Open Access Article
Open Access ArticleIdentification, characterization and resistance to digestion assessment of hemp-derived dipeptidyl-peptidase IV inhibitory peptides
Miryam
Amigo-Benavent
 abc,
Fernando
Rivero-Pino
abc,
Fernando
Rivero-Pino
 *de,
Alvaro
Villanueva-Lazo
f,
Sergio
Montserrat-de la Paz
*de,
Alvaro
Villanueva-Lazo
f,
Sergio
Montserrat-de la Paz
 de,
Richard J.
FitzGerald
de,
Richard J.
FitzGerald
 abc and
Maria C.
Millan-Linares
abc and
Maria C.
Millan-Linares
 f
f
aProteins and Peptides Research Group, Department of Biological Sciences, Faculty of Science and Engineering, University of Limerick, V94 T9PX Limerick, Ireland
bFood, Diet and Nutrition Research Group, Health Research Institute, University of Limerick, V94 T9PX Limerick, Ireland
cFood Science and Technology Group, Bernal Institute, University of Limerick, V94 T9PX Limerick, Ireland
dDepartment of Medical Biochemistry, Molecular Biology, and Immunology, School of Medicine, University of Seville, Spain. E-mail: frivero1@us.es
eInstituto de Biomedicina de Sevilla, IBiS/Hospital Universitario Virgen del Rocio/CSIC/Universidad de Sevilla, 41013 Seville, Spain
fFood Protein and Immunonutrition Group. Department of Food and Health, Institute of Fats, CSIC, Seville, Spain
First published on 24th November 2025
Abstract
Hemp seeds (Cannabis sativa L.) are an adequate source of protein. Food-derived peptides exert bioactivity depending on the sequences. In this work, enzymatic hydrolysis with Alcalase (subtilisin) and Flavourzyme was carried out to obtain hemp protein hydrolysates with increased dipeptidyl-peptidase IV (DPP-IV) inhibitory activity. In addition, the hydrolysates were subjected to digestion following the INFOGEST protocol. The serial hydrolysis with both proteases showed the highest inhibition compared to the hydrolysis only with Alcalase. The peptides sequences contained in the samples and the digested samples were identified and characterized employing different bioinformatics tools. The sequences TNGPQLIH (released after addition of Flavourzyme), and GKLDLVKPQ (from Alcalase-treated samples) were proposed as the most active peptides. The peptides were chemically synthesized, and TNGPQLIH showed an IC50 value of 1.70 mg mL−1. The DPP-IV inhibitory activity was generally conserved or improved after digestion. These peptides could be employed as constituents in foods helping to prevent the development of diabetes.
1. Introduction
Bioactive peptides can be obtained from several food sources, ideally enriched in the protein content such as concentrates or isolates, by enzymatic hydrolysis or fermentation. Numerous reports, from in vitro to human studies have proved the benefits of these components in modulating different metabolic pathways (antihypertensive, antidiabetic, anti-inflammatory) in the organism, by targeting receptors or enzymes.1–4 Their efficacy as bioactive agents is dependent on their molecular weight and amino acid profile, and consequently the treatment applied to the native protein (e.g., enzyme, operating conditions of the reaction, etc.) are crucial in obtaining a bioactive pool of peptides.5,6 In addition to that, the ONE health principles and the worldwide aim to achieve a sustainable world led to a necessity of introducing environmentally friendly sources into the food system, and ideally, capable to be used in foods to prevent some disorders due to the bioactivity.7 One such interesting source of protein is hemp seeds, as it has been described that production of hemp could promote circular economy, being an interesting, environmental and cost-effective agent for phytoremediation, while its protein being of high quality and containing several bioactive peptides.8–10Concerning antidiabetic peptides, there are several metabolic routes in which peptides can have a functional role, including the inhibition of the enzyme dipeptidyl-peptidase IV (DPP-IV). This DPP-IV is in charge of modulating the levels of incretins (e.g., glucagon-like peptide-1, GLP-1), and inhibiting DPP-IV has as consequence, an increased half-life of this hormone, responsible for insulinotropic effects.11 Existing drugs used as inhibitors, show undesirable effects in humans, such as hypoglycemia, pancreatitis or gastrointestinal issues.12 As consequence, the use of food-derived peptides with bioactivity appears as a sustainable option for the prevention and pre-treatment of diabetes.13
The use of hemp protein hydrolysates (HPHs) as possible antidiabetic agents has been scarcely evaluated. In this regard, analysis of hemp-derived peptides has been described previously employing proteases such as Corolase L10, Promod 144MG and Protamex, leading to values of inhibition, as half-maximal inhibitory concentration (IC50) values, ranging from 1.84 to 5.71 mg mL−1.14,15 hydrolysed proteins from Cannabis sativa seeds with gastrointestinal enzymes and evaluated in vitro, in situ and ex vivo the activity towards DPP-IV in Caco-2 cell culture. In addition, hemp seed protein was hydrolysed with Alcalase, and more than 500 short peptides with multifunctional activity, including inhibition of DPP-IV were identified.16 In addition to that, using bioinformatics,17,18 identified 16 peptides by in silico enzymatic hydrolysis (with proteases such as trypsin, chymotrypsin, pepsin, proteinase K, and thermolysin) of several hemp seeds protein, and the in vitro DPP-IV inhibition assay yielded 8 peptides having IC50 values lower than 0.5 mM and cell assays demonstrated also increased insulin and GLP-1 concentrations. Nonetheless, it has been widely reported in several food sources, that the use of Flavourzyme is highly adequate in releasing DPP-IV inhibitory peptides.19–21 To the authors’ knowledge, no information employing Flavourzyme to obtain DPP-IV inhibitory peptides from hemp has been published so far, and the validation on whether this enzymatic treatment is adequate in releasing a highly bioactive protein hydrolysate is lacking. The hypothesis is that the use of Flavourzyme after the hydrolysis with Alcalase (subtilisin) will enhance the DPP-IV inhibitory potential of the sample.
Hence, in this work, hemp protein was enzymatically hydrolysed employing an endopeptidase (subtilisin, of broad spectrum) at different reaction times, as well as to evaluate whether the addition of an exopeptidase (Flavourzyme) for 15 min increase the activity. The hydrolysates were characterized, and their DPP-IV inhibitory activity was analyzed. Then, the peptidome of the hydrolysates was determined by liquid chromatography – trapped ion mobility spectrometry – tandem mass spectrometry. In addition, the hydrolysates were subjected to simulated gastrointestinal digestion (SGID) following the INFOGEST protocol, and the peptidome of these samples was also characterized. In silico analysis aiming to characterize the physicochemical properties and the DPP-IV inhibitory activity of peptides were used to suggest the most active ones, whose bioactivity was validated in vitro with the chemically-synthesized pure peptide sequences.
2. Materials and methods
2.1. Materials
Cannabis sativa L. seeds were offered by Sensi Seeds Bank. The proteases employed in the hydrolysis reactions (i.e., Alcalase 2.4 L and Flavourzyme (1000 L)) were obtained from Novozymes (Bagsvaerd, Denmark). All the chemicals (reagents and solvents) were of analytical grade, either from Sigma Chemical Co., Bachem AG (Bubendorf, CH, EU) or Gibco (Waltham, MA, USA).2.2. Hydrolysis procedure
At first, a hemp protein isolate (HPI) was produced following the protocol of Montserrat de La Paz.22 Then, a hydrolysis reaction was carried out at fixed temperature (50 °C) and pH, in a jacketed stirred vessel. To the HPI diluted in distilled water (10% w/v), Alcalase was used at 0.3 AU g−1 of protein at pH 8, and the reaction lasted one hour, while aliquots were sample at 10 (HPH10A), 20 (HPH20A), 30 (HPH30A), 45 (HPH45A), and 60 min (HPH60A). After that, by adding Flavourzyme at 60 LAPU per g of protein at pH 7, leaving the enzyme act for 15 min, the sample HPH60A + 15F was obtained. Enzyme was deactivated with heat for 15 min at 85 °C.2.3. In vitro DPP-IV inhibition assay
DPP-IV inhibitory activity analysis (of hydrolysates, digested and synthesized peptides) was carried out as described by Harnedy and Fitzgerald,23 using Diprotin A™ at 5 μM as the positive control. Experiments were conducted in triplicate. The values were expressed as the mean half maximal inhibitory concentration (IC50) ± standard deviation (n = 3).2.4. Peptide identification
Peptides of both the hydrolysates and the digested samples analyses were prepared and analyzed in the Proteomics Facility at Research Support Central Service, University of Cordoba. Samples were acidified with 0.5% Trifluoroacetic acid. Desalting and concentration step was performed with ZipTip C18 (Millipore) and the digested samples were finally Speedvac dried and resuspended in 5 μl of mobile phase (2% CAN, 0.05% TFA, 200 ng per sample) for HPLC injection. Sequences were identified by liquid chromatography followed by high resolution trapped ion mobility spectrometry-mass spectrometry (LC-TIMS-MS/MS) following the methodology of Montserrat de La Paz et al.,22 PEAKS Studio ProX (Bioinformatics Solutions Inc) was used to process the raw data, with the reference library of UniProt, with parent mass error tolerance set to 15 ppm and a fragment mass error tolerance of 0.05 Da. Protein unique peptides was set to larger than 1 and a high confidence score of −10 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 15 was applied to indicate an accurately identified protein.
P > 15 was applied to indicate an accurately identified protein.
2.5. In silico characterization of bioactive peptides
To identify the peptides most likely to be bioactive in the samples, various bioinformatic tools were utilized:(a) Peptide Property Calculator (https://pepcalc.com/) was employed to determine the net charge of identified peptides at neutral pH, as well as solubility and isoelectric point.
(b) ToxinPred software, to estimate physicochemical properties;24
(c) PeptideRanker, to estimate the likelihood of being bioactive.
(e) Three different tools aiming to predict whether a peptide is able to inhibit DPP-IV were used: iDPPIV-SCM25 (https://camt.pythonanywhere.com/iDPPIV-SCM), StackDPPIV26 (https://pmlabstack.pythonanywhere.com/StackDPPIV) and AntiDMPpred27 (https://i.uestc.edu.cn/AntiDMPpred/cgi-bin/AntiDMPpred.pl).
The peptides subjected to the analyses were the first five with the highest value of −10 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, with a molecular weight <1000 Da, as DPP-IV inhibitory peptides have been highly reported as low-molecular weight peptides, and a lower molecular weight is correlated with increased bioavailability.
P, with a molecular weight <1000 Da, as DPP-IV inhibitory peptides have been highly reported as low-molecular weight peptides, and a lower molecular weight is correlated with increased bioavailability.
2.6. Simulated gastrointestinal digestion (SGID)
The simulated gastrointestinal digestion was conducted according to the INFOGEST protocol28 with slight modifications, including the following proteases: α-amylase (EC 3.2.1.1), pepsin (EC 3.4.23.1), pancreatic lipase (EC 3.1.13) and bile salts (according to supplier's protocol). The digests obtained were also evaluated for their DPP-IV inhibitory activity as described in section 2.3.2.7. In silico digestion of bioactive peptides
The tool BIOPEP, which can be found https://biochemia.uwm.edu.pl/biopep/rec_pro1.php?x=72&y=0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 was used to subject the peptides to an in silico gastrointestinal digestion, to forecast the resulting peptides after the cleavages. Then, these fragments were screened whether they are DPP-IV inhibitory sequences according to the database.
29 was used to subject the peptides to an in silico gastrointestinal digestion, to forecast the resulting peptides after the cleavages. Then, these fragments were screened whether they are DPP-IV inhibitory sequences according to the database.
2.8. Chemical synthesis of peptides
The chemical synthesis of the TNGPQLIH and GKLDLVKPQ peptides from hemp protein hydrolysates (released after addition of Flavourzyme and Alcalase-treated samples respectively), have been performed by the ICTS “NANBIOSIS”, more specifically, by the Synthesis of Peptides Unit at Institute of Advanced Chemistry of Catalonia from Consejo Superior de Investigaciones Científicas (IQAC-CSIC), at >95% purity, measured by HPLC–PAD at 220 nm, using a XBridge® C18 3.5 μm, 4.6 × 100 mm column and a gradient of 5% to 100% of B in 8 min at 30 °C (1 mL min−1). These peptides were subjected to in vitro DPP-IV inhibitory activity as described in section 2.32.9. Statistical analysis
All values are presented as the means ± standard deviations (SD). Data were evaluated using Graph Pad Prism version 9.1.2 (San Diego, CA, USA). To DPP-IV inhibitory activity, the statistics between the groups was done using two-way analysis of variance (ANOVA), followed by Tukey's multiple-comparison test or independent sample t-test, where applicable. P values less than 0.05 were considered statistically significant.3. Results and discussion
3.1. DPP-IV inhibitory activity from hydrolysates
The protein content of hempseed usually ranges from 20 to 25%, whereas this content is maximized up to a value of 90–95% after isolation. Among the different proteins identified in hemp seed (>180), 60–80% accounts for edestin (legumin-type globulin), 13% of albumin (globular-type) and in minor proportion, β-conglycinin.17,30,31 The characteristics of the hydrolysates have been previously reported, including protein content (around 80–85%, dry weight) and amino acid profile (high content of arginine, glutamic acid, and aspartic acid).22In this study, hydrolysates exhibited a dose-dependent DPP-IV inhibition behavior, allowing to determine the IC50 values of each sample (Fig. 1). In the case of HPI, an IC50 value could not be determined (>5 mg mL−1). The hydrolysates obtained exclusively with Alcalase showed an IC50 value ranging from 2.29 to 2.54 mg mL−1, whereas after the action of Flavourzyme, the value was lowered up to 1.59 mg mL−1. According to the results obtained, the hypothesis was confirmed. The statistically significant differences among the samples obtained by Alcalase were not relevant, as overall, all the samples could be grouped under the same range of inhibition. After addition of Flavourzyme, the value was highly lowered, leading to statistically significant differences. The cleavage of the protein during the Alcalase hydrolysis most likely have changed the conformational structure of the protein, allowing Flavourzyme to exert the activity. Flavourzyme is usually employed along with, or after an endopeptidase, in order to fully take advantage of its potential as protease. The accessibility of N-terminal sites is higher after the hydrolysis with Alcalase 2.4 L because of its endo-peptidase high-spectrum activity,32 paving the way to an efficient hydrolysis by Flavourzyme, which would change the profile of peptides in the samples based on its ability to cleave low molecular weight peptides.33,34
The values obtained are in the same order as values obtained in other sources with similar enzymatic treatments, such as boarfish,35Porphyra dioica19 or other vegetables,36 with values ranging from 0.5 to 5 mg mL−1 generally. The relevance of hemp protein as antidiabetic has assessed up to human studies. In this regard,37 carried out two acute randomized repeated-measures crossover studies in which the subjects ingested hemp protein (doses of 20 or 40 g), soy protein (20 or 50 g) or carbohydrates as control. The objective was to measure and compare glycemic response (glucose and insulin levels) and satiety following the different ingestions. According to the authors, both test items could decrease postprandial blood glucose response. A nutritional intervention with hemp peptides, more digestible and potentially with improved DPP-IV inhibitory activity might result in a more biologically relevant result in terms of antidiabetic potential.
To sum up, the significance of Flavourzyme in the liberation of DPP-IV inhibitory peptides has been extensively reported related to its exopeptidase activity, able to release small peptides. However, a proper identification of the sequences responsible for the bioactivity demonstrated in vitro is needed.
3.2. Identification of peptides in the hydrolysates and in silico characterization
The peptidome for the six hydrolysates was fully characterized, and the amount of peptides ranged from 1185 (HPH60A + 15F) to 2113 (HPH10A). A more detailed description and characterization of the peptidome can be found elsewhere. Overall, in the samples enzymatically hydrolyzed exclusively with Alcalase, peptides had higher molecular weight than in the sample obtained with Flavourzyme.22In the scope of this study, the peptides with higher quality of the match in the spectrum having a molecular weight below <1000 Da were selected and reported. Among the +200 peptides assessed by,38 around 90% had a molecular weight inferior to 1000 Da, and in more than half of them, the value was less than 500 Da. Based on this hypothesis, the in silico analyses to determine and propose the active sequences were exclusively performed to the lower molecular weight peptides (Table 1).
| Sample | Number of peptides identified | % of <1 kDa peptides |
|---|---|---|
| HPI | 3353 | 3.9 |
| HPH10A | 2113 | 6.6 |
| HPH20A | 2707 | 8.4 |
| HPH30A | 1704 | 9.6 |
| HPH45A | 1632 | 10.2 |
| HPH60A | 1525 | 11.6 |
| HPH60A + 15F | 1185 | 14.5 |
In Table 2, the physical–chemical characterization, the likelihood to be bioactive and the DPP-IV inhibition score according to three different tools of each peptide are shown. In addition, the results from the in silico digestion are also depicted, discussed in section 3.4.
| Peptide sequence | Sample (HPH) | −10 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
Physical and chemical characteristics | Self-aggregation-prone region & Amyloidsc | Disorder Probability (%)c | Probability in secondary structure (%)c | Prediction of bioactivity | BIOPEP SGIDh | Sequence in digested HPH | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Res. Length | Molec. Weight | Calculated net chargea | Predicted pIa | Estimated solubility in pure watera | Hydrophobicityb | Steric hindranceb | Amphipathicityb | α-helix | β-strand | coil | PeptideRanker scored | iDPPIV–SCMe | StackDPPIVf | AntiDMPpredg | Digested | Active fragments | ||||||
| a Peptides were subjected to calculation viahttps://pepcalc.com/, where the net charge at neutral pH was calculated. Meanwhile, peptide solubility in pure water was estimated on this web server based on the combined result of isoelectric point (pI), the number of charged residues and the peptide length. b Peptides were subjected to calculation viahttps://webs.iiitd.edu.in/raghava/toxinpred/design.php/, where the hydrophobicity, steric hinderance, and amphipathicity was calculated. c The web server PASTA 2.0 (https://protein.bio.unipd.it/pasta2/) was implicated to compute the tendency of peptide self-aggregation specific to the possible region at sequence (with the recorded number starting from N-terminus). For peptide discrimination, the optimal thresholds were switched as Top = 1 and Energy <−5 PEU (1 PEU (pasta energy unit) = 1.192 kcal mol−1). NI: no amyloid predicted; PA: parallel aggregation computed. The probability of intrinsic disorder and portion of estimated secondary structure that complement the aggregation data were also reported. d The likelihood for the peptides as bioactive was evaluated by PeptideRanker (https://bioware.ucd.ie/~compass/biowareweb), a server to predict bioactive peptides based on a novel N-to-1 neural network, by giving scores ranging from 0 to 1. Higher score indicated the greater the likelihood of the peptide being bioactive. e Scores obtained by the DPP-IV inhibitory peptides predictor tool: iDPPIV-SCM (values higher than 294 are considered as positive result). f Scores obtained by the DPP-IV inhibitory peptides predictor tool: StackDPPIV (threshold = 0.5). g Scores obtained by the DPP-IV inhibitory peptides predictor tool: AntiDMPpred (threshold = 0.5). h Outcomes from the in silico analyses carried out with BIOPEP (digestion refers to the fragments indicated as products from the digestion with pepsin, trypsin and chymotrypsin); active fragments are those among the products originated, which are considered DPP-IV inhibitory fragments according to the information contained in the database. | ||||||||||||||||||||||
| KNGMMAPH | 10A | 52.09 | 8 | 884.39 | 1.1 | 9.91 | Good | −0.16 | 0.57 | 0.64 | 2–5 (NI) | 100 | 0 | 0 | 100 | 0.443 | 318 | 0.39 | 0.41 | K–N–GM–M–APH | No | YES |
| 30A | 43.80 | NO | ||||||||||||||||||||
| GKLDLVKPQ | 10A | 48.68 | 9 | 996.60 | 1 | 9.93 | Good | −0.21 | 0.62 | 0.95 | 3–6 (NI) | 100 | 0 | 0 | 100 | 0.196 | 252.88 | 0.45 | 0.61 | GK–L–DL–VK–PQ | PQ VK | YES |
| PQNFAVVK | 10A | 48.28 | 8 | 901.50 | 1 | 10.57 | Poor | −0.07 | 0.64 | 0.61 | 4–7 (NI) | 100 | 0 | 25 | 75 | 0.318 | 309.14 | 0.39 | 0.35 | PQN–F–AVVK | No | YES |
| 20A | 49.07 | NO | ||||||||||||||||||||
| 30A | 48.13 | NO | ||||||||||||||||||||
| 45A | 45.37 | NO | ||||||||||||||||||||
| 60A | 47.51 | NO | ||||||||||||||||||||
| PQNHAVVK | 10A | 46.15 | 8 | 891.49 | 1.1 | 10.57 | Good | −0.20 | 0.55 | 0.80 | 4–7 (NI) | 100 | 0 | 0 | 100 | 0.154 | 311.86 | 0.23 | 0.47 | PQN–H–AVVK | No | NO |
| 20A | 47.14 | YES | ||||||||||||||||||||
| AMRNPLAGK | 10A | 45.99 | 9 | 956.52 | 2 | 11.42 | Good | −0.24 | 0.61 | 0.68 | 1–4 (NI) | 100 | 0 | 0 | 100 | 0.469 | 266.25 | 0.13 | 0.39 | AM–R–N–PL–AGK | PL | NO |
| PQLVYIVK | 20A | 51.08 | 8 | 958.59 | 1 | 10.09 | Poor | 0.06 | 0.63 | 0.61 | 3–7 (PA) | 100 | 0 | 62.5 | 37.5 | 0.206 | 310 | 0.24 | 0.46 | PQL–VY–IVK | VY | NO |
| 30A | 47.84 | NO | ||||||||||||||||||||
| 45A | 44.58 | NO | ||||||||||||||||||||
| 60A | 45.32 | NO | ||||||||||||||||||||
| VKEPVFSF | 20A | 47.78 | 8 | 951.51 | 0 | 6.82 | Good | 0.03 | 0.63 | 0.62 | 5–8 (NI) | 100 | 0 | 0 | 100 | 0.442 | 276.43 | 0.36 | 0.45 | VK–EPVF–SF | SF VK | NO |
| KNAIYTPH | 20A | 47.36 | 8 | 942.49 | 1.1 | 9.55 | Poor | −0.17 | 0.53 | 0.64 | 3–6 (NI) | 100 | 0 | 0 | 100 | 0.241 | 295.29 | 0.09 | 0.31 | K–N–AIY–TPH | No | YES |
| 30A | 58.92 | YES | ||||||||||||||||||||
| 45A | 59.54 | YES | ||||||||||||||||||||
| 60A | 58.67 | YES | ||||||||||||||||||||
| MRNPLAGK | 30A | 41.73 | 8 | 885.49 | 2 | 11.39 | Good | −0.3 | 0.62 | 0.77 | 5–8 (NI) | 100 | 0 | 0 | 100 | 0.39 | 273.43 | 0.28 | 0.35 | M–R–N–PL–AGK | PL | NO |
| 45A | 46.52 | NO | ||||||||||||||||||||
| DDRNSIIR | 45A | 44.52 | 8 | 987.51 | 0 | 6.83 | Good | −0.55 | 0.7 | 0.61 | 4–7 (NI) | 100 | 0 | 0 | 100 | 0.323 | 197.29 | 0.3 | 0.33 | DDR–N–SIIR | No | NO |
| 60A | 44.12 | NO | ||||||||||||||||||||
| TDHYLPIH | 60A | 61.15 | 8 | 994.49 | −0.8 | 6.04 | Poor | −0.06 | 0.45 | 0.36 | 3–8 (NI) | 100 | 0 | 0 | 100 | 0.447 | 332.29 | 0.35 | 0.48 | TDH–Y–L–PIH | No | NO |
| TNGPQLIH | 60A + 15F | 43.2 | 8 | 878.46 | 0.1 | 7.5 | Poor | −0.07 | 0.53 | 0.34 | 5–8 (NI) | 100 | 0 | 0 | 100 | 0.23 | 334.71 | 0.53 | 0.51 | TN–GPQL–IH | IH TN | YES |
| SAERGFLY | 60A + 15F | 33.46 | 8 | 941.46 | 0 | 6.58 | Good | −0.13 | 0.63 | 0.47 | 4–8 (NI) | 100 | 0 | 0 | 100 | 0.615 | 233 | 0.17 | 0.56 | SAER–GF–L–Y | GF | NO |
| DDNGRNVF | 60A + 15F | 31.19 | 8 | 935.41 | −1 | 3.71 | Good | −0.4 | 0.73 | 0.31 | 5–8 (NI) | 100 | 0 | 0 | 100 | 0.424 | 258.29 | 0.45 | 0.27 | DDN–GR–N–VF | VF | NO |
| RNIFKGF | 60A + 15F | 28.14 | 7 | 880.49 | 2 | 11.39 | Good | −0.2 | 0.7 | 0.87 | 1–4 (NI) | 100 | 0 | 0 | 100 | 0.798 | 219.67 | 0.26 | 0.15 | R–N–IF–K–GF | GF | NO |
| ADIFNPR | 60A + 15F | 27.18 | 7 | 831.42 | 0 | 6.71 | Good | −0.23 | 0.64 | 0.35 | 2–5 (NI) | 100 | 0 | 0 | 100 | 0.798 | 296 | 0.13 | 0.3 | ADIF–N–PR | No | NO |
The analysis conducted with the Toxin Pred software indicated that none of the peptides were toxic. No correlation has been described linking the pI, solubility, and charge of peptides with the potential DPP-IV inhibitory activity.39–41
A score >0.5 from PeptideRanker would firmly indicate that the sequence is bioactive.42 Consequently, the peptides that could be considered potentially bioactive according to this in silico tool would only be the sequences SAERGFLY, RNIFKGF and ADIFNPR, found in the sample hydrolysed with Flavourzyme. This in silico prediction is in line with the results obtained in vitro, since this sample was the one with the lowest IC50 value obtained.
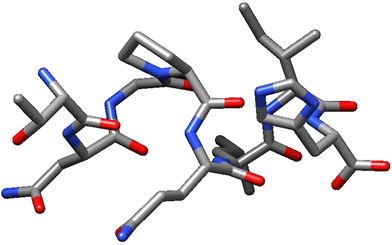
Nonetheless, it must be noticed that the sequence TNGPQLIH, showing a score of 0.23, is the only one showing inhibition of DPP-IV activity according to the StackDPPIV tool, thus, validation of its activity is needed. The structure of this peptide can be seen in Fig. 2, retrieved from USCF Chimera.43 According to the StackDPPIV tool, the other sequences identified would not exert the bioactivity claimed. According to this iDPPIV-SCM score, also the peptides KNGMMAPH, PQNFAVVK, PQNHAVVK, PQLVYIVK, TDHYLPIH and TNGPQLIH would exert DPP-IV inhibitory activity, as the score obtained is higher than 300, being the threshold established by the tool developers of 294. The values obtained are promising and are in line with values obtained in peptides derived from other vegetable source such as DLLGLW from soy or RMGPL from quinoa.44
Concerning the AntiDMPpred tool, the peptides in which the score was higher than the threshold were only three sequences: GKLDLVKPQ, TNGPQLIH and SAERGFLY. There are differences found in the in silico prediction outcomes, which are mainly due to the negative and positive controls that the authors have used, considering that for instance, 60% of the peptides employed to develop the iDPPIV-SCM tool had a peptide length of 5 residues or smaller, adding uncertainty to the predictions carried out to longer peptides.
The in silico tools are useful in screening the activity of peptides, although researchers have to take into consideration the limitations and the small weight of evidence that these bioinformatic tools provide when used alone.45In silico prediction tools are valuable as a first-pass screening method to identify potential bioactive peptides, but the outcomes are highly dependent on the algorithms and training datasets applied, which may lead to discrepancies across different platforms. For this reason, computational results should be interpreted as preliminary, guiding the selection of promising candidates rather than serving as conclusive evidence.
The ability of a peptide to inhibit DPP-IV activity depends on the residues within its sequence and their interactions with the enzyme. According to Mora et al.,46 low steric hindrance values and high amphiphilicity has been correlated with a more bioactive peptide, based on a better stabilization of the interaction with the enzyme conformation. The sequence with the highest amphiphilic nature was GKLDLVKPQ (value of 0.95), followed by PQNHAVVK and RNIFKGF. Among these peptides, the one with the lowest steric hindrance are PQNHAVVK and GKLDLVKPQ.
According to Berraquero-Garcia et al.,47 out of 230 sequences identified as DPP-IV inhibitory peptides to which an experimental IC50 value was calculated, the average length of the peptides of the 40 most active was 6, being more than 70% of this length or smaller. It has been reported the importance of the amino acid proline (P) in these type of peptides in the N-terminal, specifically in the second position gaining more importance.48 This statement is in line with several of the peptides hereby identified such as PQNFAVVK, PQNHAVVK and PQLVYIVK, having P in the first position. VKEPVFSF and TNGPQLIH on the other hand, have the residue on the fourth position, being the latter one the only one found in the Flavourzyme-treated samples. Similarly, a higher bioactivity in inhibiting DPP-IV has been correlated with the presence of alanine (A) in the first two positions of the N-terminal, as found in the peptide AMRNPLAGK, from Alcalase-treated samples and ADIFNPR, potentially released due to Flavourzyme.
Concerning the C-terminal, it has been suggested the importance of aromatic amino acids in the last two positions, such as phenylalanine (F), tryptophan (W) and tyrosine (Y) in Ct in peptides capable of inhibit DPP-IV.49 In this regard, the peptide VKEPVFSF appears as promising active peptide, as well as DDNGRNVF and RNIFKGF, although these two do not possess the N-terminal with P or A, as it does VKEPVFSF. Some similarities of specific fragments in the peptides hereby identified with already demonstrated DPP-IV peptides were found, such as the fragments DL and KP, found in the hemp peptide GKLDLVKPQ as well as in the whey peptides LKPTPEGDLE, having an IC50 value of 42 µM;50 or also PQ from this sequence, which is also found in LPQPPQE from Velvet aqueous extract51 or VPYPQ from casein, among others.52 One of the peptides hereby considered as highly bioactive based on the previous discussion, TNGPQLIH, also has the GPQ fragment from GPAGPQGPR, which in vitro possess an IC50 value of 0.51 µM, also extracted from the Velvet aqueous extract or in GPGA from Salmon.53 In addition, recently,54 reported the discovery of a hemp seed protein hydrolysate-derived peptide as highly inhibitory towards DPP-IV. The peptide VAMP has been reported to interact with DPP-IV, leading to enhanced glucose metabolism by increasing active GLP-1 levels in obese mouse models, as well as promotes an improvement of the intestinal barrier function and insulin resistance.
The similarity of fragments in the sequences indicate the similarity of the structure that these peptides might have, and consequently, correlation structure and activity, the activity is more likely to be similar. Overall, some of the di and tri-peptides contained in previously reported as DPP-IV inhibitory sequences are also inside the sequences of the hemp-derived peptides hereby identified.
Taking into account all the parameters discussed, the most active sequences proposed as responsible for the high bioactivity were proposed to be VKEPVFSF, GKLDLVKPQ and TDHYLPIH in the Alcalase-treatment samples and TNGPQLIH and SAERGFLY in the Flavourzyme-treated hydrolysate, based on the scores obtained in some of the tools employed and the molecular features of the sequences. TNGPQLIH (derived from 60A + 15F) and GKLDLVKPQ (derived from HPH20A) were chosen to be synthesized chemically and subjected to validation of their activity (section 3.4) because of the outcomes of the in silico analyses, and because they were gastro-resistant (sequences were identified in the corresponding digested sample of each of the hydrolysates).
Different recently developed prediction tools were employed in order to rank and compare the likelihood of the sequences to be responsible for the DPP-IV inhibitory activity of the protein hydrolysates evaluated in vitro. To the author's knowledge, these hemp peptides proposed as antidiabetic (capable of inhibiting the enzyme DPP-IV) have not been identified before. It is relevant to mention that the beneficial properties of protein hydrolysates arise from consuming the complete pool of peptides, not just one particular peptide. It has been reported that the presence of specific amino acids, such as arginine at the sequence terminal, helps enhance the peptide's resistance to cleavage during the proteolytic activity of digestive enzymes, but this resistance should be evaluated.
3.3. DPP-IV inhibitory activity from digested hydrolysates
The action of the enzymes during the gastrointestinal digestion might be responsible for the further cleavage of peptides. This cleavage is specific sites according to the specificity of the enzymes (e.g., trypsin, mainly in residues or arginine or lysine) would release new sequences which can be equally, more or less active, and as consequence, the activity of the hydrolysates after SGID might be different.As observed in Fig. 1, the DPP-IV inhibitory activity of the hydrolysates after being subjected to SGID ranged from 1.53 to 2.19 mg mL−1. In addition, the HPI, which originally was reported not to exert DPP-IV inhibitory activity on the conditions assayed, displayed activity with an IC50 value of 1.74 ± 0.03 mg mL−1. The samples HPH10A, 20A, 30A and 45A displayed a similar inhibition value, based on the statistical analysis, whereas the HPH60A displayed lower activity compared to these, not significantly different from its original hydrolysate. In the case of the sample obtained with Flavourzyme, the value was not significantly different from its original, indicating maintenance of the activity in an adequate range compared to other sources. In some of these digested, there were differences compared to the hydrolysates, which are likely to occur because more active peptides have been released due to the action of the digestive proteases.
For instance, Tenenbaun et al.,55 evaluated the antidiabetic activity of whey protein (isolate and hydrolysate) after SGID using the INFOGEST protocol. As expected, the digested samples were more hydrolyzed and as consequence, the amount of low molecular weight peptides was increased. In addition to that, after the SGID, the hydrolysate induced a greater secretion of GLP-1 in the enteroendocrine STC-1 cell line compared to the original sample. These results support the advantages of hydrolyzing proteins to obtain a more bioactive pool of peptides. Similarly, an increased in the DPP-IV inhibitory activity was reported in the duodenal digest of oat kernels, having an IC50 value of 0.51 mg mL−1 compared to 10.82 mg mL−1 of the intact protein.56
This challenge of potential loss of bioactivity that the field of bioactive peptides faces during gastrointestinal digestion, due to the activity of the proteases have been investigated in order to ensure the efficacy of these protein hydrolysates as bioactive agents. Among the different possibilities to overcome this issue, the encapsulation of the protein hydrolysates appears as a promising strategy contributing not to loss functionality while improving bioaccessibility.3
3.4. Peptidome of digested samples and in silico digestion of identified peptides
The identified peptides were subjected, in addition to the in vitro SGID previously described, to an in silico digestion of the sequences identified as most bioactive from the results discussed in section 3.2. In addition, to compare with the experimental data, the peptidome of the digested samples was also analyzed (SI Table S1). The results obtained from the hydrolysis with the digestive proteases: pepsin, trypsin and chymotrypsin can be found in Table 2 (column BIOPEP-SGID), together with information on whether the sequence in each hydrolysate was found (or not) in their corresponding digested sample analysed by LC-MS (last column in Table 2). The tool is limited by the lack of amylase or bile salts, which are not included, although in the scope of peptides, only the action of proteases is of relevance.The peptidome of the digested samples revealed that most of the identified sequences in the hydrolysates were not maintained after the digestion, since most of them were not identified in the digested sample (compared to their hydrolysate). In some cases, such as KNGMMAPH or PQNFAVVK, identified in the HPH10A and the digested HPH10A, and identified in other HPHs (30A, 45A, 60A), but not in the digested samples of these samples. The fact that a sequence is digested in a specific hydrolysate, but not in others can be attributed to differences in peptide accessibility and structural context resulting from the varying degrees of hydrolysis. Longer hydrolysis times modify the peptide profile, affecting sequence length, terminal residues, and exposure of cleavage sites to gastrointestinal enzymes. Consequently, some peptides may become more susceptible to digestion, while others are stabilized or protected within peptide aggregates or resistant motifs.
On the contrary, the sequence KNAIYTPH, found in all the Alcalase hydrolysates, was not digested, as it was also identified in all the digested samples. This reveals that this sequence is highly resistant to the activity of the digestive proteases, which makes it a suitable candidate for functional foods, since it could potentially reach the target organs.
However, it must be noted that these analyses are not accounting for the amount of peptide sequences in the sample, which is also a limitation in drawing conclusions. However, for the patterns of how the sequences are digested, and which amino acids, di or tri-peptides are released, the BIOPEP tool was used, since the mass spectrometry analyses is unable to identiy <4 aa sequences.
As it can be observed, for the in silico prediction, some of the fragments which are hypothetically released after the action of the proteases, have been also reported as DPP-IV inhibitory sequences, thus, would potentially exert the activity after being orally ingested. In addition, these dipeptides are likely to be absorbed through the PepT1 transporter, based on the features, and consequently, would be bioavailable as such.57 For instance, from the peptide TNGPQLIH, the fragments IH and TN have been indicated as DPP-IV inhibitory sequences.58 In the same line, other fragments such as GF, PL, VG or PQ have been reported as DPP-IV inhibitory peptides, for instance as aforementioned PQ as part of the fragment GPQ, already reported in other peptides as well. Nonetheless, for some of the peptides, no information on the potential DPP-IV inhibitory potential of the fragments was retrieved from the BIOPEP database. This outcome does not necessarily mean that the digested products are not active, as the bioactivity was reported by in vitro assays. The activity might be because part of the peptides has not been digested and the original sequence is maintained, or because the new sequences have not been reported as DPP-IV inhibitory in literature, while they can also exert the activity.
In this regard, since the identification of bioactive peptides is not frequently reported in literature, as previously noted, there is still a research gap in the characterization of hemp peptides and how these are digested, as identifying specific sequences from in vivo digestion assays is currently not feasible. Recently,59 subjected albumin, edestin, and vicilin to in silico digestion and then screened for DPP-IV inhibitory peptides using IDPPIV-SCM, reporting that FNVDTE from edestin and EAQPST from vicilin emerged as promising candidates for DPP-IV inhibitors.
Given that protein hydrolysates contain several components, it is challenging to separate high molecular weight compounds from low molecular weight peptides, which are frequently those more prone to exert bioactivity. In this regard, bioinformatics analysis is essential for locating substances with biological activity. The in silico digestion of peptides and identification of DPP-IV inhibitory fragments among them complements the results obtained in the in vitro experiments, showing that the peptides might still be bioactive in a certain extent, and that the profile of peptides released depends on the degree of hydrolysis that the hydrolysate had.
Finally, the bioactivity of peptides should also be evaluated using synthetic peptides, to validate the results obtained by in vitro and in silico experiments, as well as their effectiveness in actual food matrices under particular processing and storage settings, and research in animal models to determine the underlying mechanisms by which these substances might have the beneficial impact.
3.4. Bioactivity of the chemically-synthesized peptides
Two peptides, TNGPQLIH and GKLDLVKPQ, present in all samples analysed (original hydrolysates and their digested), were synthesized based on in silico predictions and subsequently evaluated for their inhibitory activity against DPP-IV. As indicated previously, the hydrolysate 60A + 15F showed an IC50 of 1.59 mg mL−1, while the HPH20A hydrolysate exhibited a slightly higher IC50 of 2.23 mg mL−1.TNGPQLIH, derived from the 60A + 15F hydrolysate, displayed an IC50 of 1.70 mg mL−1, which closely matched that of the parent hydrolysate. This strong agreement suggests that TNGPQLIH is likely one of the major contributors to the DPP-IV inhibitory effect of the 60A + 15F sample and this inhibition value is within the typical range reported for moderately potent food-derived DPP-IV inhibitory peptides. The comparable activity of the individual peptide and the complex hydrolysate implies that the presence of TNGPQLIH at sufficient concentrations within the hydrolysate could account for most of its inhibitory potential. TNGPQLIH contains both hydrophobic (Leu, Ile) and proline residues that may stabilize interactions with the DPP-IV active site, contributing to its relatively high potency. However, it is equally plausible that the observed activity of the hydrolysate arises from a combination of active and inactive components. Synergistic interactions between multiple peptides can enhance apparent activity, while non-active peptides may dilute the overall inhibitory effect. Therefore, although TNGPQLIH likely represents a key active component, the contribution of other sequences—either more potent but less abundant, or less active but more prevalent—cannot be excluded.
In contrast, GKLDLVKPQ identified from the HPH20A hydrolysate, showed a much weaker inhibition, with an IC50 > 5 mg mL−1 indicating lower DPP-IV inhibitory potential. This represents a significant loss in activity compared to the corresponding hydrolysate, which had an IC50 of 2.23 mg mL−1. Such discrepancy reveals a disconnect between in silico prediction and in vitro validation. While computational screening tools can highlight promising candidates based on sequence similarity, physicochemical descriptors, or docking scores, they remain limited by the quality and scope of their training datasets and by the inherent simplifications of peptide–enzyme interaction models, as already discussed above. These algorithms often fail to account for conformational dynamics, post-translational modifications, aggregation behaviour, and assay conditions that influence binding affinity and catalytic inhibition. As a result, peptide sequences predicted to be highly active may show limited activity experimentally, as observed for GKLDLVKPQ. This outcome emphasizes the necessity of combining in silico predictions with empirical testing and highlights that predictive models should be considered as hypothesis-generating rather than definitive screening tools.
Overall, these results confirm that while computational approaches are useful for prioritizing candidates, empirical validation remains essential for identifying functionally relevant DPP-IV inhibitory peptides in complex hydrolysates.
4. Conclusions
Hemp seed protein is considered an interesting source of bioactive peptides with several properties, including the capacity of modulating the glycemic index by inhibiting the enzyme DPP-IV. The profile of peptides released after enzymatic hydrolysis highly depends on the proteases employed. In this work, the effect of Alcalase, followed by Flavourzyme was assessed for the first time in a hemp protein isolate, and the products obtained were assessed in terms of inhibition of DPP-IV. The IC50 values obtained ranged from 1.59 to 2.54 mg mL−1, similar or lower than the values obtained for other food-derived peptides subjected to the same enzymatic treatment. The addition of Flavourzyme highly increased the bioactivity of the sample. The peptides of the hydrolysates and the digested samples were identified by LC-TIMS-MS and were analyzed by different in silico prediction tools on their ability to specifically inhibit the enzyme. Sequences proposed as responsible for the high bioactivity were proposed to be VKEPVFSF, GKLDLVKPQ and TDHYLPIH in the Alcalase-treatment samples and TNGPQLIH and SAERGFLY in the Flavourzyme-treated hydrolysate, based on the prediction tools and the physicochemical characterization. The hydrolysates were also subjected to simulated gastrointestinal digestion in vitro (INFOGEST protocol) and in silico (BioPEP database). The inhibition of DPP-IV (IC50 value) ranged from 1.53 to 2.19 mg mL−1, generally improved or maintained compared to the non-digested samples, while the in silico hydrolysis showed that some of the fragments released are also considered as DPP-IV inhibitors. The activity of the sequences TNGPQLIH and GKLDLVKPQ was confirmed using chemically synthesized peptides. The inhibition of DPP-IV was substantial for TNGPQLIH, supporting its role as a key contributor to the activity of the hydrolysate. In contrast, GKLDLVKPQ showed negligible inhibition, indicating that in silico predictions do not always translate into experimental activity. These results highlight the value of computational screening for guiding peptide discovery, but also the need for experimental confirmation to identify truly bioactive sequences. This is the first publication, to the authors’ knowledge, in which hemp protein hydrolysates obtained with Alcalase and Flavourzyme and peptides’ sequences have been characterized as capable to inhibit DPP-IV.Author contributions
Conceptualization, M. C. M.-L., M. A.-B.; methodology, A. V., M. A,-B.; formal analysis, M. C. M.-L.; investigation, M. C. M.-L., R. D. F.; resources, M. C. M.-L., S. M.-d. l. P; writing – original draft preparation, F. R.-P.; writing – review and editing; supervision, M. C. M.-L.; R. D. F.; funding acquisition, M. C. M-L., S. M.-d. l. P. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5fo02421h.Acknowledgements
This research was funded by grant P20_00661 from the Andalusian Plan for Research, Development and Innovation (PAIDI) 2020, grant US-1381492 from the European Regional Development Fund (ERDF) and CYTED Network “Pro-Alt” (125RT0166). Maria C. M.-L acknowledges the international mobility grant received from the Spanish Ministry of Science, Innovation and Universities for mobility stays at foreign higher education and research centres (PRX23/00485). F. R.-P has the benefit of a Juan de la Cierva postdoctoral fellowship (FJC2022-050043-I) from the Spanish Ministry of Science and Innovation. The authors thank Carlos Fuentes from the Proteomics Unit of the Central Research Support Service of Cordoba University (UCO) and Dr Miriam Royo from Synthesis of Peptides Unit of NABIOSIS (CSIC) for their technical assistance during the fulfilment of this work. We acknowledge support from the PTI+NEUROAGING, Consejo Superior de Investigaciones Científicas (CSIC), Spain.References
- M. Samsamikor, D. Mackay, R. C. Mollard and R. E. Aluko, A double-blind, randomized, crossover trial protocol of whole hemp seed protein and hemp seed protein hydrolysate consumption for hypertension, Trials, 2020, 21, 354 CrossRef CAS.
- L. Mora and F. Toldrá, Advanced enzymatic hydrolysis of food proteins for the production of bioactive peptides, Curr. Opin. Food Sci., 2023, 49, 100973 Search PubMed.
- F. Rivero-Pino, Bioactive food-derived peptides for functional nutrition: Effect of fortification, processing and storage on peptide stability and bioactivity within food matrices, Food Chem., 2023, 406, 135046 CrossRef CAS.
- A. Rakha, H. Rasheed, A. B. Altemimi, S. Tul-Muntaha, I. Fatima, M. S. Butt, S. Hussain, Z. F. Bhat, A. Mousavi Khaneghah and R. M. Aadil, Tapping the nutraceutical potential of industrial hemp against arthritis and diabetes - A comprehensive review, Food Biosci., 2024, 59, 104195 CrossRef CAS.
- M. Cermeño, T. Kleekayai, M. Amigo-benavent, P. Harnedy-Rothwell and R. J. Fitzgerald, Current knowledge on the extraction, purification, identification and validation of bioactive peptides from seaweed, Electrophoresis, 2020, 1–42 Search PubMed.
- T. Kleekayai, M. Khalesi, M. Amigo-Benavent, M. Cermeño, P. Harnedy-Rothwell and R. J. FitzGerald, Enzyme-Assisted Extraction of Plant Proteins, ed. A. J. Hernández-Álvarez, M. Mondor and M. G. Nosworthy, Springer International Publishing, Cham, 2023, pp. 131–178 Search PubMed.
- R. Perez-Gregorio and J. Simal-Gandara, A Critical Review of Bioactive Food Components, and of their Functional Mechanisms, Biological Effects and Health Outcomes, Curr. Pharm. Des., 2017, 23, 2731–2741 CrossRef.
- G. Santos-Sánchez, A. I. Álvarez-López, E. Ponce-España, A. Carrillo-Vico, C. Bollati, M. Bartolomei, C. Lammi and I. Cruz-Chamorro, Hempseed (Cannabis sativa) protein hydrolysates: A valuable source of bioactive peptides with pleiotropic health-promoting effects, Trends Food Sci. Technol., 2022, 127, 303–318 CrossRef.
- F. Rivero-Pino, M. C. Millan-Linares and S. Montserrat-de-la-Paz, in Sustainable Food Science: A Comprehensive Approach, Elsevier, 2023 Search PubMed.
- M. C. Millan-Linares, F. Rivero-Pino, T. Gonzalez-de-la-Rosa, A. Villanueva and S. Montserrat-de-la-Paz, Identification, characterization, and molecular docking of immunomodulatory oligopeptides from bioavailable hempseed protein hydrolysates, Food Res. Int., 2024, 176, 113712 CrossRef CAS PubMed.
- A. Miguéns-Gómez, C. Grau-Bové, M. Sierra-Cruz, R. Jorba-Martín, A. Caro, E. Rodríguez-Gallego, R. Beltrán-Debón, M. T. Blay, X. Terra, A. Ardévol and M. Pinent, Gastrointestinally digested protein from the insect alphitobius diaperinus stimulates a different intestinal secretome than beef or almond, producing a differential response in food intake in rats, Nutrients, 2020, 12, 1–15 CrossRef.
- F. Wang, G. Yu, Y. Zhang, B. Zhang and J. Fan, Dipeptidyl Peptidase IV Inhibitory Peptides Derived from Oat (Avena sativa L.), Buckwheat (Fagopyrum esculentum), and Highland Barley (Hordeum vulgare trifurcatum (L.) Trofim) Proteins, J. Agric. Food Chem., 2015, 63, 9543–9549 CrossRef CAS PubMed.
- I. M. E. Lacroix and E. C. Y. Li-Chan, Inhibition of dipeptidyl peptidase (DPP)-IV and α-glucosidase activities by pepsin-treated whey proteins, J. Agric. Food Chem., 2013, 61, 7500–7506 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Investigation of the Potential of Hemp, Pea, Rice and Soy Protein Hydrolysates as a Source of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides, Food Dig., 2015, 6, 19–29 CrossRef CAS.
- C. Lammi, C. Bollati, F. Gelain, A. Arnoldi and R. Pugliese, Enhancement of the stability and anti-DPPIV activity of hempseed hydrolysates through self-assembling peptide-based hydrogels, Front. Chem., 2019, 7, 670 CrossRef.
- A. Cerrato, C. Lammi, A. L. Capriotti, C. Bollati, C. Cavaliere, C. M. Montone, M. Bartolomei, G. Boschin, J. Li, S. Piovesana, A. Arnoldi and A. Laganà, Isolation and functional characterization of hemp seed protein-derived short- and medium-chain peptide mixtures with multifunctional properties for metabolic syndrome prevention, Food Res. Int., 2023, 163, 112219 CrossRef CAS.
- H. Chen, B. Xu, Y. Wang, W. Li, D. He, Y. Zhang, X. Zhang and X. Xing, Emerging natural hemp seed proteins and their functions for nutraceutical applications, Food Sci. Hum. Wellness, 2023, 12, 929–941 CrossRef CAS.
- H.-H. H. Chen, W. Li, Y. Wang, B. Xu, X. Hu, X.-B. B. Li, J.-Y. Y. Liu, C. C.-Y. Y. Zhang, C. C.-Y. Y. Zhang and X.-H. H. Xing, Mining and Validation of Novel Hemp Seed-Derived DPP-IV-Inhibiting Peptides Using a Combination of Multi-omics and Molecular Docking, J. Agric. Food Chem., 2023, 71, 9164–9174 CrossRef CAS PubMed.
- M. Cermeño, J. Stack, P. R. Tobin, M. B. O'Keeffe, P. A. Harnedy, D. B. Stengel and R. J. Fitzgerald, Peptide identification from a: Porphyra dioica protein hydrolysate with antioxidant, angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory activities, Food Funct., 2019, 10, 3421–3429 RSC.
- P. A. Harnedy, V. Parthsarathy, C. M. McLaughlin, M. B. O'Keeffe, P. J. Allsopp, E. M. McSorley, F. P. M. O'Harte and R. J. FitzGerald, Atlantic salmon (Salmo salar) co-product-derived protein hydrolysates: A source of antidiabetic peptides, Food Res. Int., 2018, 106, 598–606 CrossRef CAS PubMed.
- F. Rivero-Pino, A. Guadix and E. M. Guadix, Identification of novel dipeptidyl peptidase IV and α-glucosidase inhibitory peptides from Tenebrio molitor, Food Funct., 2021, 12, 873–880 RSC.
- S. Montserrat-de-la-Paz, A. Villanueva-Lazo, F. Millan, V. Martin-Santiago, F. Rivero-Pino and M. C. Millan-Linares, Production and identification of immunomodulatory peptides in intestine cells obtained from hemp industrial by-products, Food Res. Int., 2023, 174, 113616 CrossRef CAS PubMed.
- P. A. Harnedy and R. J. FitzGerald, In vitro assessment of the cardioprotective, anti-diabetic and antioxidant potential of Palmaria palmata protein hydrolysates, J. Appl. Phycol., 2013, 25, 1793–1803 CrossRef CAS.
- S. Gupta, P. Kapoor, K. Chaudhary, A. Gautam, R. Kumar and G. P. S. Raghava, In Silico Approach for Predicting Toxicity of Peptides and Proteins, PLoS One, 2013, 8, e73957 CrossRef CAS PubMed.
- P. Charoenkwan, S. Kanthawong, C. Nantasenamat, M. M. Hasan and W. Shoombuatong, IDPPIV-SCM: A sequence-based predictor for identifying and analyzing dipeptidyl peptidase IV (DPP-IV) inhibitory peptides using a scoring card method, J. Proteome Res., 2020, 19, 4125–4136 CrossRef CAS PubMed.
- P. Charoenkwan, C. Nantasenamat, M. M. Hasan, M. A. Moni, P. Lio’, B. Manavalan and W. Shoombuatong, StackDPPIV: A novel computational approach for accurate prediction of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides, Methods, 2022, 204, 189–198 CrossRef CAS.
- X. Chen, J. Huang and B. He, AntiDMPpred: a web service for identifying anti-diabetic peptides, PeerJ, 2022, 10, e13581 CrossRef PubMed.
- A. Brodkorb, L. Egger, M. Alminger, P. Alvito, R. Assunção, S. Ballance, T. Bohn, C. Bourlieu-Lacanal, R. Boutrou, F. Carrière, A. Clemente, M. Corredig, D. Dupont, C. Dufour, C. Edwards, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. R. Mackie, C. Martins, S. Marze, D. J. McClements, O. Ménard, M. Minekus, R. Portmann, C. N. Santos, I. Souchon, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and I. Recio, INFOGEST static in vitro simulation of gastrointestinal food digestion, Nat. Protoc., 2019, 14, 991–1014 CrossRef CAS PubMed.
- P. Minkiewicz, A. Iwaniak and M. Darewicz, BIOPEP-UWM database of bioactive peptides: Current opportunities, Int. J. Mol. Sci., 2019, 20, 5978 CrossRef CAS.
- G. Aiello, E. Fasoli, G. Boschin, C. Lammi, C. Zanoni, A. Citterio and A. Arnoldi, Proteomic characterization of hempseed (Cannabis sativa L.), J. Proteomics, 2016, 147, 187–196 CrossRef CAS.
- B. J. W. van Klinken, M. L. Stewart, S. Kalgaonkar and L. Chae, Health-Promoting Opportunities of Hemp Hull: The Potential of Bioactive Compounds, 2024, preprint, DOI:10.1080/19390211.2024.2308264.
- C. Acquah, E. D. Stefano and C. C. Udenigwe, Role of hydrophobicity in food peptide functionality and bioactivity, J. Food Bioact., 2018, 4, 88–98 CrossRef.
- M. R. Segura Campos, L. A. Chel Guerrero and D. A. Betancur Ancona, Angiotensin-I converting enzyme inhibitory and antioxidant activities of peptide fractions extracted by ultrafiltration of cowpea Vigna unguiculata hydrolysates, J. Sci. Food Agric., 2010, 90, 2512–2518 CrossRef CAS PubMed.
- O. L. Tavano, A. Berenguer-Murcia, F. Secundo and R. Fernandez-Lafuente, Biotechnological Applications of Proteases in Food Technology, Compr. Rev. Food Sci. Food Saf., 2018, 17, 412–436 CrossRef PubMed.
- V. Parthsarathy, C. M. McLaughlin, P. A. Harnedy, P. J. Allsopp, W. Crowe, E. M. McSorley, R. J. FitzGerald and F. P. M. M. O'Harte, Boarfish (Capros aper) protein hydrolysate has potent insulinotropic and GLP-1 secretory activity in vitro and acute glucose lowering effects in mice, Int. J. Food Sci. Technol., 2018, 54, 271–281 CrossRef.
- S. Sharma, R. Pradhan, A. Manickavasagan, A. Tsopmo, M. Thimmanagari and A. Dutta, Corn distillers solubles by two-step proteolytic hydrolysis as a new source of plant-based protein hydrolysates with ACE and DPP4 inhibition activities, Food Chem., 2023, 401, 134120 CrossRef CAS.
- R. C. Mollard, A. Johnston, A. S. Leon, H. Wang, P. J. Jones and D. S. Mackay, Acute effects of hemp protein consumption on glycemic and satiety control : results of 2 randomized crossover trials, Appl. Physiol., Nutr., Metab., 2021, 46, 887–896 CrossRef CAS.
- R. Liu, J. Cheng and H. Wu, Discovery of Food-Derived Dipeptidyl Peptidase IV Inhibitory Peptides, A Review, Int. J. Mol. Sci., 2019, 20, 463 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. FitzGerald, Strategies for the discovery and identification of food protein-derived biologically active peptides, Trends Food Sci. Technol., 2017, 69, 289–305 CrossRef CAS.
- A. B. Nongonierma, S. L. Maux, C. Esteveny and R. J. FitzGerald, Response surface methodology applied to the generation of casein hydrolysates with antioxidant and dipeptidyl peptidase IV inhibitory properties, J. Sci. Food Agric., 2017, 97, 1093–1101 CrossRef CAS.
- P. Kęska and J. Stadnik, Structure–activity relationships study on biological activity of peptides as dipeptidyl peptidase IV inhibitors by chemometric modeling, Chem. Biol. Drug Des., 2020, 95, 291–301 CrossRef.
- C. Mooney, N. J. Haslam, G. Pollastri and D. C. Shields, Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity, PLoS One, 2012, 7, e45012 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, UCSF Chimera - A visualization system for exploratory research and analysis, J. Comput. Chem., 2004, 25, 1605–1612 Search PubMed.
- F. Rivero-Pino, F. J. Espejo-Carpio and E. M. Guadix, Identification of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from vegetable protein sources, Food Chem., 2021, 354, 129473 CrossRef CAS PubMed.
- F. Rivero-Pino, M. C. Millan-Linares and S. Montserrat-De-la-Paz, Strengths and limitations of in silico tools to assess physicochemical properties, bioactivity, and bioavailability of food-derived peptides, Trends Food Sci. Technol., 2023, 138, 433–440 CrossRef CAS.
- L. Mora, D. González-Rogel, A. Heres and F. Toldrá, Iberian dry-cured ham as a potential source of α-glucosidase-inhibitory peptides, J. Funct. Foods, 2020, 67, 103840 CrossRef CAS.
- C. Berraquero-García, F. Rivero-Pino, L. Ospina, R. Perez-Galvez, F. J. Espejo-Carpio, A. Guadix, P. J. García-Moreno and E. M. Guadix, Activity, structural features and in silico digestion of antidiabetic peptides, Food Biosci., 2023, 102954 CrossRef.
- M. Gonzalez-Montoya, B. Hernández-Ledesma, R. Mora-Escobedo and C. Martinez-Villaluenga, Bioactive peptides from germinated soybean with anti-diabetic potential by inhibition of dipeptidyl peptidase-IV, a-amylase, and a-glucosidase enzymes, Int. J. Mol. Sci., 2018, 19, 2883 CrossRef.
- C.-H. Hsieh, T.-Y. Wang, C.-C. Hung, C.-L. Jao, Y.-L. Hsieh, S.-X. Wu and K.-C. Hsu, In silico, in vitro and in vivo analyses of dipeptidyl peptidase IV inhibitory activity and the antidiabetic effect of sodium caseinate hydrolysate, Food Funct., 2016, 7, 1122–1128 RSC.
- I. M. E. Lacroix and E. C. Y. Li-Chan, Food-derived dipeptidyl-peptidase IV inhibitors as a potential approach for glycemic regulation – Current knowledge and future research considerations, Trends Food Sci. Technol., 2016, 54, 1–16 CrossRef CAS.
- Y. Yu, Y. Jin, F. Wang, J. Yan, Y. Qi and M. Ye, Protein digestomic analysis reveals the bioactivity of deer antler velvet in simulated gastrointestinal digestion, Food Res. Int., 2017, 96, 182–190 CrossRef CAS PubMed.
- L. Zheng, Q. Xu, L. Lin, X. A. Zeng, B. Sun and M. Zhao, In Vitro Metabolic Stability of a Casein-Derived Dipeptidyl Peptidase-IV (DPP-IV) Inhibitory Peptide VPYPQ and Its Controlled Release from Casein by Enzymatic Hydrolysis, J. Agric. Food Chem., 2019, 67, 10604–10613 CrossRef CAS.
- E. C. Y. Li-Chan, S. L. Hunag, C. L. Jao, K. P. Ho and K. C. Hsu, Peptides derived from Atlantic salmon skin gelatin as dipeptidyl-peptidase IV inhibitors, J. Agric. Food Chem., 2012, 60, 973–978 CrossRef CAS.
- H. Chen, W. Li, W. Hu, B. Xu, Y. Wang, J. Liu, C. Zhang, C. Zhang, X. Zhang, Q. Nie and X. Xing, A novel bifunctional peptide VAMP mined from hemp seed protein hydrolysates improves glucose homeostasis by inhibiting intestinal DPP-IV and increasing the abundance of Akkermansia muciniphila, 2024, preprint, DOI:10.1101/2024.07.22.604525.
- M. Tenenbaum, C. Dugardin, J. Moro, J. Auger, A. Baniel, A. Boulier, R. Ravallec and B. Cudennec, In vitro comparison of whey protein isolate and hydrolysate for their effect on glucose homeostasis markers, Food Funct., 2023, 4173–4182 RSC.
- M. Darewicz, M. Pliszka, J. Borawska-Dziadkiewicz, P. Minkiewicz and A. Iwaniak, Multi-Bioactivity of Protein Digests and Peptides from Oat (Avena sativa L.) Kernels in the Prevention of the Cardiometabolic Syndrome, Molecules, 2022, 27, 7907 CrossRef CAS.
- X. Sun, C. Acquah, R. E. Aluko and C. C. Udenigwe, Considering food matrix and gastrointestinal effects in enhancing bioactive peptide absorption and bioavailability, J. Funct. Foods, 2020, 64, 103680 CrossRef CAS.
- V. T. T. Lan, K. Ito, M. Ohno, T. Motoyama, S. Ito and Y. Kawarasaki, Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor, Food Chem., 2015, 175, 66–73 CrossRef CAS PubMed.
- A. Thongtak, K. Yutisayanuwat, N. Harnkit, T. Noikaew and P. Chumnanpuen, Computational Screening for the Dipeptidyl Peptidase-IV Inhibitory Peptides from Putative Hemp Seed Hydrolyzed Peptidome as a Potential Antidiabetic Agent, Int. J. Mol. Sci., 2024, 25, 5730 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |