Open Access Article
Open Access ArticlePrunus mume extract promoted biofilm formation of Lactiplantibacillus plantarum JB1 to improve acid and oxidative stress resistance
Teng Long
Miao
a,
Chuang
Zhang
a,
Hui
Wang
a,
Xiaohong
Chen
a,
Hong Mei
Xiao
*a and
Qiu Qin
Zhang
 *ab
*ab
aCollege of Food Science and Technology, Nanjing Agricultural University, Nanjing 210095, PR China. E-mail: zqq@njau.edu.cn; xhm@njau.edu.cn; 2022108075@njau.edu.cn; zhangchuang@njau.edu.cn; huiwang@njau.edu.cn; xhchen@njau.edu.cn
bSanya Institute of Nanjing Agricultural University, Sanya, Hainan 572024, China
First published on 29th September 2025
Abstract
Lactic acid bacteria (LAB) play a pivotal role in sustainable food fermentation systems, yet their industrial application is often limited by sensitivity to environmental stress. This study introduces a novel, green strategy utilizing Prunus mume extract (PME)—an underutilized natural resource with strong antioxidant activity—to enhance the stress resistance of L. plantarum JB1. This study explicitly investigated the role of PME pre-stress in controlling biofilm formation via the LuxS/AI-2 quorum-sensing pathway as a key mechanism for improved resistance under acid and oxidative stress. The results indicated that PME pre-stress significantly improves bacterial survival under acid (21.08% increase) and oxidative stress (20.14% increase). PME pre-stress enhanced β-galactosidase and L-LDH enzyme activities and altered the cell surface polysaccharide composition, further mitigating cell damage, improving L. plantarum JB1 stress resistance under acid and oxidative stress. By leveraging a natural agricultural by-product to fortify probiotics, this research provides a sustainable solution to reduce cell loss in fermentation, minimize the energy-intensive stabilization process, and extend the shelf life of fermented foods. This approach contributes directly to cleaner industrial practices, supports a circular economy by adding value to food waste streams, and strengthens food security through more efficient production of probiotic-fortified foods.
Sustainability spotlightPrunus mume extract (PME) is a renewable agricultural by-product. The natural antioxidant properties of PME make it have the potential to enhance the stress resistance of probiotics. The results showed that PME pre-stress improved the resistance of Lactobacillus plantarum JB1 by promoting biofilm formation. PME has great application potential in low pH fermented foods (such as pickles, yogurt, etc.), which can reduce production loss and food waste by prolonging the survival time of probiotics and the shelf life of fermented foods. This achievement not only increases the added value of agricultural by-products (bioactive plant extracts), but also transforms traditional agricultural by-products into functional fermentation additives, which is in line with the concept of circular economy. |
1. Introduction
The global shift toward sustainable food production demands innovative strategies to enhance the efficiency and resistance of microbial strains used in fermentation, a cornerstone of eco-friendly food processing. Lactic acid bacteria (LAB), widely employed as probiotics and industrial starters, face significant challenges in harsh environments, such as acidic fermentation conditions or oxidative stress during storage.1 These stressors not only reduce LAB viability but also increase energy consumption and waste in production systems. Traditional approaches to improve stress resistance, such as genetic modification or synthetic additives, often conflict with consumer preferences for natural and sustainable solutions.Biofilm formation, a natural microbial defense mechanism, presents a promising avenue for sustainable food technology. Biofilm is a mechanism by which bacteria resist adverse environments. Its formation is mainly due to the secretion of extracellular polysaccharide proteins, polysaccharides, nucleic acids, lipids, and other macromolecules by a large number of bacteria. These substances adhere to the surface of microbial cells, causing a large number of bacteria to gather and form communities.2,3 The formation of biofilms can greatly improve the stress resistance of bacterial cells and effectively enhance their resistance to harsh environments such as high temperature, low temperature, low pH, peroxide environment, and bile salts.4 Biofilms enhance bacterial survival under stress, potentially reducing the need for energy-intensive stabilization methods like freeze-drying or excessive refrigeration. LuxS/AI-2 is the most common quorum sensing system in lactic acid bacteria that regulates biofilm formation. Autoinducer-2 (AI-2) is synthesized by the LuxS protease encoded by the LuxS gene, and its concentration threshold can regulate the biofilm formation ability of the strain. AI-2 has the activity of inducing V. harveyi BB170 bioluminescence, so the concentration of AI-2 can be reflected by measuring the bioluminescence intensity of V. harveyi BB170. However, a significant research gap exists in the use of plant-derived bioactive compounds to induce and improve probiotic biofilm formation. Prunus mume extract (PME), a fruit rich in antioxidants, exemplifies a renewable resource with untapped potential in sustainable microbial engineering. Prunus mume extract has been proven to have various beneficial effects, such as improving blood flow, reducing obesity, and fighting cancer and viruses, but its benefits for probiotics are still unknown.5–7
Therefore, to address this knowledge gap, this study aims to explore the novel application of PME to improve the acid and oxidative stress resistance of L. plantarum JB1. The research specifically seeks to evaluate the effect of PME on the biofilm formation capability of the bacterium and determine its subsequent impact on survival under simulated stressful conditions. Furthermore, the study intends to elucidate the underlying mechanisms of this enhanced tolerance by analyzing changes in AI-2 activity, key enzyme functions, cell integrity, and surface properties. Ultimately, this work proposes a sustainable bio-engineering strategy using natural plant extracts to harness biofilm formation, advancing the development of robust and eco-friendly probiotic cultures for food industry applications.
2. Materials and methods
2.1. Materials
Prunus mume extract (PME) was purchased from Nanjing Longlijia Agricultural Development. Dimethylbenzene (CAS: 106-42-3), ethyl acetate (CAS: 141-78-6), trichloromethane (CAS: 67-66-3) and H2O2 (CAS: 7722-84-1) were bought from Sinopharm Chemical Reagent Co., Ltd. Crystal violet (CAS: 548-62-9) and acetic acid (CAS: 64-19-7) were procured from Shanghai Haosai Technology Co., Ltd. 2′,7′-Dichlorodihydrofluorescein diacetate (DCFH-DA, CAS: 4091-99-0) and 2-nitrophenyl-beta-D-galactopyranoside (ONPG, CAS: 369-07-3) were procured from Source Leaf Organism (Shanghai Yuanye Biotechnology Co., Ltd, Shanghai, China). Propidium iodide (PI, CAS: 25535-16-4) was procured from Yeasen Biotechnology (Shanghai) Co., Ltd. SYTO9 Green Fluorescent Nucleic Acid Stains (S34854) were bought from Invitrogen Corporation (California, USA). A L-lactate dehydrogenase activity detection kit (BC0680) was procured from Beijing Solarbio Science & Technology Co., Ltd.2.2. Culture conditions of bacterial strains
Lactiplantibacillus plantarum JB1 was from Nanjing Agricultural University, Nanjing, China.8Vibrio harveyi BB170 is a directionally mutated strain with an AI-2 receptor that can be used to measure AI-2 activity.9L. plantarum JB1 was cultured in de Man, Rogosa and Sharpe (MRS) medium (Qingdao Hope Bio Technology Co., Ltd, Qingdao, China): peptone, 10 g L−1; beef extract, 10 g L−1; yeast extract, 5 g L−1; glucose, 20 g L−1; K2HPO4, 2 g L−1; sodium acetate, 5 g L−1; MgSO4·7H2O, 0.2 g L−1; MnSO4, 0.05 g L−1; ammonium citrate, 2 g L−1; Tween-80, 1 mL L−1; and pH 6.2 ± 0.2.
Autoinducer bioassay (AB) medium was used to culture V. harveyi BB170 (Gu et al., 2021![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21). AB medium: sodium chloride, 17.5 g L−1; MgSO4·7H2O, 12.3 g L−1; acid hydrolyzed casein, 2.0 g L−1; distilled water, 960 mL; then the pH of the solution was adjusted to 7.5 with KOH (3 mol L−1). After the AB medium was sterilized at 121 °C for 20 min and cooled down, 10 mL of sterile 0.1 mol per L L-arginine (L-arginine was filtered through a 0.22 μm filter), 10 mL of sterile 1.0 mol per L potassium phosphate buffer (pH 7.0), and 20 mL of sterile 50% glycerol were added into AB medium.
21). AB medium: sodium chloride, 17.5 g L−1; MgSO4·7H2O, 12.3 g L−1; acid hydrolyzed casein, 2.0 g L−1; distilled water, 960 mL; then the pH of the solution was adjusted to 7.5 with KOH (3 mol L−1). After the AB medium was sterilized at 121 °C for 20 min and cooled down, 10 mL of sterile 0.1 mol per L L-arginine (L-arginine was filtered through a 0.22 μm filter), 10 mL of sterile 1.0 mol per L potassium phosphate buffer (pH 7.0), and 20 mL of sterile 50% glycerol were added into AB medium.
2.3. Stress treatment
In the pre-experiment, after L. plantarum JB1 was cultured in 0, 20, 40, 60, 80, and 100 mg per mL PME, it was found that L. plantarum JB1 did not grow in 60, 80, and 100 mg per mL PME, so 0–60 mg per mL PME was chosen as the concentration of the formal experiment. L. plantarum JB1 was cultured in MRS medium with 0, 10, 20, 30, 40, 50, and 60 mg per mL PME, and named as PME-0, PME-10, PME-20, PME-30, PME-40, PME-50, and PME-60. After incubation, L. plantarum JB1 cells were collected by centrifugation at 6000 rpm, 4 °C, for 6 min. The bacterial cells were washed twice with 0.9% NaCl for stress treatment.To verify whether PME pre-stress can improve the digestion of probiotics in the stomach and intestines, MRS medium with pH2 was used to simulate the gastric acid environment, MRS medium with 10 mmol per L H2O2 was used to simulate the reactive oxygen species in the intestine, and MRS medium with 0.4% bile salts was used to simulate the intestinal environment. In order to simulate the transport time of probiotics in human gastrointestinal digestion, a 3 h stress treatment was used. In the preliminary experiment, PME could significantly improve the survival rate and biofilm formation ability of L. plantarum JB1 under acid stress and oxidative stress, but it did not improve the resistance of L. plantarum JB1 under bile salt stress (Supplement Fig. 1). Finally, the stress treatment medium for acid and oxidative stress was MRS medium with pH 2 and with 10 mmol per L H2O2, respectively.
L. plantarum JB1 before and after stress treatments was centrifuged at 6000 rpm, 4 °C, for 6 min, and collected. The bacterial cells were washed twice with 0.9% NaCl. The post-stress bacterial suspension was used to determine survival rate, biofilm formation ability, AI-2 activity, auto-aggregation ability, surface properties, FTIR, enzyme activity of β-galactosidase and L-LDH, flow cytometry detection, cell membrane damage and cell wall damage.
2.4. Determination of the survival rate
During the stress treatment of bacteria at 37 °C for 3 h, the bacterial solution was collected every 1 h and the viable cell count was obtained.10 The formula for calculating the survival rate is shown in eqn (1):Survival rate = (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N1/log N1/log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N2) × 100% N2) × 100% | (1) |
2.5. Measurement of biofilm formation ability
The biofilm formation ability was determined by crystal violet staining according to the method of Nouraei et al.11 with some modifications. L. plantarum JB1 (2%, v/v) was inoculated into MRS medium with or without PME, transferred 200 μL per well into a 96-well cell culture plate covered with a sealing membrane and incubated at 37 °C for 24 h. The culture medium was discarded from the plate, 200 μL stress treatment medium was added to the plate for stress, and then the culture medium was discarded from the plate. The 96-well cell culture plate was washed 3–4 times with 0.9% NaCl to remove the non-adherent cells, ensuring that the assay measures only the adhering biofilm, and fixed in a 60 °C oven for 30 min. Crystal violet (200 μL) staining solution (0.1%, m/v) was added to each well for 15 min, then the crystal violet staining solution was discarded. Each well was washed 3–4 times with 0.9% NaCl, and 200 μL glacial acetic acid solution (33%, v/v) was added to each well for 15 min of elution. Eluent (200 μL) was added to a new 96-well cell culture plate, and the absorbance value was measured at 560 nm.2.6. Determination of L. plantarum JB1 AI-2 activity
The AI-2 activity was determined according to the method of Gu et al.12 with some modifications. The bacterial solution processed was centrifuged at 6000 rpm for 6 min and filtered through a 0.22 μm filter to obtain the cell-free supernatant (CFS). V. harveyi BB170 was inoculated into AB medium at a 2% inoculation rate and incubated at 30 °C for 8 h. The V. harveyi BB170 culture was diluted 2000-fold with fresh AB medium. CFS and diluted V. harveyi BB170-AB medium were mixed (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) and incubated with shaking at 100 rpm at 30 °C. Activated V. harveyi BB170, AB, and MRS were used to replace CFS as positive control, negative control, and medium control, respectively. Multifunctional enzyme-linked immunosorbent assay with imaging (PerkinElmer EnSight, PerkinElmer, Singapore) was used to measure the luminescence intensity of the reaction solution in bioluminescence mode every 30 min within 0–6 h. When the luminescence intensity of the negative control reaches the lowest point, the sample of relative luminescence intensity (RLI) measured was equal to the luminescence intensity of the sample divided by the luminescence intensity of the medium control.
100) and incubated with shaking at 100 rpm at 30 °C. Activated V. harveyi BB170, AB, and MRS were used to replace CFS as positive control, negative control, and medium control, respectively. Multifunctional enzyme-linked immunosorbent assay with imaging (PerkinElmer EnSight, PerkinElmer, Singapore) was used to measure the luminescence intensity of the reaction solution in bioluminescence mode every 30 min within 0–6 h. When the luminescence intensity of the negative control reaches the lowest point, the sample of relative luminescence intensity (RLI) measured was equal to the luminescence intensity of the sample divided by the luminescence intensity of the medium control.
2.7. Determination of L. plantarum JB1 auto-aggregation ability and surface properties
The absorbance of the bacterial suspension was adjusted to 0.6 ± 0.02 with 0.9% NaCl, denoted as A0.The auto-aggregation of L. plantarum JB1 was determined according to the method proposed by Krausova et al.13 The post-stress bacterial suspension (4 mL) was added into each test tube, incubated at 37 °C for 1 and 2 h. The upper layer solution (200 μL) was measured at 600 nm and denoted as A1. The formula for calculating the auto-agglutination ability of L. plantarum JB1 is shown below in eqn (2):
| Auto-aggregation (%) = (1 − A1/A0) × 100% | (2) |
The surface properties of L. plantarum JB1 were determined according to the method proposed by Xiao et al.14 Dimethylbenzene (1 mL, non-polar solvent), ethyl acetate (1 mL, electron donor), and trichloromethane (1 mL, electron acceptor) were mixed with 3 mL of the bacterial suspension, respectively, incubated at 37 °C for 20 min and the value of the aqueous phase was measured at 600 nm, denoted as A2. The formula for calculating the hydrophobicity of L. plantarum JB1 is shown below in eqn (3):
| Hydrophobicity (%) = (1 − A2/A0) × 100% | (3) |
2.8. Fourier transform infrared spectroscopy (FTIR) analysis of L. plantarum JB1 surface properties
The post-stress bacterial cells were vacuum freeze-dried to obtain bacterial powder. Under infrared light, the bacterial powder (1–2 mg) was mixed with KBr, and the ratio of KBr to sample is 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w
1 (w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) w), installed into a quartz grinder for grinding, and pressed into molds. Each sample was analyzed with a Fourier transform infrared spectrometer (Nicolet iS10, Thermo Fisher Scientific, Massachusetts, USA), with a detection wavelength of 4000–400 cm−1, a resolution of 4 cm−1, and 64 scans.
w), installed into a quartz grinder for grinding, and pressed into molds. Each sample was analyzed with a Fourier transform infrared spectrometer (Nicolet iS10, Thermo Fisher Scientific, Massachusetts, USA), with a detection wavelength of 4000–400 cm−1, a resolution of 4 cm−1, and 64 scans.
2.9. Determination of enzyme activity of β-galactosidase and L-lactate dehydrogenase (L-LDH) of L. plantarum JB1
Determination of the enzyme activity of β-galactosidase: the post-stress bacterial cells (5 mL) were taken into a test tube and broken by ice bath ultrasonic waves. The ultrasonic power is 200 W, the ultrasound lasts for 5 s, the gap is 5 s, and the fragmentation lasts for 30 min. The bacterial solution was centrifuged at 6000 rpm, 4 °C for 6 min to obtain the supernatant, which is the crude enzyme solution. ONPG used as the enzyme substrate, the colored product ONP (ortho-nitrophenol) generated was determined by the colorimetric method. Crude enzyme solution (1 mL) and ONPG solution (1 mL, 10 mmol L−1) were mixed and reacted in a 37 °C water bath for 30 min. Na2CO3 (2 mL, 1 mol L−1) was added into the reaction solution to terminate the reaction, and the reaction solution absorbance was measured at 420 nm. The standard curve of ONP was used to calculate the concentration of ONP in the reaction solution. Enzyme activity is defined as the production of 1 μmol ONP from the decomposition of ONPG by 1 mL of enzyme solution at 37 °C for 1 min. The formula for calculating β-galactosidase activity is shown below in eqn (4):| β-Galactosidase activity (U mL−1) = (X1 × 4)/(30 × 1000) | (4) |
The activity of L-LDH was detected with the L-lactate dehydrogenase activity detection kit (BC0680, Beijing Solarbio Science & Technology Co., Ltd, Beijing). The enzyme activity unit is defined as the catalytic production of 1 nmol pyruvate per minute by every 10000 bacteria.15,16
2.10. Flow cytometry detection of L. plantarum JB1
SYTO 9 (5 μM mL−1) and PI (60 μM mL−1) were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in a centrifuge tube. A dye mixture (100 μL) was added to 1 mL of the post-stress bacterial suspension, which was incubated at 37 °C in the dark for 20 min.17 The bacterial suspension was washed twice with 0.9% NaCl and detected by flow cytometry (Model CytoFLEX, Beckman Coulter Inc., California, USA). The detailed flow cytometry configuration is as follows: blue laser (488 nm/483 nm); detection channels FITC (503/40 nm), PE (585/42 nm). PI is excited by the 488 nm blue laser and detected in the PE (585/42 nm) channel. SYTO 9 is excited by the 483 nm blue laser and detected in the FITC (525/40 nm) channel.
1 ratio in a centrifuge tube. A dye mixture (100 μL) was added to 1 mL of the post-stress bacterial suspension, which was incubated at 37 °C in the dark for 20 min.17 The bacterial suspension was washed twice with 0.9% NaCl and detected by flow cytometry (Model CytoFLEX, Beckman Coulter Inc., California, USA). The detailed flow cytometry configuration is as follows: blue laser (488 nm/483 nm); detection channels FITC (503/40 nm), PE (585/42 nm). PI is excited by the 488 nm blue laser and detected in the PE (585/42 nm) channel. SYTO 9 is excited by the 483 nm blue laser and detected in the FITC (525/40 nm) channel.
2.11. Determination of cell membrane damage and cell wall damage of L. plantarum JB1
| Bacterial loss rate in NaCl-MRS medium = 1 − (N3/N4) × 100% | (5) |
| Bacterial cell loss rate in lysozyme-MRS medium = 1 − (N5/N6) × 100% | (6) |
2.12. Statistical analyses
All data are expressed as the mean values ± standard deviations (SD). Statistical comparisons between different times were performed with a one-way analysis of variance, followed by Duncan's test at p < 0.05, using SPSS 25.0 software (Statistical Product and Service Solutions, USA). Drawing was performed with Origin 2025 (OriginLab, USA). The number of parallel samples for all measurement indicators is 3, repeated twice.3. Results and discussion
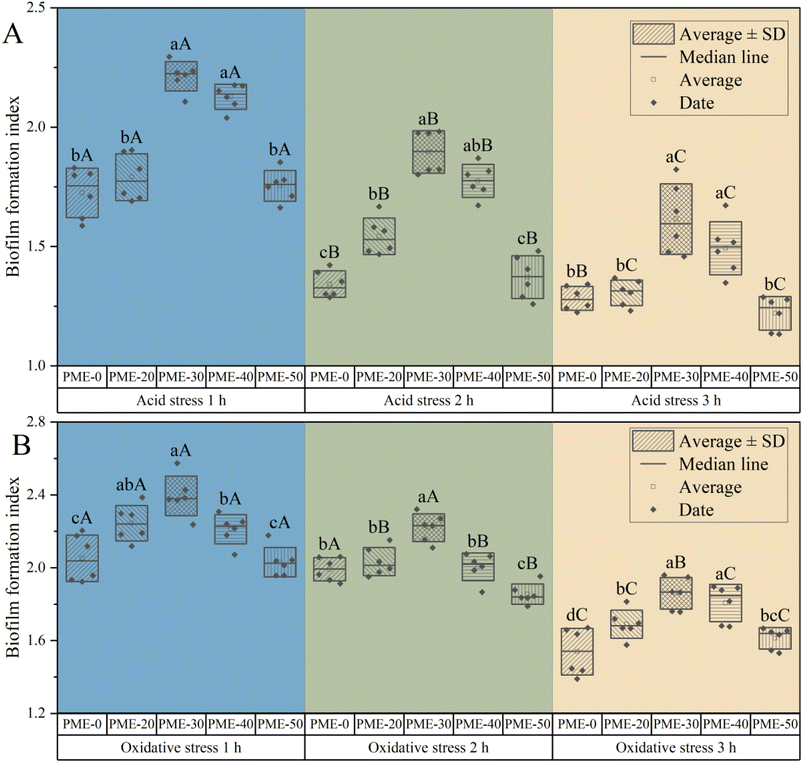
The survival rate of L. plantarum JB1 was measured under acid and oxidative stress after different concentrations of PME pre-stress (Fig. 1). After different stresses, the survival rate of each group had similar trends. There was no significant difference (p > 0.05) between PME-30, PME-40, and PME-50 survival rates within the 1 h of stress, while PME-0 and PME-20 survival rates were significantly lower than other groups (p < 0.05). It may be that the low concentration of PME does not significantly improve the resistance of probiotics. However, with the increase of PME concentration, there was no significant difference between the high concentration groups (PME-30, PME-40, and PME-50), which may be that the ability of PME to improve the resistance of probiotics reaches the maximum. As the duration of stress was prolonged, the differences between different concentrations gradually became significant (p < 0.05). After 3 h of stress treatment, the difference in survival rate among the groups was the most obvious. There was no significant difference in the survival rate of the high concentration group in the short treatment time, but there was also a significant difference between the high concentration groups with the extension of time. It may be that the high concentration of PME leads to stress in the bacteria, which temporarily improves the resistance. With the extension of stress time, the stress effect decreases, and the survival rate will decline. Among all concentrations of PME, PME-30 had the highest survival rate under 3 h of acid and oxidative stress. However, PME-60 did not grow, possibly due to the high concentration of PME, which caused the culture medium to be too acidic and inhibited the growth of L. plantarum JB1.20Biofilm formation index of L. plantarum JB1 was measured under acid (Fig. 2A) and oxidative (Fig. 2B) stress after different concentrations of PME pre-stress. The biofilm formation index ability of PME-30 within 1 h of stress was significantly higher than those of other groups (p < 0.05), followed by PME-20 and PME-40, and finally PME-0 and PME-50. As the duration of stress was prolonged, PME-30 still had the highest biofilm formation ability. PME-30 with a concentration of 30 mg mL−1 PME was selected as the culture medium for the L. plantarum JB1, and 3 h of acid and oxidative stress was used in subsequent experiments.
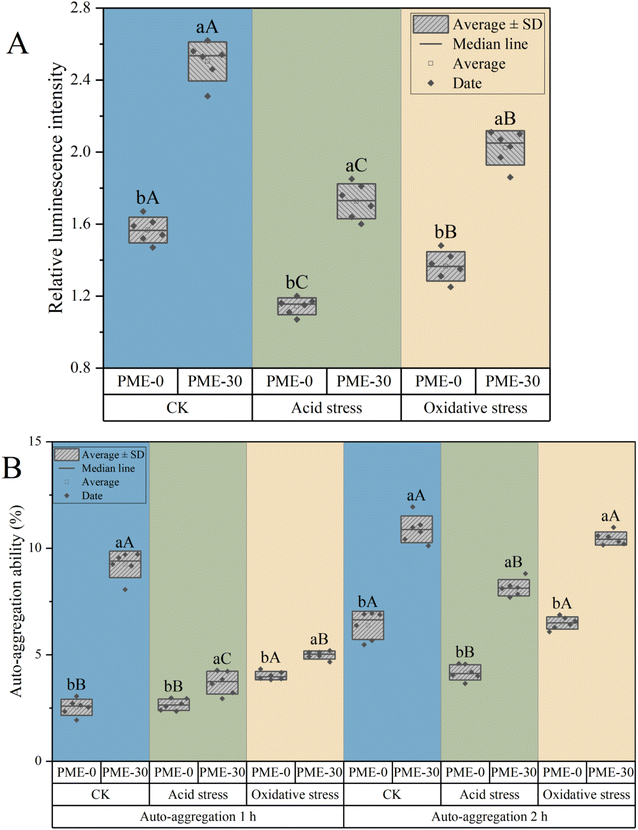
In order to further verify the regulatory pathway of PME in increasing the biofilm, the AI-2 content of lactic acid bacteria was measured under acid and oxidative stress (Fig. 3A). AI-2 is an important substance in the mediated quorum sensing system (QS) between bacterial cells, and its content is closely related to the ability of bacterial cells to produce biofilms.21 The higher the AI-2 content, the stronger the ability of bacterial cells to produce biofilms. Fig. 3A shows the relative luminescence intensity (RLI) of signal molecule AI-2 in L. plantarum JB1 under acid and oxidative stress after PME pre-stress. There was a significant difference (p < 0.05) in the activity of signal molecule AI-2 of PME-0 and PME-30 between the CK group (without stress). After acid and oxidative stress, the activity of signal molecule AI-2 in L. plantarum JB1 was significantly reduced (p < 0.05). The activity of signal molecule AI-2 in all groups of PME-30 was significantly higher than that in PME-0 (p < 0.05), and oxidative stress resulted in a higher degree of signal molecule reduction than acid stress. The results indicate that PME cultivation facilitates the production of signal molecule AI-2 by L. plantarum JB1 to promote biofilm formation.21 The research results of Gu et al.22 indicate that the decrease in AI-2 activity is induced by the stress response of bacterial cells to acid and oxidative stress, which is consistent with the results of this study.
Probiotics need to adhere to and colonize in the host's intestinal system before they can exert their probiotic properties. Auto-aggregation ability and surface hydrophobicity are important indicators reflecting the non-specific adhesion of probiotics to the intestine. In order to further investigate the effects of PME pre-stress on bacterial characteristics and the non-specific adhesion of probiotics to the intestine, the auto-aggregation of bacterial cells under acid and oxidative stress was observed after PME pre-stress (Fig. 3B). The auto-aggregation ability of PME-30 at 1 h and 2 h was significantly higher than that of PME-0 (p < 0.05). After acid and oxidative stress, the auto-aggregation ability of the L. plantarum JB1 decreased slightly (p < 0.05). Overall, the auto-aggregation ability of PME-30 decreased slightly and was still significantly higher than that ofPME-0 (p < 0.05). Auto-aggregation ability refers to the ability of bacterial cells to bind together and form cell aggregates, which helps probiotics form biofilms and adhere and colonize in the host intestine.23 PME pre-stress could enhance the auto-aggregation ability of the L. plantarum JB1 and improve its intestinal adhesion ability.
Dimethylbenzene is used to represent the surface hydrophobicity of bacterial cells; ethyl acetate is used to indicate the ability of bacterial cells to obtain electrons; trichloromethane is used to represent the electron-supplying ability of bacterial cells.24,25 As shown in Table 1, after acid stress, the affinity of PME-0 and PME-30 for dimethylbenzene increased, while their affinity for ethyl acetate and trichloromethane decreased. This indicates that the hydrophobicity of L. plantarum JB1 has been improved, but its ability to obtain/supply electrons has decreased. After oxidative stress, the affinity of PME-0 and PME-30 for dimethylbenzene, ethyl acetate, and trichloromethane was significantly increased (p < 0.05), indicating that the hydrophobicity and electron-obtaining/-supplying ability of the cells were improved. Compared with PME-0 and PME-30, as well as after acid and oxidative stress, the affinity of PME-30 for dimethylbenzene and trichloromethane was significantly lower than that of PME-0 (p < 0.05), while its affinity for ethyl acetate was higher than that of PME-0, indicating that PME pre-stress did not improve the hydrophobicity and electron supplying ability of the cells, but increased their electron obtaining ability. It could be seen that L. plantarum JB1 has certain hydrophobicity and electron-accepting ability, and its electron-accepting ability is much greater than its electron-supplying ability. Surface hydrophobicity refers to the unstable state of bacteria in polar water, which leads to the rearrangement of bacterial cells. It is related to the surface proteins, phospholipids, carbohydrate compounds, biofilm and other structures of bacterial cells, mainly involving electrostatic interactions and hydrophobic forces. Strong surface hydrophobicity also helped bacterial cells adhere and colonize on the surface of the host's intestine. Previously, Wang et al.26 found that protein substances have the ability to enhance the surface hydrophobicity of bacterial cells, while polysaccharide substances have the ability to enhance the surface hydrophilicity of bacterial cells. Therefore, it could be inferred that PME pre-stress may change the surface hydrophobicity of bacterial cells by altering the protein or polysaccharide composition of bacterial surfaces and biofilms.27
| Dimethylbenzene | Ethyl acetate | Trichloromethane | ||
|---|---|---|---|---|
| a Lowercase letters represent the same surface characteristics, and there are significant differences (p < 0.05) between different PME concentrations under the same stress. Capital letters represent the same surface characteristics, and there are significant differences (p < 0.05) between different stresses under the same PME concentration. | ||||
| Control group (CK) | PME-0 | 2.05 ± 0.31aC | 12.58 ± 0.31bB | 5.12 ± 0.34aB |
| PME-30 | 1.12 ± 0.42bC | 14.02 ± 0.56aB | 3.67 ± 0.33bB | |
| Acid stress | PME-0 | 4.33 ± 0.37aB | 7.72 ± 0.17bC | 4.99 ± 0.36aB |
| PME-30 | 1.68 ± 0.36bB | 13.13 ± 0.61aC | 1.77 ± 0.19bC | |
| Oxidative stress | PME-0 | 5.14 ± 0.35aA | 25.15 ± 0.17aA | 10.76 ± 0.38aA |
| PME-30 | 2.36 ± 0.35bA | 23.54 ± 0.24bA | 7.26 ± 0.19bA | |
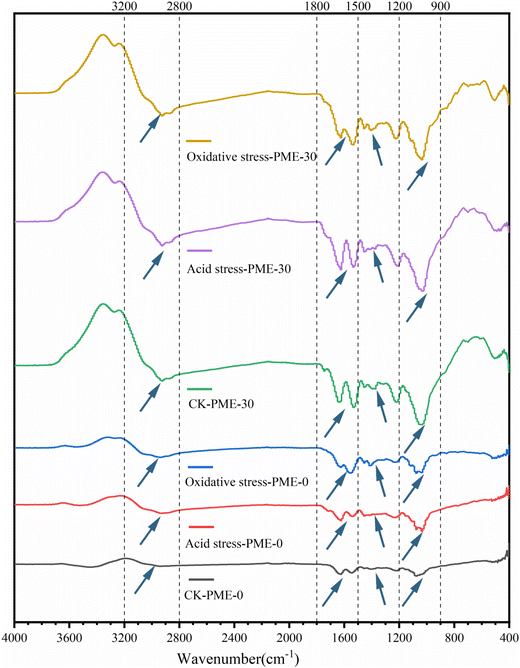
FT-IR can measure changes in specific functional bonds to determine changes in the interactions between polysaccharides and proteins on the surface of bacterial cells. In order to further understand the changes in the surface properties of L. plantarum JB1, FT-IR was used to characterize the surface properties of L. plantarum JB1. The FT-IR results are shown in Fig. 4, and the peak values of each segment correspond to specific components, for example: 3100–2800 cm−1 composed of the functional group CH3, CH2, CH is the characteristic peak generated by the main stretching vibration, usually corresponding to the fatty acid composition of cell membranes; 1800–1500 cm−1 corresponding to the characteristic peaks generated by the amide I and amide II bands of proteins and peptides; 1500–1200 cm−1 corresponding to the mixed region, which contains stretching vibrations of proteins, fatty acids, and phosphate compounds; the characteristic peak of 1200–900 cm−1 mainly reflecting the composition of carbohydrates in the cell wall.28,29 The peaks of PME-30 in all characteristic regions were more tortuous than PME-0, indicating that PME altered the composition of fatty acids, proteins, and carbohydrates in the bacterial cells, thereby changing the surface properties of the bacterial cells. The CK-PME-0 showed significant changes in the characteristic peaks at 1800–1500 cm−1, 1100–1200 cm−1, and 1100–900 cm−1 after acid and oxidative stress, indicating that the damage caused by acid and oxidative stress to the bacterial cells was mainly on the surface proteins and polysaccharides. Compared with acid and oxidative stress, there were significant differences in the characteristic peaks of CK-PME-30 at 1800–1500 cm−1 and 1500–1200 cm−1, while there was no significant difference at 1200–900 cm−1. This indicates that PME mainly improves the resistance of bacterial cells to acid and oxidative stress by changing the composition of polysaccharides. In the wavelength range of 1200–900 cm−1, there are significant differences between PME-30 and PME-0. However, after acid stress and oxidative stress, there is no significant difference in the PME-30 spectrum, while the PME-0 spectrum showed significant changes. This further indicated that PME pre-stress mainly improved the stability of bacterial polysaccharides under acid and oxidative stress, thereby enhancing the resistance of bacterial cells to acid and oxidative stress.
In order to further investigate the effect of PME pre-stress on bacterial activity, the enzyme activity of β-galactosidase and L-LDH of L. plantarum JB1 was measured under acid and oxidative stress after PME pre-stress (Fig. 5). β-Galactosidase is a key enzyme for lactic acid bacteria to utilize carbon sources, and its activity reflects the physiological activity and fermentation capacity of the bacterial strain.30Fig. 5 shows the changes in enzyme activity of β-galactosidase of L. plantarum JB1 under acid and oxidative stress after PME pre-stress. After acid stress, the enzyme activity of PME-0 decreased by 0.73 U, while the enzyme activity of PME-30 bacteria increased by 0.51 U. After oxidative stress, the enzyme activity significantly decreased (p < 0.05), and the enzyme activity of PME-0 and PME-30 decreased by 1.32 U and 0.88 U, respectively. After stress treatment, the enzyme activity of PME-30 was significantly higher than that of PME-0 (p < 0.05). In the CK group (without stress), the enzyme activity of PME-30 was significantly higher than that of PME-0 (p < 0.05). The above results indicate that PME pre-stress could directly increase the enzyme activity of β-galactosidase of L. plantarum JB1 and indirectly improve the physiological activity of L. plantarum JB1 to ensure its activity under acid and oxidative stress and improve its resistance.
L-LDH is a key enzyme in the glycolysis pathway that catalyzes the conversion of lactate to pyruvate. The activity of this enzyme directly affects the metabolic capacity of bacterial strains, with higher L-LDH activity leading to stronger metabolic capacity.31 As shown in Fig. 5, enzyme activity significantly decreased under acid and oxidative stress (p < 0.05). The enzyme activity of PME-0 decreased by 2.26 U and 5.01 U, while that of PME-30 decreased by 1.38 U and 2.25 U, respectively. This indicates that both acid and oxidative stress have adverse effects on enzyme activity, with oxidative stress having a greater adverse effect on enzyme activity. The enzyme activity of PME-30 in the CK group (without stress) and after acid and oxidative stress was significantly higher than that of PME-0 (p < 0.05), indicating that PME pre-stress could improve the enzyme activity of L-LDH of L. plantarum JB1 and maintain it in adverse environments, thereby enhancing their resistance to acid and oxidative stress.
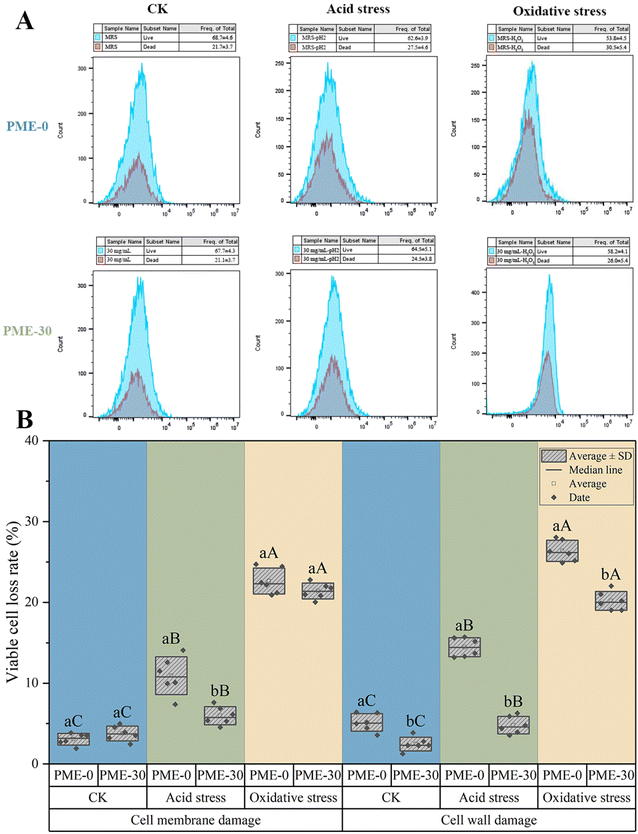
In order to investigate the damage of L. plantarum JB1 under acid and oxidative stress after PME pre-stress, the integrity of the cell membrane of L. plantarum JB1 was detected using a fluorescent probe. SYTO-9 can penetrate bacteria with intact cell membranes and stain them green, while PI can only penetrate bacteria with damaged cell membranes and stain them red. The integrity of the cell membrane of L. plantarum JB1 can be determined by using SYTO-9 and PI. As shown in Fig. 6A, the bacterial red fluorescence intensity of PME-0 and PME-30 in the CK group (without stress) was the lowest, accounting for 21.7% and 21.1%, respectively, indicating that PME pre-stress did not have a significant effect on the cell membrane integrity of L. plantarum JB1. After 3 h acid stress, the proportion of red fluorescent labeled bacteria in PME-0 and PME-30 was 27.5% and 24.5%, respectively. After 3 h oxidative stress, the proportion of red fluorescent labeled dead bacteria in PME-0 and PME-30 was 30.5% and 26.0%, respectively. Compared with all groups of PME-0, the increase in red fluorescence of PME-30 bacteria was alleviated, and the damage to the cell membrane was significantly reduced. The bacteria, after PME pre-stress, did indeed alleviate the cell membrane damage caused by acid and oxidative stress, which is one of the reasons why the bacterial cells improved their resistance to acid and oxidative stress.
When the cell membrane is damaged, the permeability of the cell membrane increases, and Na+ and Cl− will enter the interior of the cell through the cell membrane, causing an increase in osmotic pressure inside the bacterial body, thereby inhibiting bacterial growth and reproduction, ultimately leading to cell death. The higher the rate of bacterial loss, the more severe the damage to the cell membrane.32Fig. 6B shows the changes in bacterial loss rate of L. plantarum JB1 on NaCl-MRS solid medium after PME pre-stress under acid and oxidative stress. The loss rate of bacteria after acid and oxidative stress significantly increased (p < 0.05), and the loss rate of PME-0 bacteria increased to 10.93% and 22.64%, respectively; the bacterial loss rates of PME-30 increased to 5.97% and 20.41%, respectively. The experimental results indicated that both acid and oxidative stress caused damage to bacterial cell membranes, with oxidative stress causing greater damage. There was no significant difference (p < 0.05) in the bacterial loss rate of PME-0 and PME-30 between the CK group (without stress) and the oxidative stress group, indicating that PME pre-stress cannot protect the cell membrane. After acid stress, the bacterial loss rate of PME-30 was significantly lower than the PME-0 (p < 0.05), indicating that PME pre-stress could prevent sodium and chloride ions from entering the interior of the bacterial cells, thereby better maintaining the integrity of the cell membrane and protecting the bacterial cells, maintaining their activity under acid stress.
Strains with damaged cell walls exhibit increased sensitivity to lysozyme. Lysozyme at sublethal concentrations can inhibit the synthesis of peptidoglycans, thereby preventing cell wall repair and ultimately leading to cell death. Similar to the bacterial loss rate of the cell membrane, the higher the bacterial loss rate, the more severe the cell wall damage.33Fig. 6B shows the changes in the bacterial loss rate of L. plantarum JB1 on lysozyme-MRS solid medium after PME pre-stress under acid and oxidative stress. The bacterial loss rate significantly increased after acid and oxidative stress (p < 0.05), and the bacterial loss rate of PME-0 increased to 14.45% and 26.37%, respectively; the bacterial loss rate of PME-30 increased to 4.80% and 20.19%, respectively. The experimental results indicated that both acid and oxidative stress caused damage to bacterial cell walls, with oxidative stress causing greater damage.34 The bacterial loss rate of PME-30 in the CK group (without stress), acid stress group and oxidative stress group was significantly lower than that of PME-0 (p < 0.05). PME pre-stress could improve the integrity of the bacterial cell wall. Biofilm is an important protective barrier for lactobacilli,26 so PME pre-stress may protect bacteria and reduce cell wall damage by increasing the synthesis of biofilm (Fig. 2).
4. Conclusions
In order to meet the sustainability challenges of energy intensive processing and loss of probiotic activity in the food fermentation system, this study used Prunus mume extract (PME), a renewable agricultural by-product, to explore its role in improving the stress resistance of L. plantarum JB1. The results showed that PME pretreatment promoted biofilm formation by activating AI-2 signaling, enhanced the activities of β-galactosidase and L-LDH to maintain metabolic homeostasis under stress, and reduced acid and oxidative stress damage by regulating cell surface polysaccharides, which significantly increased the survival rate of the strain under acid stress by 21.08%.This work distinguishes itself from existing probiotic stabilisation strategies by moving beyond mere outcome-based protection to a mechanism-driven, pre-adaptation approach. Unlike energy-intensive physical methods or the addition of synthetic chemicals, our strategy leverages a natural, renewable by-product to intrinsically enhance the probiotic's own stress resistance through physiological pathways. This mechanism reduces the dependence on synthetic preservatives and high energy consumption stabilization technologies (such as deep freezing or frequent pH adjustment). This achieves stabilisation in a way that is both clean-label and energy-efficient.
The study further found that the improvement effect of PME on acid stress was better than that of oxidative stress, suggesting that PME has more application potential in low pH fermented foods (such as pickles and yogurt). It is important to acknowledge that this study was conducted in a laboratory model system. Future research should focus on validating these protective effects in real food fermentation and under storage conditions, and exploring the efficacy of PME across a broader spectrum of probiotic species to assess its universal applicability. Furthermore, the economic feasibility of large-scale PME extraction and integration into industrial processes warrants thorough investigation.
This study provides a new strategy for the development of clean and low-energy sustainable food production technology and promotes the coordinated development of high-value utilization of agricultural by-products and green food manufacturing.
Author contributions
Teng Long Miao: data curation, investigation, formal analysis, writing – original draft; Chuang Zhang: investigation, writing – review & editing; Hui Wang: methodology, validation; Xiaohong Chen: resources, writing – review & editing; Hong Mei Xiao and Qiu Qin Zhang: funding acquisition, project administration, resources, supervision, writing – review & editing.Conflicts of interest
The work described has not been published previously, and it is not under consideration for publication elsewhere. The authors declare that they have no conflict of interests.Data availability
The manuscript contains all data necessary to support the findings, statements, and conclusions. Data will be made available on request.Supplementary Information (SI) is available. See DOI: https://doi.org/10.1039/d5fb00241a.
Acknowledgements
This research was supported by Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF), Grant No: CX (22)3189; National Natural Science Foundation of China Grant No: 32272350 and 32060547; the Project of Sanya Yazhou Bay Science and Technology City, Grant No: SCKJ-JYRC-2022-69.References
- Q. Li, H. Y. Lin, J. Li, L. Liu, J. L. Huang, Y. Cao, T. T. Zhao, D. J. McClements, J. Chen, C. M. Liu, J. Y. Liu, P. Y. Shen and M. Z. Zhou, Improving probiotic (Lactobacillus casei) viability by encapsulation in alginate-based microgels: Impact of polymeric and colloidal fillers, Food Hydrocolloids, 2023, 134, 108028 CrossRef CAS.
- F. L. Dong, S. Liu, D. H. Zhang, J. C. Zhang, X. L. Wang and H. Zhao, Osmotic pressure induced by extracellular matrix drives Bacillus subtilis biofilms' self-healing, Comput. Biol. Chem., 2022, 97, 107632 CrossRef CAS PubMed.
- R. M. Donlan and J. W. Costerton, Biofilms: survival mechanisms of clinically relevant microorganisms, Clin. Microbiol. Rev., 2002, 15(2), 167–193 CrossRef CAS PubMed.
- G. Frakolaki, V. Giannou, D. Kekos and C. Tzia, A review of the microencapsulation techniques for the incorporation of probiotic bacteria in functional foods, Crit. Rev. Food Sci. Nutr., 2021, 61(9), 1515–1536 CrossRef CAS PubMed.
- C. Bailly, Anticancer properties of Prunus mume extracts (Chinese plum, Japanese apricot), J. Ethnopharmacol., 2020, 246, 112215 CrossRef CAS PubMed.
- S. Bu, C. Yuan, F. Cao, Q. Xu, Y. Zhang, R. Ju, L. Chen and Z. Li, Concentrated extract of Prunus mume fruit exerts dual effects in 3T3-L1 adipocytes by inhibiting adipogenesis and inducing beiging/browning, Food Nutr. Res., 2021, 65, 5492 CAS.
- D. Liang and Z. J. Liu, Research on inhibition rate of plum essence to the proliferation of breast cancer cells MCF-7 and liver cancer cells HepG2, Hans J. Food Nutr. Sci., 2019, 8(1), 75–79 CrossRef.
- S. Qin, X. M. Zeng, M. Jiang, X. Rui, W. Li, M. S. Dong, X. H. Chen and Q. Q. Zhang, Genomic and biogenic amine-reducing characterization of Lactiplantibacillus plantarum JB1 isolated from fermented dry sausage, Food Control, 2023, 154, 109971 CrossRef CAS.
- B. L. Bassler, E. P. Greenberg and A. M. Stevens, Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi, J. Bacteriol., 1997, 179(12), 4043–4045 CrossRef CAS.
- D. L. Xu, X. G. Zhao, G. C. Mahsa, K. Ma, C. L. Zhang, X. Rui, M. S. Dong and W. Li, Controlled release of Lactiplantibacillus plantarum by colon-targeted adhesive pectin microspheres: Effects of pectin methyl esterification degrees, Carbohydr. Polym., 2023, 313, 120874 CrossRef CAS PubMed.
- H. Nouraei, S. Zare, M. Nemati, N. Amirzadeh, M. Motamedi, S. Shabanzadeh, K. Zomorodian and K. Pakshir, Comparative analysis of enzymatic profiles and biofilm formation in clinical and environmental Candida kefyr isolates, Environ. Microbiol. Rep., 2024, 16(3), e13282 CrossRef CAS.
- Y. Gu, B. J. Zhang, J. J. Tian, L. J. Li and Y. F. He, Physiology, quorum sensing, and proteomics of lactic acid bacteria were affected by Saccharomyces cerevisiae YE4, Food Res. Int., 2023, 166, 112612 CrossRef CAS.
- G. Krausova, I. Hyrslova and I. Hynstova, In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains, Fermentation, 2019, 5(4), 100 CrossRef CAS.
- L. Y. Xiao, Y. Yang, S. Han, X. Rui, K. Ma, C. L. Zhang, G. X. Wang and W. Li, Effects of genes required for exopolysaccharides biosynthesis in Lacticaseibacillus paracasei S-NB on cell surface characteristics and probiotic properties, Int. J. Biol. Macromol., 2023, 224, 292–305 CrossRef CAS PubMed.
- C. Papaneophytou, M. E. Zervou and A. Theofanous, Optimization of a colorimetric assay to determine lactate dehydrogenase b activity using design of experiments, SLAS Discovery, 2021, 26(3), 383–399 CrossRef CAS PubMed.
- J. J. Tian, X. M. Wang, X. Zhang, C. P. Zhang, X. H. Chen, M. S. Dong, X. Rui, Q. Q. Zhang, Y. Fang and W. Li, Isolation, structural characterization and neuroprotective activity of exopolysaccharide from Paecilomyces cicadae TJJ1213, Int. J. Biol. Macromol., 2021, 183, 1034–1046 CrossRef CAS.
- L. L. Zhao, J. Wang, X. W. Sheng, S. R. Li, W. J. Yan, J. Qian, J. H. Zhang and V. Raghavan, Non-thermal plasma inhibited the growth and aflatoxins production of Aspergillus flavus, degraded aflatoxin B1 and its potential mechanisms, Chem. Eng. J., 2023, 475, 146017 CrossRef CAS.
- M. A. Golowczyc, J. Silva, P. Teixeira, G. L. De Antoni and A. G. Abraham, Cellular injuries of spray-dried Lactobacillus spp. isolated from kefir and their impact on probiotic properties, Int. J. Food Microbiol., 2011, 144(3), 556–560 CrossRef.
- P. Gong, J. Sun, K. Lin, W. Di, L. Zhang and X. Han, Changes process in the cellular structures and constituents of Lactobacillus bulgaricus sp1.1 during spray drying, LWT–Food Sci. Technol., 2019, 102, 30–36 CrossRef CAS.
- P. Zhao, C. Liu, S. Qiu, K. Chen, Y. Wang, C. Hou, R. Huang and J. Li, Flavor profile evaluation of soaked Prunus mume wine with different base liquor treatments using principal component analysis and heatmap analysis, Foods, 2023, 12(10), 2016 CrossRef CAS.
- Y. Gu, J. J. Tian, Y. Zhang, R. Wu, L. J. Li, B. J. Zhang and Y. F. He, Dissecting signal molecule AI-2 mediated biofilm formation and environmental tolerance in Lactobacillus plantarum, J. Biosci. Bioeng., 2021, 131(2), 153–160 CrossRef CAS PubMed.
- Y. Gu, B. Li, J. J. Tian, R. Wu and Y. F. He, The response of LuxS/AI-2 quorum sensing in Lactobacillus fermentum 2-1 to changes in environmental growth conditions, Ann. Microbiol., 2018, 68, 287–294 CrossRef CAS.
- M. C. Collado, J. Meriluoto and S. Salminen, Measurement of aggregation properties between probiotics and pathogens: In vitro evaluation of different methods, J. Microbiol. Methods, 2007, 71(1), 71–74 CrossRef CAS PubMed.
- A. Nivoliez, P. Veisseire, E. Alaterre, C. Dausset, F. Baptiste, O. Camarès, M. Paquet-Gachinat, M. Bonnet, C. Forestier and S. Bornes, Influence of manufacturing processes on cell surface properties of probiotic strain Lactobacillus rhamnosus Lcr35®, Appl. Microbiol. Biotechnol., 2015, 99(1), 399–411 CrossRef CAS PubMed.
- H. Xu, H. S. Jeong, H. Y. Lee and J. Ahn, Assessment of cell surface properties and adhesion potential of selected probiotic strains, Lett. Appl. Microbiol., 2009, 49(4), 434–442 CrossRef CAS PubMed.
- H. H. Wang, X. J. Wang, L. L. Yu, F. Gao, Y. Jiang and X. L. Xu, Resistance of biofilm formation and formed biofilm of Escherichia coli O157:H7 exposed to acid stress, LWT–Food Sci. Technol., 2020, 118, 108787 CrossRef CAS.
- A. S. Naidu, W. R. Bidlack and R. A. Clemens, Probiotic spectra of lactic acid bacteria (LAB), Crit. Rev. Food Sci. Nutr., 1999, 39(1), 13–126 CrossRef CAS PubMed.
- C. J. Boonaert and P. G. Rouxhet, Surface of lactic acid bacteria: relationships between chemical composition and physicochemical properties, Appl. Environ. Microbiol., 2000, 66(6), 2548–2554 CrossRef CAS.
- B. Dziuba, A. Babuchowski, D. Nałęcz and M. Niklewicz, Identification of lactic acid bacteria using FTIR spectroscopy and cluster analysis, Int. Dairy J., 2007, 17(3), 183–189 CrossRef CAS.
- Z. S. Xu, C. Li, Y. X. Ye, T. Wang, S. S. Zhang and X. L. Liu, The β-galactosidase LacLM plays the major role in lactose utilization of Lactiplantibacillus plantarum, LWT–Food Sci. Technol., 2022, 153, 112481 CrossRef CAS.
- I. Hironaka, T. Iwase, S. Sugimoto, K. Okuda, A. Tajima, K. Yanaga and Y. Mizunoe, Glucose triggers ATP secretion from bacteria in a growth-phase-dependent manner, Appl. Environ. Microbiol., 2013, 79(7), 2328–2335 CrossRef CAS PubMed.
- Y. Yang, R. X. Wang, Y. X. Yang, J. J. E and J. G. Wang, Effects of different prefreezing temperatures on the freeze-drying survival rate and stability during room temperature storage of Lactiplantibacillus plantarum LIP-1, Food Biosci., 2022, 50, 102087 CrossRef CAS.
- M. M. Hlaing, B. R. Wood, D. McNaughton, D. Ying, G. Dumsday and M. A. Augustin, Effect of drying methods on protein and DNA conformation changes in Lactobacillus rhamnosus GG cells by fourier transform infrared spectroscopy, J. Agric. Food Chem., 2017, 65(8), 1724–1731 CrossRef CAS PubMed.
- M. José Martín, F. Lara-Villoslada, M. Adolfina Ruiz and M. Encarnación Morales, Microencapsulation of bacteria: A review of different technologies and their impact on the probiotic effects, Innovative Food Sci. Emerging Technol., 2015, 27, 15–25 CrossRef.
| This journal is © The Royal Society of Chemistry 2026 |