterized material no longer matches the material structure employed in biological studies, leading to inaccurate conclusions. Because phase governs properties central to gene and drug delivery (loading, stability, release, cytocompatibility), a clearer understanding of how routine solvent washes influence Zn–mIM biocomposites is needed.
For gene delivery, liposomes have been widely used in mRNA vaccines and siRNA therapeutics. However, colloidal instability, rapid degradation, and cargo leakage behavior in physiological fluids underscore the need to find alternative nonviral carriers. The tunable physicochemical properties of MOFs enable controlled drug release and improved storage stability.17 A concise comparison is provided in SI (see Table S1). ZIF-based carriers have been explored in cancer therapy, gene therapy, photodynamic/photothermal therapy, theranostic nanomedicines, and antimicrobial applications.6,18–20 However, successful translation depends on reliable synthesis and processing conditions. Prior studies have shown that phase outcome can depend on precursor concentrations, biomolecule identity, and washing solvent (water versus ethanol). The mechanistic understanding of solvent-induced transitions has remained incomplete. In DNA-loaded ZIFs (DNA@ZIF), precursor mass fractions and metal-to-ligand molar ratio influence crystal topology and DNA loading.21,22 While the washing solvent was preliminarily observed influencing the phase of DNA@ZIF systems,21 a systematic study of how ethanol/water wash mixtures control phase formation, recrystallization, nucleic acid retention, and biocompatibility in plasmid-loaded ZIF biocomposites is still lacking. As highlighted in recent critical review studies, progress toward clinical use will depend on closing the reproducibility gap: developing robust synthetic protocols, standardized preparation/characterization, and systematic structure–function studies that validate MOF carriers under biologically relevant conditions.23,24
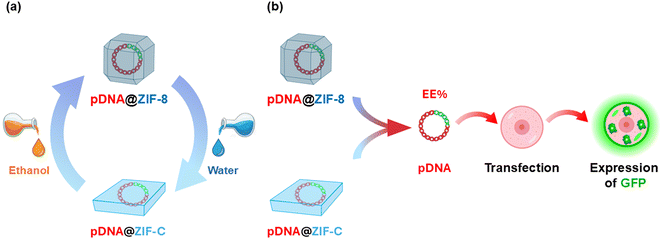
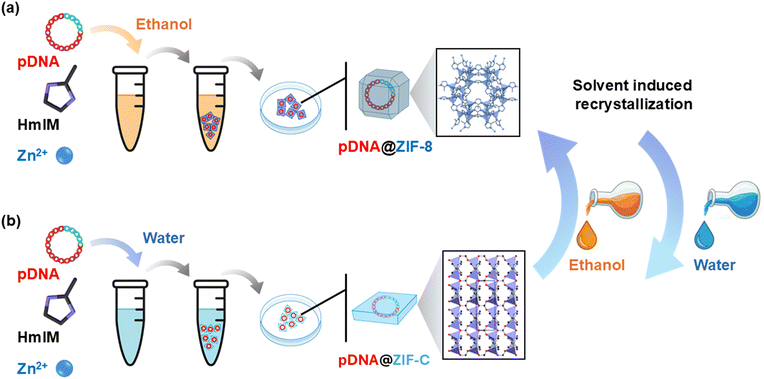
Here, for the first time, we systematically investigate the influence of mixtures of ethanol/water (EtOH/H2O) in the synthesis of plasmid DNA-loaded ZIF particles (pDNA@ZIF) and as wash mixtures. Interestingly, while the ZIF-C to sod ZIF-8 is the main recrystallization process documented in the literature, here we observe the unnoticed recrystallization from sod ZIF-8 to ZIF-C. The concept of solvent-dependent phase transitions is summarized in Scheme 1. We observed the effect of carbonate ion and solvent polarity in directing the transformation of ZIF phases. Specifically, we (i) systematically map the crystalline phase mixtures as a function of EtOH/H2O wash ratio, (ii) identify and quantify a mixed sod/ZIF-C phase window, (iii) show that exchanging wash solvents post-synthesis can drive reversible sod ⇄ ZIF-C transformations (Scheme 1a) and (iv) assess plasmid DNA loading/retention/integrity (Scheme 1b) and cytotoxicity across sod and ZIF-C phases. Importantly, we found that plasmids recovered from these recrystallized framework materials retain functional integrity for subsequent transfection into mammalian cells. The PC-3 prostate cancer cell line was selected as a representative in vitro model to evaluate the cytocompatibility and gene delivery efficiency. Collectively, our results demonstrate that solvent washing, as a post-synthetic treatment, is a simple and effective procedure for tuning phase and properties in pDNA@ZIF systems relevant to gene delivery and broader nanomedicine applications. A final section summarizes the results and provides guidelines addressing the development of the ZIF-based materials.23,24
2. Results and discussion
The molar ratio between 2-methylimidazole (HmIM) and Zn2+ and the surface charge of biomolecules play a significant role in the formation of ZIF.25 A typical biomimetic mineralization approach was employed to encapsulate plasmid DNA into two distinct ZIFs (i.e., sod ZIF-8 and ZIF-C) according to our previous studies.16,26,27 A fixed 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand-to-metal ratio was used (160 mM HmIM to 40 mM Zn(OAc)2, total volume 200 μL) to encapsulate 5 μg of pDNA. Two distinct batches were prepared to perform the synthesis: (i) water (aqueous conditions from HmIM and Zn2+ dissolved in deionized (DI) H2O, see Scheme 1a) and (ii) ethanol, EtOH (HmIM dissolved in absolute EtOH, and Zn2+ dissolved in 50% v/v EtOH/H2O, see Scheme 1b).
1 ligand-to-metal ratio was used (160 mM HmIM to 40 mM Zn(OAc)2, total volume 200 μL) to encapsulate 5 μg of pDNA. Two distinct batches were prepared to perform the synthesis: (i) water (aqueous conditions from HmIM and Zn2+ dissolved in deionized (DI) H2O, see Scheme 1a) and (ii) ethanol, EtOH (HmIM dissolved in absolute EtOH, and Zn2+ dissolved in 50% v/v EtOH/H2O, see Scheme 1b).
Here, pDNA was encapsulated within ZIF-based materials in two different conditions: H2O and EtOH. The MOF precursors (zinc acetate and HmIM) were dissolved in H2O and EtOH, respectively.25 Right after mixing the Zn2+ solution with the DNA/ligand solution, the suspension turned turbid, indicating rapid coordination-driven self-assembly of the Zn–mIM network.28 After 10 minutes, high-speed centrifugation was used to collect the biocomposites (see SI for details). To investigate the effect of post-synthesis solvent processing, the as-formed pellets were washed three times with EtOH/H2O mixtures containing 0–100% H2O (v/v), using the same solvent composition for all three washes. The washed biocomposites were then vacuum-dried and are referred to in the text by the final wash solvent composition (e.g., 50% H2O wash). This systematic preparation of solvent-wash samples (see Scheme 2) allowed us to identify the distinct contributions of (i) the solvent present during the in situ formation of pDNA@ZIF and (ii) the solvent during washes as post-synthetic treatments before characterization and biological testing. The sections below examine how solvent exposures can tune the crystal phase, morphology, DNA loading, and cytotoxicity.
2.1. Crystallinity and morphology analysis
The crystalline nature of the pDNA@ZIF was studied by XRD. pDNA@ZIF was prepared from an aqueous synthesis (Zn(OAc)2 + HmIM in H2O; L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M = 4
M = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). When this powder sample was washed with anhydrous ethanol (i.e., 0% H2O), diffraction patterns characteristic of pure sod were obtained, confirming results from prior reports.26 To examine the previously unexplored relative contribution of EtOH/H2O mixtures on the pDNA@ZIF (sod) samples, these samples were washed with different EtOH/H2O solutions with a progressive increase of H2O (10% v/v increments). During this screening, an unexpected sod → ZIF-C phase transition was observed (Fig. 1a). Biocomposites washed with up to 60% water retained the characteristic sod topology of ZIF-8. This indicates that the sod phase is preserved when the washing solvent is predominantly EtOH. However, a small fraction of ZIF-C phase began to emerge for EtOH/H2O = 50/50, indicating a partial sod → ZIF-C transformation and a mixed-phase structure. Starting from a H2O = 70% or higher, all pDNA@ZIF composites transform completely into ZIF-C. These data indicate that the washing solvent composition regulates the ZIF crystal structure in H2O-synthesized pDNA@ZIF (sod) samples. For comparison, pDNA@ZIF (sod) samples were synthesized by using ethanolic precursors (Scheme 1b) and exposed to the same EtOH/H2O wash series. For these samples, the sod diffraction patterns indicate that the original topology was retained as the dominant topology for all washes (Fig. S1). Even at the previously observed most aggressive wash (100% H2O), there is no appearance of the ZIF-C (110) peak at 11.0°. The relative sod peak intensities and full-width-at-half-maximum (FWHM) remain almost unchanged; only a slight overall intensity drop is evident at ≥80% H2O, implying a trace loss of crystallinity, but the phase transition is suppressed. SEM analyses showed that pDNA@ZIF particles prepared by using EtOH as a solvent possess the classical rhombic dodecahedral morphology (Fig. S2).
1). When this powder sample was washed with anhydrous ethanol (i.e., 0% H2O), diffraction patterns characteristic of pure sod were obtained, confirming results from prior reports.26 To examine the previously unexplored relative contribution of EtOH/H2O mixtures on the pDNA@ZIF (sod) samples, these samples were washed with different EtOH/H2O solutions with a progressive increase of H2O (10% v/v increments). During this screening, an unexpected sod → ZIF-C phase transition was observed (Fig. 1a). Biocomposites washed with up to 60% water retained the characteristic sod topology of ZIF-8. This indicates that the sod phase is preserved when the washing solvent is predominantly EtOH. However, a small fraction of ZIF-C phase began to emerge for EtOH/H2O = 50/50, indicating a partial sod → ZIF-C transformation and a mixed-phase structure. Starting from a H2O = 70% or higher, all pDNA@ZIF composites transform completely into ZIF-C. These data indicate that the washing solvent composition regulates the ZIF crystal structure in H2O-synthesized pDNA@ZIF (sod) samples. For comparison, pDNA@ZIF (sod) samples were synthesized by using ethanolic precursors (Scheme 1b) and exposed to the same EtOH/H2O wash series. For these samples, the sod diffraction patterns indicate that the original topology was retained as the dominant topology for all washes (Fig. S1). Even at the previously observed most aggressive wash (100% H2O), there is no appearance of the ZIF-C (110) peak at 11.0°. The relative sod peak intensities and full-width-at-half-maximum (FWHM) remain almost unchanged; only a slight overall intensity drop is evident at ≥80% H2O, implying a trace loss of crystallinity, but the phase transition is suppressed. SEM analyses showed that pDNA@ZIF particles prepared by using EtOH as a solvent possess the classical rhombic dodecahedral morphology (Fig. S2).
SEM micrographs provide complementary morphological evidence of the phase transition (Fig. 1b). Truncated cube particles, as a typical shapes that eventually result in rhombic dodecahedral particles,29 have sod topology that dominates at ≤40% H2O; at 50–60% H2O their facets become roughened and tiny plate-like crystallites nucleate on the surface, consistent with the nascent ZIF-C phase detected by XRD analysis. When the H2O fraction reaches 70% or higher, the original cubic morphology disappears, and the crystals are replaced entirely by larger, plate-like particles, which show the typical ZIF-C morphology.
Combined XRD and SEM analyses suggest a solution-mediated recrystallization mechanism. Peak-width evolution indicates that at 50% H2O the nascent ZIF-C (110) reflection is broad (FWHM ≈ 0.43°, calculated average crystallite size: 18 nm) with a low intensity, then the (110) reflection sharpens to 0.27° (calculated average crystallite size: 30 nm) as the H2O fraction reaches 70%, while sod peaks disappear. SEM shows plate-like ZIF-C crystallites growing on dissolving the rhombic dodecahedral sod particles. These data support the dissolution of the original sod framework followed by re-precipitation of ZIF-C, rather than a topotactic solid-state rearrangement.30 As a weaker coordinating solvent, ethanol promotes faster nucleation by limiting Zn2+ solvation and yields small crystals. In contrast, water strongly coordinates with Zn2+ through its lone pair electrons and favors the crystal growth of ZIF-8.
2.2. Phase quantification and reactivity under washes
A web application named “ZIF Phase Analysis” was used to quantify the relative amounts of sod and ZIF-C in the biocomposites.31 The tool deconvolutes XRD patterns and ascribes diffraction peaks to the proper phases, examining the related intensities.32 With this, we analyzed all XRD patterns collected from the different pDNA@ZIF samples (Fig. 2). Mixed sod/ZIF-C profiles appeared only for the aqueous syntheses washed with 50% or 60% H2O, whereas ≥70% H2O yielded phase-pure ZIF-C and ≤40% H2O preserved phase-pure sod. In contrast, particles grown in ethanol remained >99% sod regardless of wash composition, confirming the qualitative XRD trends discussed in section 2.1. | ||
| Fig. 2 Calculated phase of pDNA@ZIF prepared in (a) ethanol and (b) aqueous ZIF precursors and subsequent washing with different % of water in ethanol. | ||
The markedly higher reactivity of the aqueous-route sod toward the sod → ZIF-C transformation can be explained by a greater density of structural defects (e.g., missing linkers and/or under-coordinated Zn nodes) originating during rapid nucleation in water. Specifically, water-based pDNA@ZIF (sod) (hereafter simply pDNA@ZIF-8) samples show broader reflections, a diffuse hump at 18–20° 2θ, and dodecahedra with a rougher surface, all supporting the presence of defects (see Fig. 2a). MOF lattice defects enhance the material network hydrophilicity and reduce the framework stability, providing vacant sites for carbonate uptake, lowering the barrier for sod → ZIF-C recrystallization.33–35 We note that defect-assisted phase transitions in crystalline materials have been documented.36
By comparison, EtOH-based syntheses yield sod with narrower diffraction peaks and higher relative intensities. SEM micrographs show sharp, truncated cubes (Fig. 2b and S2). Collectively, these data indicate the formation of crystalline particles with fewer defects. The enhanced lattice regularity, combined with the hydrophobic nature of sod ZIF-8,37,38 further retards water penetration, carbonate diffusion in porous sod, and Zn ion leaching. This is consistent with the literature that experimentally demonstrates how sod ZIF-8 prepared in organic solvents is robust to water exposure.39 Overall, our data support a defect-mediated, solution-assisted recrystallization mechanism for the sod → ZIF-C transition in pDNA@ZIF-8 particles prepared in water. In contrast, an EtOH-based synthesis yields particles with stable sod topology.
2.3. Particle growth kinetics and encapsulation efficiency
A spectroscopic study was performed to compare the growth kinetics of pDNA@ZIF prepared in H2O and EtOH (Fig. 3a). Immediately after mixing precursors with pDNA, the solution turbidity at 355 nm was recorded to examine the particle formation.16,40 Real-time absorption measurements reveal that particles nucleate rapidly in both solvents, but a clear change in slope separates an initial rapid nucleation phase from a slower particle growth phase. The higher maximum turbidity and steeper initial slope in water indicate faster nucleation/growth and a greater particle concentration. This solvent effect is synergistic to the enhanced pDNA solubility in water: specifically, H2O fully solvates the plasmid.41 Under these conditions, all charged seeds (pDNA) can promote the biomimetic mineralization of ZIF materials. Conversely, nucleic acids are rather insoluble in EtOH,42 thus the partial precipitation of pDNA reduces the heterogeneous nucleation of ZIF particles. The obtained precipitates were collected and digested with 2% nitric acid to analyze the Zn content of the biocomposites via microwave plasma–atomic emission spectrometer (MP-AES). A significant four-fold increase in yield was observed when biomimetic mineralization was conducted under aqueous conditions compared to ethanolic conditions, confirming the beneficial role of choosing a H2O/pDNA system to increase the yield of pDNA@ZIF (Fig. S2).The encapsulation efficiency (EE) of pDNA in both alcoholic and aqueous conditions was verified by agarose gel electrophoresis. After washing with different % of H2O in EtOH, the biocomposites were digested with 20 mM EDTA to release the loaded pDNA completely. The intensity of the nucleic acid band was quantified using ImageJ and then plotted for comparison alongside other data points (Fig. 3b and S3). pDNA@ZIF prepared in H2O possesses an average EE of 80 ± 4% across the full EtOH/H2O wash series (Fig. 2b and S4). Ethanolic syntheses showed consistently lower EE (∼45–55%); this observation agrees with the limited DNA solubility during nucleation. H2O is therefore the preferred medium for the preparation of pDNA@ZIF, providing higher yield and plasmid content. While the subsequent EtOH washes control the ZIF phase, this post-synthetic procedure does not influence the cargo content. With these experiments, we therefore identify guidelines specifically relevant for the preparation of pDNA@ZIF with controlled properties.
2.4. Impact of washing solvent on ZIF crystallinity
Given that pDNA@ZIF composites are routinely isolated, purified, or resuspended through multiple washes and/or solvent exchanges,43–45 we hypothesized that each cycle may further promote lattice reorganization. To examine possible phase transitions, we vacuum-dried EtOH-washed pDNA@ZIF-8 and H2O-washed pDNA@ZIF-C and subjected each to five additional washes performed with pure EtOH or pure H2O. After each washing cycle, the stability of the crystalline phase and particle morphology were investigated by PXRD and SEM, respectively (Fig. 3 and S5–S8).The diffraction pattern of pDNA@ZIF-8 (initially washed with EtOH three times) matched the theoretical sod ZIF-8 diffraction pattern (see characteristic peaks at 7.34°, 10.4°, 12.8°, 16.5°, and 18.1°).46–48 The diffraction pattern remained unchanged after five additional washes with 100% EtOH (Fig. 4a and S4a). However, the diffraction pattern changed when pDNA@ZIF-8 was washed with H2O (Fig. 4a and S4b). While the sod topology withstands the first wash, beginning with the second water rinse and continuing thereafter, a new peak at the 2θ value of 11.04° emerged. The position of this new peak can be ascribed to the (110) ZIF-C crystalline plane.15,49,50 While the intensity of the (110) ZIF-C diffraction peak increased with subsequent washes, the intensity of the (110) sod diffraction peak showed an opposite trend, indicating that sod progressively re-crystallized into ZIF-C. Thus, the diffraction patterns indicate that multiple H2O washes result in the coexistence of sod and ZIF-C. Next, the morphologies of pDNA@ZIF-8 after five washes with pure EtOH or H2O were investigated by SEM (Fig. 4b and S5). EtOH washes (from one to five) do not influence the truncated cube morphology, confirming the stability of pDNA@ZIF-8 particles. However, in the case of H2O washing, the SEM images revealed that, for five washes, some original truncated cubes became holed and mixed with plate-like particles typically ascribed to ZIF-C. This supports the hypothesis that the phase transformation occurs via dissolution-recrystallization mechanisms. During synthesis, 2-methylimidazolate (mIM−) coordinates with Zn2+ cations to form the framework structure. Here, the polarity of the solvent facilitates the deprotonation of HmIM and promotes the hydrolysis of Zn2+ coordination. Carbonate ions of water play the role of phase transformation. Falcaro and co-workers showed that washing treatment induces phase transformation of ZIF biocomposite.30 Low CO2 solubility in water allows the incorporation of one CO32− with one Zn2+ per formula unit to form ZIF-C (Zn2(mIM)2CO3). On the other hand, solubilization of atmospheric CO2 during ethanol washing modifies the coordination equilibrium (Zn–mIM–CO32−) in ZIF-C by removing CO32− to yield a carbonate-free sod framework (Zn(mIM)2).
Then, we examined pDNA@ZIF-C (initially washed with water, 3 times) by using the same procedure. During EtOH washing, the diffractogram of pDNA@ZIF-C did not show changes until the 2nd wash (Fig. 4c and S6). However, after the 3rd EtOH diffraction peaks of sod ZIF-8 emerged, subsequent EtOH washes resulted in a progressive increase of the intensity of the sod peaks and a decrease of (110) ZIF-C. This trend highlights how EtOH washes can trigger the progressive phase transformation of pDNA@ZIF-C into pDNA@ZIF-8. Conversely, water washes do not trigger any effect on the original ZIF-C phase (Fig. S6). Then, the morphology of pDNA@ZIF-C after this washing procedure was examined via SEM (Fig. 4d and S7). The pDNA@ZIF-C maintained typical plate-like morphology up to 2nd EtOH wash, while subsequent washes yielded a drastic change in morphology (formation of truncated cubes), indicating the ZIF-C → sod recrystallization. We note that this phase transformation is followed by a variation of the original particle size, a particle property that in drug delivery systems should be judiciously related to the planned administration route.51 The plasmid encapsulated sod ZIF-8 and ZIF-C followed phase transformation due to the dissolution–recrystallization mechanism.30 Competitive coordination of water and carbonate ions can destabilize the sod framework and promote the formation of the denser ZIF-C phase.
2.5. Stability of ZIF crystalline phase
After synthesizing pDNA@ZIF particles, the investigation of their stability is critical to ensure the consistency of their properties over time. Despite the standardization of protocols being an urgent matter in MOF biomedicine23,24 and the wide attention given to ZIF systems as drug carriers, at the moment there are no studies examining the stability of pDNA@ZIF systems. Motivated by this gap and by some early reports showing how some pure sod ZIF-8 is converted by water to particles with an unknown crystalline phase with leaf-like morphology,52 we examine the stability of pDNA@ZIF-8 and pDNA@ZIF-C when stored in EtOH and DI H2O (static conditions in sealed vials) for hours (i.e., 1, 3, 5, and 24 h). After incubation, the particles were centrifuged, collected, and dried for XRD analysis (Fig. 5a and S9). When immersed in EtOH (Fig. 5a and S9a), the diffraction pattern of pDNA@ZIF-8 remained invariant, thus fully retaining sod phase within 24 h. However, when pDNA@ZIF-8 was immersed in H2O (Fig. 5b and S9b), a 1-hour exposure produced a weak peak of (110) ZIF-C (2θ = 11.04°), indicating the beginning of a sod → ZIF-C transformation. The intensity of such a peak increases over time and becomes 20-times more intense after 24 h immersion.The electron micrograph of representative pDNA@ZIF-8 particles exposed to EtOH revealed a truncated cube morphology (Fig. 5b and S10a). This observation suggests the long-term stability of the sod phase in EtOH. Conversely, when the same pDNA@ZIF-8 particles were incubated in H2O for more than 3 h (Fig. 5b and S10b), the emergence of the typical plate-like ZIF-C shape appeared alongside the feedstock cubic sod ZIF-8. This observation is consistent with the changes observed in the XRD patterns.
Next, we focused on examining the effect of EtOH on pDNA@ZIF-C particles and we performed identical solvent/time exposures (pure H2O and EtOH for 1, 3, 5, and 24 h). While pDNA@ZIF-C maintained its crystallinity in H2O for the entire period (i.e., 24 h), during EtOH incubation (Fig. 5c and S11b) the (110) ZIF-C peak showed a gradual intensity reduction with the immersion time. The decrease in intensity coincided with a broadening of the FWHM, indicating a reduction of crystalline domain size. The appearance of rising peaks at 7.34° and 33° indicated the formation of sod (ZIF-8) and zinc hydroxide.53,54 The detailed mechanism behind this phase transformation and the formation of Zn byproducts requires further investigation. Electron micrographs show a change in pDNA@ZIF-C morphology that is triggered by EtOH exposure (Fig. 5d and S12a). Plate-like ZIF-C particles decreased in number and size, transforming into cubic crystals. Interestingly, pDNA@ZIF-C in water rearranged into an interconnected particle network (Fig. 5d and S12b).
2.6. Encapsulated DNA content in the phase transformed ZIF
ZIF has been confirmed to be an effective therapeutic carrier for DNA delivery.55 However, as we show here that sod ⇄ ZIF-C transformations occur via dissolution and recrystallization, it is crucial to determine whether the carrier reorganization compromises the pDNA payload and integrity. Thus, we tested both washed and prolonged exposures to EtOH and H2O.2.7. Cytotoxicity
A viable gene-delivery vehicle combines efficient payload delivery with minimal cytotoxicity. We therefore evaluated the cytotoxicity of the pDNA-loaded biocomposites against PC-3 cells using the MTT reduction assay. In living cells, the MTT, a yellow tetrazole, is reduced to purple formazan by the activity of mitochondrial dehydrogenases. Therefore, the MTT reduction assay reflects the redox activity of living cells. Fig. 6 shows the percentage of cell viability after a 24 h exposure to pDNA@ZIF-8 and pDNA@ZIF-C. This duration is used in preliminary cytotoxicity assessments of MOFs, and it is sufficient for materials to interact with cells for transfection studies that typically use 1–3 h of exposure time for cellular uptake.16,56Fig. 6a shows the effect of successive washes with H2O and EtOH. When pDNA@ZIF-8 was synthesized and repeatedly washed in H2O, cell viability remained ≥85% through all five cycles. An almost identical profile was obtained for the EtOH-washed series, with the lowest value of 82% recorded after the fifth wash. When referring to the aforementioned structural characterization, the 5th wash correlates with sod ZIF-8 with fewer defects. Thus, when considering preliminary reports on the cytotoxicity of pure ZIF-C and sod ZIF-8,15,49 we hypothesize that the higher the content of highly crystalline sod ZIF-8 with respect to ZIF-C, the higher the cytotoxicity.
Fig. 6b examines the effect of incubation time on cytotoxicity. When for pDNA@ZIF-8 the process of synthesis, wash, and aging was entirely performed in H2O, cell viability remained ≥85% across all steps. A slightly enhanced cell viability was observed after the third and fifth washes (sod → ZIF-C), while an additional 5% drop was observed for the samples stored in EtOH (more defective sod → less defective sod). These data further support the lower cytotoxicity of ZIF-C compared to sod ZIF-8 in mixed ZIFs phases and show how defects in sod ZIF-8 can slightly enhance cell viability. Despite these measurable changes in viability, all the ZIF-based composites tested in Fig. 6 exhibited viability values above 70%, a threshold defined by ISO 10993-5 standard to formally distinguish non-cytotoxic from cytotoxic materials.57
2.8. Bioactivity analysis
To verify that pDNA retained its functional integrity after encapsulation, solvent handling, and phase conversion, we performed a transfection in PC-3 cells to express green fluorescent protein (GFP). Because the different MOF matrices (sod ZIF-8, ZIF-C, or their mixtures) could themselves influence cellular uptake, we first released pDNA from the ZIF-based particles with EDTA and then loaded pDNA in Lipofectamine to use a standard transfection protocol. This release and reload approach aims at the assessment of the pDNA functionality by removing any carrier-specific effects.The transfection efficiency was quantified by examining GFP-positive PC-3 cells 24 h after transfection with a confocal laser scanning microscope (CLSM). Fluorescence data for transfections by plasmid recovered from particles that had undergone (i) one to five sequential washes in H2O or EtOH and (ii) 1, 3, 5 or 24 h aging in the same solvents is shown in Fig. 7. Untreated cells (negative control) showed no green fluorescence, whereas all cells receiving released plasmid emitted the typical GFP signals (Fig. 7 and S16). The fluorescence intensity and the fraction of positive cells (≈40% across the data set) were statistically indistinguishable from those obtained with pDNA extracted from freshly synthesized and never-washed pDNA@ZIF-8. The experiment demonstrates that the encapsulated plasmid retained full transcriptional and translational functionality irrespective of solvent history. Here, the release-and-reload strategy confirmed the functional integrity of encapsulated nucleic acid, which remained transcriptionally competent after solvent-mediated phase change. Therefore, this approach provides an unambiguous indication that solvent treatments did not cause loss of function of encapsulated nucleic acid.
2.9. Comparative summary and recommendations
When comparing water- and ethanol-based syntheses, an EtOH solvent yields pDNA@ZIF-8 with a higher crystallinity, a much lower encapsulation efficiency (EE ≈ 45–55%), and an inferior yield of solid material. Thus, the following comparison focuses on carriers prepared in water, where fully dissolved pDNA acts as an effective nucleation seed for biomimetic mineralization. When pDNA@ZIF is prepared in water at a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 = L
1 = L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M ratio (i.e., 40 mM Zn(OAc)2·2H2O and 640 mM HmIM added to 1 mL pDNA solution), a defect-rich, cubic sod ZIF-8 biocomposite with EE = 80 ± 4% is obtained. Storage in water (≤1 h) preserves the sod topology and yields the maximum plasmid density per gram of ZIF. If the suspension is then brought into contact with a water predominant environment (H2O ≥ 70%), defects favor a solution-mediated re-crystallization into the denser ZIF-C polymorph. In 20 h the recrystallization is complete. The phase change releases no more than 12% of the initially captured pDNA (payload retained ≈ 88%). However, such a pDNA release that accompanies sod → ZIF-C can afford the delivery of higher absolute amounts of pDNA during the first hours after administration. On the other hand, sod-based carriers that remain suspended in pure water without completing the transition lose up to 20% pDNA over the same interval, most likely through enhanced diffusion in defects rather than wholesale lattice rearrangement. Exposure to ethanol converts ZIF-C (Zn2(mIM)2CO3) into the carbonate-free sod phase (Zn(mIM)2) by removing Zn2+ and CO32− from the framework, a process influenced by how carbonate ions interact with each solvent. Ethanol, despite the higher CO2 solubility compared to water, does not generate carbonate ions due to its weaker basicity, which helps to drive the recrystallization toward the sod phase.30 On the other hand, water dissolves CO2 to form carbonate and bicarbonate ions. In this case, the thermodynamics of the system is driving the sod to ZIF-C conversion. In fact, Navrotsky and co-workers evaluated the thermodynamic driving force for sod and ZIF-C topologies.58ZIF-C (ΔH = −87.88 kJ mol−1) is thermodynamically more stable than the sod (−21.04 kJ mol−1) phase and the transformation between ZIF phases (e.g., sod ⇄ ZIF-C) is driven by Gibbs free energy.
M ratio (i.e., 40 mM Zn(OAc)2·2H2O and 640 mM HmIM added to 1 mL pDNA solution), a defect-rich, cubic sod ZIF-8 biocomposite with EE = 80 ± 4% is obtained. Storage in water (≤1 h) preserves the sod topology and yields the maximum plasmid density per gram of ZIF. If the suspension is then brought into contact with a water predominant environment (H2O ≥ 70%), defects favor a solution-mediated re-crystallization into the denser ZIF-C polymorph. In 20 h the recrystallization is complete. The phase change releases no more than 12% of the initially captured pDNA (payload retained ≈ 88%). However, such a pDNA release that accompanies sod → ZIF-C can afford the delivery of higher absolute amounts of pDNA during the first hours after administration. On the other hand, sod-based carriers that remain suspended in pure water without completing the transition lose up to 20% pDNA over the same interval, most likely through enhanced diffusion in defects rather than wholesale lattice rearrangement. Exposure to ethanol converts ZIF-C (Zn2(mIM)2CO3) into the carbonate-free sod phase (Zn(mIM)2) by removing Zn2+ and CO32− from the framework, a process influenced by how carbonate ions interact with each solvent. Ethanol, despite the higher CO2 solubility compared to water, does not generate carbonate ions due to its weaker basicity, which helps to drive the recrystallization toward the sod phase.30 On the other hand, water dissolves CO2 to form carbonate and bicarbonate ions. In this case, the thermodynamics of the system is driving the sod to ZIF-C conversion. In fact, Navrotsky and co-workers evaluated the thermodynamic driving force for sod and ZIF-C topologies.58ZIF-C (ΔH = −87.88 kJ mol−1) is thermodynamically more stable than the sod (−21.04 kJ mol−1) phase and the transformation between ZIF phases (e.g., sod ⇄ ZIF-C) is driven by Gibbs free energy.
Immediate solvent exchange to anhydrous EtOH (three consecutive 1 mL washes, same centrifugation parameters) removes water from the porous frameworks, recovers defects, and locks the framework in the sod topology. The resulting particles are smaller, with sharper cubic features, and exhibit a slightly lower PC-3 cell viability floor (≈82%) than their water-aged, ZIF-C counterparts (≥85%), yet they retain the full plasmid payload as long as ethanol remains the continuous phase. Notably, re-introducing ≥ 70% water at any later point restarts the sod → ZIF-C conversion and again generates a rapid plasmid-release pulse, demonstrating full reversibility of the solvent trigger.
3. Conclusion
This study addresses reproducibility issues in ZIF-based gene delivery by demonstrating that the crystal structure, defects, and biological properties of plasmid-loaded zinc–imidazolate frameworks (pDNA@ZIF composites), for the same initial stoichiometry, can be controlled by the solvent exposure protocol applied. This includes the post-preparation washes and storage conditions. Using a single, scalable aqueous biomimetic mineralization protocol at a fixed 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 HmIM
1 HmIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Zn2+ ratio, we establish a solvent–phase map in which routine washing and storage solvents alone define the final material: media that remain ≤60% water preserve a defect-rich sod-ZIF-8, whereas ≥70% water drives a previously unreported, vacancy-assisted reconstruction into carbonate-containing ZIF-C, and ethanol reverses the process on demand. These sod ⇄ ZIF-C transitions occur with low plasmid loss (≤12% during sod → ZIF-C and ≤20% for the reverse step), whereas pDNA maintains transcriptional activity. By correlating X-ray patterns, SEM micrographs, and MP-AES data, we show that the higher vacancy density from the aqueous synthesis facilitates the solution-mediated transformation, while ethanol-grown sod particles with fewer defects remain phase-invariant. Biological assays reveal complementary advantages: post-synthesis ethanol-washed sod-ZIF-8 offers the greatest pDNA density and a solvent-triggered burst release, whereas H2O-aged ZIF-C provides superior colloidal stability and slightly higher cell viability. Because both Zn–mIM polymorphs originate from the same precursors and reaction conditions, researchers can now choose the washing solvent or procedure as a facile approach to finely tune the particle properties (e.g., morphology, DNA loading, cytocompatibility) without altering the synthesis. Therefore, these findings shed light on solvent washes, as a step often relegated to a one-line procedural detail that is here revealed to be a critical design parameter for ZIF biocomposites. Reporting details about washing conditions is therefore essential for the reproducible development, comparison, and clinical translation of future ZIF-based nucleic-acid delivery systems.
Zn2+ ratio, we establish a solvent–phase map in which routine washing and storage solvents alone define the final material: media that remain ≤60% water preserve a defect-rich sod-ZIF-8, whereas ≥70% water drives a previously unreported, vacancy-assisted reconstruction into carbonate-containing ZIF-C, and ethanol reverses the process on demand. These sod ⇄ ZIF-C transitions occur with low plasmid loss (≤12% during sod → ZIF-C and ≤20% for the reverse step), whereas pDNA maintains transcriptional activity. By correlating X-ray patterns, SEM micrographs, and MP-AES data, we show that the higher vacancy density from the aqueous synthesis facilitates the solution-mediated transformation, while ethanol-grown sod particles with fewer defects remain phase-invariant. Biological assays reveal complementary advantages: post-synthesis ethanol-washed sod-ZIF-8 offers the greatest pDNA density and a solvent-triggered burst release, whereas H2O-aged ZIF-C provides superior colloidal stability and slightly higher cell viability. Because both Zn–mIM polymorphs originate from the same precursors and reaction conditions, researchers can now choose the washing solvent or procedure as a facile approach to finely tune the particle properties (e.g., morphology, DNA loading, cytocompatibility) without altering the synthesis. Therefore, these findings shed light on solvent washes, as a step often relegated to a one-line procedural detail that is here revealed to be a critical design parameter for ZIF biocomposites. Reporting details about washing conditions is therefore essential for the reproducible development, comparison, and clinical translation of future ZIF-based nucleic-acid delivery systems.
4. Materials and methods
4.1. Required chemicals
Zinc acetate dihydrate (Zn(OAc)2·2H2O) and HmIM were purchased from Sigma-Aldrich (Australia). PureLink™ plasmid kit, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and Lipofectamine 3000 were purchased from ThermoFisher Scientific. PC-3 prostate cancer cells were kindly provided by Prof John Mariadoson's lab in the Olivia Newton-John Cancer Research Institute. A 6.5 kbp length plasmid DNA was collected from Sir Ian Potter NanoBiosensing Facility, RMIT University, Melbourne, Australia.4.2. Preparation of pDNA@ZIF
Organic preparation includes the addition of 5 μg of plasmid DNA into 100 μL of HmIM (160 mM) in ethanol. A separate solution of Zn(OAc)2 (100 μL, 40 mM) in 50% ethanol (v/v) was also prepared and added. The mixture was aged for 10 minutes at room temperature. The obtained precipitate was recovered by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rcf for 12
rcf for 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) minutes.
minutes.
Similarly, inorganic preparation includes the addition of 5 μg plasmid DNA to 100 μL HmIM (160 mM) in deionized water. A separate solution of Zn(OAc)2 (100 μL, 40 mM) in water was also prepared and added. The mixture was aged for 10 minutes at room temperature. The obtained precipitate was recovered by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rcf for 12 minutes.
rcf for 12 minutes.
4.3. Preparation of washing solvents and washing treatment
A series of washing solutions were prepared by increasing the water% in ethanol (v/v). The DNA, HmIM, and Zn(OAc)2 mixtures were aged for 10 minutes at room temperature. The formed biocomposites were recovered by centrifugation at 13000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rcf for 12
rcf for 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) minutes. The supernatant was separated to remove unreacted precursors and ions. Then the pellet was repeatedly washed with 200 μL of washing solvents (0–100% H2O) and centrifuged thrice in the same solvents to examine the influence of water content in the washing solvent on determining the crystal fate of the plasmid@ZIF biocomposite. Finally, the biocomposites were vacuum-dried overnight.
minutes. The supernatant was separated to remove unreacted precursors and ions. Then the pellet was repeatedly washed with 200 μL of washing solvents (0–100% H2O) and centrifuged thrice in the same solvents to examine the influence of water content in the washing solvent on determining the crystal fate of the plasmid@ZIF biocomposite. Finally, the biocomposites were vacuum-dried overnight.
4.4. Characterization
X-ray diffraction (XRD) was performed in Bruker D8 General Area Detector Diffraction System (GADDS) using Cu Kα (λ = 1.54056 Å) radiation at 40 kV generator intensity and 40 mA current. The 2θ scanning range was 5–60° and the step size was 0.01°. The vacuum-dried samples were mounted on a surface clean silicon wafer attached to a sample holder and placed into the instrument. All the spectra were recorded at room temperature. The acquired (.raw) data was converted to a UXD file using the File Exchange Program XCH (Ver. 5.0.10, 2004, Bruker AXS, Socabim, Karlsruhe, Germany). Following that, the file was converted to Text Documents (.txt) and plotted using the OriginPro software. Crystal structure data (.cif) of simulated sod ZIF-8 and ZIF-C patterns were collected from previous literature and processed in Mercury (3.8).For electron microscopy, a diluted biocomposite solution was dropped on a clean silicon wafer surface and air-dried to remove the solvent. Then 5 nm iridium coating was applied using Leica EM ACE600 Sputter Coater to enhance conductivity under Scanning Electron Microscope (SEM) measurements. FEI Verios 460 L SEM was used to characterise the morphology of samples. The accelerating voltage and current of the electron beam were 2 kV and 0.2 nA, respectively. All tests were performed in triplicate to quantify the variability and reproducibility.
4.5. Growth kinetics study
To gain additional insight into the growth kinetics of pDNA@ZIF, we conducted turbidity measurements using UV-vis absorption spectroscopy. The optical turbidity of the mixture, both in organic (e.g., ethanol) and inorganic (i.e., aqueous) conditions, was measured by recording the absorption value at 350 nm position for up to 600 seconds.4.6. Encapsulation efficiency determination
The DNA loading in the water- and ethanol-based preparation of ZIF was investigated by agarose gel electrophoresis. The prepared pDNA@ZIF (both in ethanol and aqueous ZIF precursors) was washed with a range of solvents with different water % in ethanol (v/v). The final biocomposites were dissolved in 200 μL of aqueous EDTA (20 mM) to break the ZIF shell and release DNA in the solution. All samples were loaded on pre-heated 1% agarose gel containing SYBR Safe stain alongside an equal quantity of DNA used during synthesis as control. The electrophoresis was carried out in 1 × TAE buffer for 120 min at 90 V and bands were visualized in the GelDoc® system (BioRad, Hercules, USA). The intensity of DNA bands was calculated using densitometry analysis by ImageJ software, and encapsulation efficiency (EE) of DNA was determined by the following formula:4.7. Zn yield determination
The centrifuged pDNA@ZIF pellet was separated and then dried under a vacuum. Next, 2% nitric acid was applied to dissolve the ZIF carrier and to quantify the Zn content via an Agilent 4200 Microwave Plasma-Atomic Emission Spectrometer.4.8. Effect of washing solvent on ZIF phases
The effect of the washing solvent on ZIF-8 particles was investigated by rinsing the prepared pDNA@ZIF-8 five times with either 500 μL ethanol or deionized water. Similarly, the effect of washing solvent on ZIF-C particles was investigated by washing the prepared pDNA@ZIF-C five times with either deionized water or ethanol. At each washing step, the ZIF pellet was recovered by centrifugation and dried for further analysis.4.9. Stability of ZIF phases
ZIF biocomposites were prepared by mixing 5 μg plasmid DNA into 100 μL aqueous HmIM (160 mM) and 100 μL aqueous Zn(OAc)2 (40 mM). After 10 minutes the obtained precipitate was recovered by centrifugation and washed with absolute ethanol and deionized water to form pDNA@ZIF-8 and pDNA@ZIF-C. The stability of the biocomposites was investigated by keeping the ZIF particles in 500 μL ethanol and deionized water in static conditions. At each time interval (i.e., 1, 3, 5, and 24 h) the ZIF pellet was recovered by centrifugation and dried for further analysis.4.10. Cytotoxicity assessment
PC-3 cells were seeded in a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 96 well plates and incubated for 24 h at 37 °C. Later, cells were treated with 25 and 50 μg mL−1 of pDNA@ZIF in a fresh RPMI medium containing 10% fetal bovine serum and 1% penicillin–streptomycin. The cells were then incubated at 37 °C and then the media was aspirated after 24 h. Immediately, 100 μL of serum-free media containing MTT reagent were further incubated at 37 °C in the dark. After 4 h, the MTT media was aspirated carefully without disturbing purple formazan crystals formed at the bottom of the well. The purple formazan crystals were dissolved in 100 μL DMSO, and the absorbance was measured by a microplate reader at 570 nm. Pristine sod ZIF-8 and ZIF-C were used as negative controls. The percentage of cell viability was calculated by the formula:
000 cells per well in 96 well plates and incubated for 24 h at 37 °C. Later, cells were treated with 25 and 50 μg mL−1 of pDNA@ZIF in a fresh RPMI medium containing 10% fetal bovine serum and 1% penicillin–streptomycin. The cells were then incubated at 37 °C and then the media was aspirated after 24 h. Immediately, 100 μL of serum-free media containing MTT reagent were further incubated at 37 °C in the dark. After 4 h, the MTT media was aspirated carefully without disturbing purple formazan crystals formed at the bottom of the well. The purple formazan crystals were dissolved in 100 μL DMSO, and the absorbance was measured by a microplate reader at 570 nm. Pristine sod ZIF-8 and ZIF-C were used as negative controls. The percentage of cell viability was calculated by the formula:4.11. Bioactivity assessment
The 6.5 kbp plasmid DNA used in this study contains a gene responsible for producing a green fluorescent protein (GFP). While the plasmid itself doesn't glow, once it undergoes transcription and translation processes within the cell's machinery, it generates a harmless GFP. When exposed to blue light, this GFP emits a vibrant green fluorescence. Thus, expression of green fluorescence under blue light indicates successful uptake of material followed by transcription and translation of the coded gene. A cancerous human prostate cell line (PC-3) was used in this study to investigate the bioactivity of the encapsulated nucleic acid in ZIF. The PC-3 cells were treated with pDNA@ZIFs and were examined live to investigate the expression of GFP in cells using CLSM (NSTORM SuperResolution/Confocal microscope, Nikon). Prior to treatment, the pDNA@ZIFs were treated with sterile EDTA (0.5 M, pH 8.0) and the released DNA was encapsulated in the Lipofectamine 3000 reagent. After 24 h of transfection, cells were washed with sterile 1× PBS (pH 7.4) thrice. Live cell imaging was employed to record the expressed GFP in untreated and treated cells using CLSM.4.12. Statistical analysis
All tests were performed in triplicate. Statistical differences in the properties of different samples were analyzed using One-Way ANOVA in GraphPad Prism.Conflicts of interest
There are no conflicts to declare.Data availability
On behalf of all authors, I'm confirming that all the data presented in this work are original and the data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5lf00306g.Acknowledgements
S. A. P. acknowledges the Australian Government for the RMIT Research Stipend Scholarship to support the PhD program. R. S. acknowledges the Ian Potter Foundation for support in establishing the Sir Ian Potter NanoBiosensing Facility. The authors acknowledge technical support from the RMIT Microscopy and Microanalysis Facility and Micro Nano Research Facility respectively for conducting materials characterization and cell-based assays. The graphical abstract and schemes were partly generated by Gemini.References
-
A. R. Gascón, A. del Pozo-Rodríguez and M. Á. Solinís Non-viral delivery systems in gene therapy, in Gene Therapy - Tools and Potential Applications, IntechOpen, 2013 Search PubMed
.
- S. D. Li and L. Y. Huang, Nonviral gene therapy: promises and challenges, Gene Ther., 2000, 7, 31–34 CrossRef CAS PubMed
.
- Y. K. Sung and S. W. Kim, Recent advances in the development of gene delivery systems, Biomater. Res., 2019, 23, 8 CrossRef CAS PubMed
.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discovery, 2021, 20(2), 101–124 CrossRef CAS PubMed
.
- J. Zhuang, A. P. Young and C. K. Tsung, Integration of biomolecules with metal-organic frameworks, Small, 2017, 13, 1700880 CrossRef PubMed
.
- H. Wang, Y. Chen, H. Wang, X. Liu, X. Zhou and F. Wang, DNAzyme-loaded metal–organic frameworks (MOFs) for self-sufficient gene therapy, Angew. Chem., Int. Ed., 2019, 58, 7380–7384 CrossRef CAS PubMed
.
- Z. Wang, J. Niu, C. Zhao, X. Wang, J. Ren and X. Qu, A bimetallic metal–organic framework encapsulated with DNAzyme for intracellular drug synthesis and self-sufficient gene therapy, Angew. Chem., Int. Ed., 2021, 60, 12431–12437 CrossRef CAS PubMed
.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, The chemistry and applications of metal-organic frameworks, Science, 2013, 341, 1230444 CrossRef
.
- H. N. Abdelhamid, Zeolitic imidazolate frameworks (ZIF-8) for biomedical applications: A review, Curr. Med. Chem., 2021, 28, 7023–7075 CAS
.
- J. A. Allegretto, M. Arcidiácono, P. Y. Steinberg, P. C. Angelomé, O. Azzaroni and M. Rafti, Impact of chemical primers on the growth, structure, and functional properties of ZIF-8 films, J. Phys. Chem. C, 2022, 126, 6724–6735 CrossRef CAS
.
- M. Hoop, C. F. Walde, R. Riccò, F. Mushtaq, A. Terzopoulou, X. Z. Chen, A. J. deMello, C. J. Doonan, P. Falcaro, B. J. Nelson and J. Puigmarti-Luis, Biocompatibility characteristics of the metal organic framework ZIF-8 for therapeutical applications, Appl. Mater. Today, 2018, 11, 13–21 CrossRef
.
- S. Kumari, T. S. Howlett, R. N. Ehrman, S. Koirala, O. Trashi, I. Trashi, Y. H. Wijesundara and J. J. Gassensmith,
In vivo biocompatibility of ZIF-8 for slow release via intranasal administration, Chem. Sci., 2023, 14, 5774–5782 RSC
.
- Y. Liu, X. Cao and J. Ge, Antioxidative composites based on multienzyme systems encapsulated in metal–organic frameworks, ACS Appl. Mater. Interfaces, 2021, 13, 46431–46439 CrossRef CAS PubMed
.
- M. D. J. Velásquez-Hernández, R. Ricco, F. Carraro, F. T. Limpoco, M. Linares-Moreau, E. Leitner, H. Wiltsche, J. Rattenberger, H. Schröttner and P. Frühwirt, Degradation of ZIF-8 in phosphate buffered saline media, CrystEngComm, 2019, 21, 4538–4544 RSC
.
- A. Poddar, S. Pyreddy, F. Carraro, S. Dhakal, A. Rassell, M. R. Field, T. S. Reddy, P. Falcaro, C. M. Doherty and R. Shukla, ZIF-C for targeted RNA interference and CRISPR/Cas9 based gene editing in prostate cancer, Chem. Commun., 2020, 56, 15406–15409 RSC
.
- S. Pyreddy, A. Poddar, F. Carraro, S. A. Polash, C. Dekiwadia, B. Murdoch, Z. Nasa, T. S. Reddy, P. Falcaro and R. Shukla, Targeting telomerase utilizing zeolitic
imidazole frameworks as non-viral gene delivery agents across different cancer cell types, Biomater. Adv., 2023, 149, 213420 CrossRef CAS PubMed
.
- F. C. Herbert, S. S. Abeyrathna, N. S. Abeyrathna, Y. H. Wijesundara, O. R. Brohlin, F. Carraro, H. Amenitsch, P. Falcaro, M. A. Luzuriaga and A. Durand-Silva, Stabilization of supramolecular membrane protein–lipid bilayer assemblies through immobilization in a crystalline exoskeleton, Nat. Commun., 2021, 12, 2202 CrossRef CAS PubMed
.
- H. Chu, J. Shen, C. Wang and Y. Wei, Biodegradable iron-doped ZIF-8 based nanotherapeutic system with synergistic chemodynamic/photothermal/chemo-therapy, Colloids Surf., A, 2021, 628, 127388 CrossRef CAS
.
- Q. Wang, Y. Sun, S. Li, P. Zhang and Q. Yao, Synthesis and modification of ZIF-8 and its application in drug delivery and tumor therapy, RSC Adv., 2020, 10, 37600–37620 RSC
.
- H. Xie, X. Liu, Z. Huang, L. Xu, R. Bai, F. He, M. Wang, L. Han, Z. Bao, Y. Wu, C. Xie and Y. Gong, Nanoscale zeolitic imidazolate framework (ZIF)-8 in cancer theranostics: Current challenges and prospects, Cancers, 2022, 14, 3935 CrossRef CAS PubMed
.
- S. A. Polash, A. Poddar, S. Pyreddy, F. Carraro, A. M. D'Angelo, G. Bryant, P. Falcaro and R. Shukla, Phase characterization and bioactivity evaluation of nucleic acid-encapsulated biomimetically mineralized ZIF-8, ACS Appl. Mater. Interfaces, 2025, 17, 3002–3012 CrossRef CAS PubMed
.
- S. A. Polash, A. Poddar, F. Ahmady, G. Kannourakis, A. Jayachandran and R. Shukla, Impact of ligand concentration on the properties of nucleic-acid-encapsulated MOFs and inflammation modulation in prostate cancer cells, ACS Appl. Bio Mater., 2024, 7, 7635–7645 CrossRef CAS PubMed
.
- Y. Guo, Y. Li, S. Zhou, Q. Ye, X. Zan and Y. He, Metal-organic framework-based composites for protein delivery and therapeutics, ACS Biomater. Sci. Eng., 2022, 8(10), 4028–4038 CrossRef CAS PubMed
.
- A. Wang, M. Walden, R. Ettlinger, F. Kiessling, J. J. Gassensmith, T. Lammers, S. Wuttke and Q. Peña, Biomedical metal-organic framework materials: Perspectives and challenges, Adv. Funct. Mater., 2024, 34(43), 2308589 CrossRef CAS
.
- M. Malekmohammadi, S. Fatemi, M. Razavian and A. Nouralishahi, A comparative study on ZIF-8 synthesis in aqueous and methanolic solutions: Effect of temperature and ligand content, Solid State Sci., 2019, 91, 108–112 CrossRef CAS
.
- A. Poddar, J. J. Conesa, K. Liang, S. Dhakal, P. Reineck, G. Bryant, E. Pereiro, R. Ricco, H. Amenitsch, C. Doonan, X. Mulet, C. M. Doherty, P. Falcaro and R. Shukla, Encapsulation, visualization and expression of genes with biomimetically mineralized zeolitic imidazolate framework-8 (ZIF-8), Small, 2019, 15, e1902268 CrossRef PubMed
.
- S. A. Polash, S. Pyreddy, A. N. Abraham, S. Mahasivam, V. Bansal, L. Varadi, G. Bryant and R. Shukla, Impact of nucleic acid encapsulated MOF crystal phase on protein corona formation, Mater. Adv., 2023, 4, 4761–4774 RSC
.
- Z. Öztürk, M. Filez and B. M. Weckhuysen, Decoding nucleation and growth of zeolitic imidazolate framework thin films with atomic force microscopy and vibrational spectroscopy, Chem. – Eur. J., 2017, 23, 10915–10924 CrossRef PubMed
.
- J. Troyano, A. Carné-Sánchez, C. Avci, I. Imaz and D. Maspoch, Colloidal metal-organic framework particles: the pioneering case of ZIF-8, Chem. Soc. Rev., 2019, 48(23), 5534–5546 RSC
.
- M. R. Hafner, F. Carraro, B. Abbasgholi-NA, S. Dal Zilio, M. Schmallegger, H. Wiltsche, L. Ge, M. R. Reithofer, E. Astria and L. Villanova, 4D mapping of ZIF biocomposites for high protein loading and tunable release profiles, Adv. Funct. Mater., 2025, e18940 CrossRef
.
- M. R. Hafner, L. Villanova and F. Carraro, App-based quantification of crystal phases and amorphous content in ZIF biocomposites, CrystEngComm, 2022, 24, 7266–7271 RSC
.
- F. Carraro, M. D. J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Phase dependent encapsulation and release profile of ZIF-based biocomposites, Chem. Sci., 2020, 11, 3397–3404 RSC
.
- S. Jamdade, Z. Yu, S. E. Boulfelfel, X. Cai, R. Thyagarajan, H. Fang and D. S. Sholl, Probing Structural Defects in MOFs Using Water Stability, J. Phys. Chem. C Nanomater. Interfaces, 2024, 128(9), 3975–3984 CrossRef CAS PubMed
.
- J. Choi, L. C. Lin and J. C. Grossman, Role of structural defects in the water adsorption properties of MOF-801, J. Phys. Chem. C, 2018, 122(10), 5545–5552 CrossRef CAS
.
- R. Han and D. S. Sholl, Computational model and characterization of stacking faults in ZIF-8 polymorphs, J. Phys. Chem. C, 2016, 120(48), 27380–27388 CrossRef CAS
.
- X. Zhang, J. Zhang, H. Wang, J. Rogal, H.-Y. Li, S. H. Wei and T. Hickel, Defect-characterized phase transition kinetics, Appl. Phys. Rev., 2022, 9(4), 041311 CAS
.
- W. W. Zhan, Q. Kuang, J. Z. Zhou, X. J. Kong, Z. X. Xie and L. S. Zheng, Semiconductor@metal-organic framework core-shell heterostructures: a case of ZnO@ZIF-8 nanorods with selective photoelectrochemical response, J. Am. Chem. Soc., 2013, 135(5), 1926–1933 CrossRef CAS PubMed
.
- X. Su, T. Zheng, Y. Zhu, X. Tao, K. Yu, Z. Zhao, Z. Wu, J. Lu, C. Gao and D. Zhao, Enhanced n-butanol permselective vapor permeation by incorporating ZIF-8 into a polydimethylsiloxane composite membrane: Effect of filler loading contents, Z. Anorg. Allg. Chem., 2022, 648(12), e202200022 CrossRef CAS
.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O'Keeffe and O. M. Yaghi, Exceptional chemical and thermal stability of zeolitic imidazolate frameworks, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186–10191 CrossRef CAS PubMed
.
- M. Taheri and T. Tsuzuki, Photo-accelerated hydrolysis of metal organic framework ZIF-8, ACS Mater. Lett., 2021, 3, 255–260 CrossRef CAS
.
- S. I. Nakano and N. Sugimoto, The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents, Biophys. Rev., 2016, 8, 11–23 CrossRef CAS PubMed
.
- S. He, B. Cao, Y. Yi, S. Huang, X. Chen, S. Luo, X. Mou, T. Guo, Y. Wang, Y. Wang and G. Yang, DNA precipitation revisited: A quantitative analysis, Nano Sel., 2022, 3, 617–626 CrossRef CAS
.
- A. U. Ortiz, A. P. Freitas, A. Boutin, A. H. Fuchs and F. X. Coudert, What makes zeolitic imidazolate frameworks hydrophobic or hydrophilic? The impact of geometry and functionalization on water adsorption, Phys. Chem. Chem. Phys., 2014, 16, 9940–9949 RSC
.
- E. E. Sann, Y. Pan, Z. Gao, S. Zhan and F. Xia, Highly hydrophobic ZIF-8 particles and application for oil-water separation, Sep. Purif. Technol., 2018, 206, 186–191 CrossRef CAS
.
- S. Tanaka and Y. Tanaka, A simple step toward enhancing hydrothermal stability of ZIF-8, ACS Omega, 2019, 4, 19905–19912 CrossRef CAS PubMed
.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed
.
- N. K. Maddigan, O. M. Linder-Patton, P. Falcaro, C. J. Sumby, S. G. Bell and C. J. Doonan, Influence of the synthesis and storage conditions on the activity of Candida antarctica lipase B ZIF-8 biocomposites, ACS Appl. Mater. Interfaces, 2021, 13, 51867–51875 CrossRef CAS
.
- H. Kaur, G. C. Mohanta, V. Gupta, D. Kukkar and S. Tyagi, Synthesis and characterization of ZIF-8 nanoparticles for controlled release of 6-mercaptopurine drug, J. Drug Delivery Sci. Technol., 2017, 41, 106–112 CrossRef CAS
.
- Z. Huang, M. Ge, F. Carraro, C. Doonan, P. Falcaro and X. Zou, Can 3D electron diffraction provide accurate atomic structures of metal-organic frameworks?, Faraday Discuss., 2021, 225, 118–132 RSC
.
- S. A. Basnayake, J. Su, X. Zou and K. J. Balkus Jr., Carbonate-based zeolitic imidazolate framework for highly selective CO2 capture, Inorg. Chem., 2015, 54, 1816–1821 CrossRef CAS PubMed
.
- T. Simon-Yarza, A. Mielcarek, P. Couvreur and C. Serre, Nanoparticles of metal-organic frameworks: On the road to in vivo efficacy in biomedicine, Adv. Mater., 2018, 30(37), e1707365 CrossRef PubMed
.
- H. Zhang, M. Zhao and Y. S. Lin, Stability of ZIF-8 in water under ambient conditions, Microporous Mesoporous Mater., 2019, 279, 201–210 CrossRef CAS
.
- C. F. Holder and R. E. Schaak, Tutorial on powder X-ray diffraction for characterizing nanoscale materials, ACS Nano, 2019, 13, 7359–7365 CrossRef CAS PubMed
.
- V. N. Kalpana, B. A. S. Kataru, N. Sravani, T. Vigneshwari, A. Panneerselvam and V. D. Rajeswari, Biosynthesis of zinc oxide nanoparticles using culture filtrates of Aspergillus niger: Antimicrobial textiles and dye degradation studies, OpenNano, 2018, 3, 48–55 CrossRef
.
- A. Poddar, S. Pyreddy, S. A. Polash, C. M. Doherty and R. Shukla, A quest for cytocompatible metal organic frameworks in non-viral gene therapy: relevance of zeolitic imidazolate framework-8, Biomater. Biosyst., 2022, 100065 CAS
.
- B. Liu, L. Sun, X. Lu, Y. Yang, H. Peng, Z. Sun, J. Xu and H. Chu, Real-time drug release monitoring from pH-responsive CuS-encapsulated metal-organic frameworks, RSC Adv., 2022, 12, 11119–11127 RSC
.
- A. Sharma and G. Luthra, Significance of ISO 10993 Standards in ensuring biocompatibility of medical devices: A review, J. Pharm. Res. Int., 2023, 35, 23–34 CrossRef
.
- G. J. Leonel, C. B. Lennox, Y. Xu, M. Arhangelskis, T. Friscic and A. Navrotsky, Experimental and Theoretical evaluation of the thermodynamics of the carbonation reaction of ZIF-8 and its close-packed polymorph with carbon dioxide, J. Phys. Chem. C, 2023, 127, 19520–19526 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |