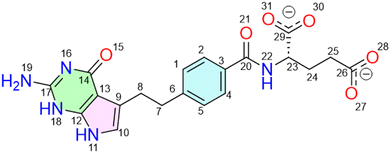
Molecular insight into pemetrexed as a partial agonist of PPARγ through molecular dynamics simulations
Yifan
She
a,
Jiasheng
Zhao
a,
Shunlin
Ren
b,
Lei
Zhang
a,
Shengli
Zhang
a and
Zhiwei
Yang
 *a
*a
aMOE Key Laboratory for Nonequilibrium Synthesis and Modulation of Condensed Matter, School of Physics, Xi’an Jiaotong University, Xi’an 710049, China. E-mail: yzws-123@xjtu.edu.cn
bDepartment of Medicine, Veterans Affairs McGuire Medical Center/Department of Medicine, Virginia Commonwealth University, Richmond, Virginia, USA
First published on 25th November 2025
Abstract
Pemetrexed (PMX), a first-line chemotherapeutic for non-small cell lung cancer (NSCLC), has recently been identified as a ligand of peroxisome proliferator-activated receptor gamma (PPARγ). However, the structural and dynamic basis of this interaction remains unclear. In this study, docking was combined with microsecond-scale molecular dynamics (MD) simulations to characterize PMX binding to the PPARγ ligand-binding domain (LBD). PMX was observed to adopt a binding pose resembling known partial agonists, stabilized by hydrogen bonds with residues Ser289, Tyr327, and Tyr473. Energetic and conformational analyses revealed that PMX avoids deep engagement with the AF-2 surface, which is a region critical for coactivator recruitment and full transcriptional activation. Free energy landscapes, principal component analysis, and dynamic cross-correlation maps further demonstrated that PMX induces conformational dynamics consistent with a partial agonist profile. This study provides an atomistic perspective on the recognition mechanism of PMX as a PPARγ partial agonist, offering a structural foundation for designing multitarget agents that simultaneously disrupt nucleotide metabolism and transcriptional regulation in NSCLC.
1. Introduction
Lung cancer persists as the leading cause of cancer-related mortality globally, with non-small cell lung cancer (NSCLC) representing approximately 85% of cases.1 Although targeted therapies and immunotherapies have advanced treatment options, conventional chemotherapy remains essential for inoperable advanced disease.2 Pemetrexed (PMX, Scheme 1), a multi-targeted antifolate agent, is a first-line and maintenance therapy for non-squamous NSCLC.3 Its primary mechanism involves inhibiting key folate-dependent enzymes—thymidylate synthase (TS), dihydrofolate reductase (DHFR), and glycinamide ribonucleotide formyltransferase (GARFT)—thereby disrupting de novo synthesis of purines and pyrimidines required for the DNA/RNA replication.4,5Beyond this established antiproliferative effect, emerging evidence indicated that the clinical efficacy of PMX may involve additional cellular pathways. A recent study by Hang et al. demonstrated a significant correlation between PPARγ expression and prolonged progression-free survival in PMX-treated NSCLC patients.6 PPARγ, a ligand-activated nuclear receptor, regulates adipogenesis, lipid metabolism, and insulin sensitization.7 Its agonists, such as thiazolidinediones (TZDs), exhibit antiproliferative and pro-apoptotic effects in various cancers, including NSCLC, highlighting PPARγ as a potential therapeutic target.8,9 PPARγ activation downregulates hypoxanthine-guanine phosphoribosyl transferase (HGPRT), a key enzyme in the nucleotide salvage pathway.6 This creates a synergistic “dual blockade” of nucleotide biosynthesis when combined with the inhibition of de novo synthesis, thereby enhancing cancer cell sensitivity to PMX.6 Furthermore, experimental data suggest that PMX may act as a partial agonist of PPARγ, capable of suppressing the NF-κB signaling pathway and inhibiting tumor growth in vivo.6
The Protein Data Bank (PDB) contains multiple crystal structures of the PPARγ ligand-binding domain (PPARγ-LBD), including complexes with agonists and apo forms.10,11 PPARγ-LBD consists of 270 amino acids and features dimerization motifs and a ligand-dependent activation function (AF-2 surface), located between Helix 3 (H3) and Helices 7 and 10 (H7 and H10) at the C-terminus (Fig. S1A).12,13 Agonist binding induces conformational changes in the AF-2 region, promoting corepressor displacement and coactivator recruitment.12,13 Full and partial agonists adopt distinct binding modes within the PPARγ-LBD. Full agonists such as rosiglitazone (RSG) are positioned among three α-helices (H3, H7, and H10), with acidic head groups interacting with residues Ser289, His323, His449, and Tyr473.14 Residue Tyr473, part of the AF-2 surface, is critical for the activities of full agonists.15–17 In contrast, partial agonists such as (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid (LRG) bind to a different region of LBD, engaging primarily in hydrophobic interactions with residues Ser342 and Ser289 (Fig. S1B).15–17
However, the atomistic details of the PMX-PPARγ interaction remain elusive. Understanding how the structurally unique PMX binds to and allosterically modulates the receptor is fundamental to deciphering its polypharmacology and to guiding the design of novel synergistic agents. Computational approaches, particularly docking and molecular dynamics (MD) simulations, serve as powerful tools for characterizing protein–ligand interactions at high resolution, thereby complementing experimental findings.18–20 These methods enable the elucidation of binding poses, key residue contributions, and complex stability over time. In this study, an integrated computational strategy was adopted to investigate PMX binding to the PPARγ-LBD. By comparing its interactions with two receptor conformational states, one stabilized by the full agonist rosiglitazone and the other by a partial agonist, (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, we deciphered the energetic and structural basis of PMX recognition. Subsequent microsecond-scale MD simulations, binding free energy calculations, and dynamic analyses collectively characterized the stability, conformational landscape, and functional implications of the PMX-PPARγ complexes. Our study provides an atomistic view of PMX as a PPARγ partial agonist, bridging cellular efficacy with molecular recognition and offering a theoretical foundation for designing dual-pathway inhibitors against NSCLC.
2. Methodology
2.1. System preparation
The coordinates of the PPARγ ligand-binding domain (LBD) in complex with a full agonist (rosiglitazone, RSG) and a coactivator peptide (PDB ID: 1FM621), as well as with a partial agonist ((2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, LRG) (PDB ID: 3B3K22) were respectively retrieved from the Protein Data Bank (Fig. S1). All the hetero-atoms of non-protein parts were removed, and missing hydrogen atoms were added at the physiological pH.23–25 The obtained two structures are referred to as “Full” and “Partial” throughout this study. Energy minimization was performed using a 1000-step steepest descent algorithm, followed by conjugate gradient minimization until convergence to 0.01 kcal mol−1 Å−1. The energy-minimized systems were further refined by 1000 ns molecular dynamics (MD) simulations, using AMBER1826 and ff14SB Force field.27 The setup for MD simulations is provided in Section 2.2. The geometries and partial atomic charges of pemetrexed (PMX, Scheme 1) were handled by the “Minimize Ligands” tools, with a convergence criterion of 0.001 kcal mol−1 Å−1.232.2. Docking and MD simulations
Based on our previously established ensemble-based docking method that accounts for the ligand-induced conformational plasticity of PPARγ,24,25,28 docking was performed using a 10.0 Å radius binding site sphere centered on the location of either the full agonist RSG or the partial agonist LRG, as shown in Fig. S1. The optimal binding orientations of PMX within both “Full” and “Partial” were explored using a combination of random rotations and simulated annealing, based on interaction energies and geometric complementarity.29,30 For details of docking refer to previous studies.25,31Each docked system was subsequently refined through explicit-solvent MD simulations using AMBER18,26 with the ff14SB27 and GAFF32 force fields. Each complex was solvated in a periodic box of TIP3P water molecules, ensuring a minimum clearance of 10.0 Å from any solute atom.33 Energy minimization was carried out in a stepwise manner: first, 500 steps of steepest descent followed by 500 steps of conjugate gradient minimization, with a restraint of 20 kcal mol−1 Å−2 applied to the protein and ligand; next, the same number of steps with a reduced restraint of 10 kcal mol−1 Å−2 applied to the protein backbone and ligand atoms; and finally, an unrestrained minimization using 1000 steps of steepest descent and 4000 steps of conjugate gradient minimization. The system was gradually heated from 0 to 310 K over 0.1 ns in the NVT ensemble with a Langevin thermostat,34 and subsequently equilibrated in the NPT ensemble (T = 310 K, P = 1 bar). Production MD simulations were conducted for 1000 ns under NPT conditions using a 2.0 fs time step, with coordinates recorded every 10.0 ps. Note that the apo systems (Full and Partial) were subjected to 1000 ns MD simulations under the same conditions as their PMX-bound counterparts (PMX-Full and PMX-Partial complexes), thereby enabling the conformational comparison.
2.3. Binding free energy calculation
The binding free energies (ΔGbind) for protein–ligand interactions were calculated using the molecular mechanics generalized Born surface area (MM/GBSA) approach in AmberTools18,26 which has been validated for predicting binding affinities.19 Briefly, ΔGbind was computed as:| ΔGbind = Gcomplex − Gprotein − Gligand | (1) |
The MM-GBSA decomposition method was used to evaluate the contribution of each residue to the binding energy, which includes van der Waals (ΔEvdw), electrostatic (ΔEele), and solvation (ΔGsolvation) terms:
| ΔGresidue = ΔEMM + ΔGGB + ΔGsurf − TΔS | (2) |
2.4. Structural analysis
The root-mean-square deviation (RMSD) of backbone atoms and the root-mean-square fluctuation (RMSF) of residues were analyzed using the cpptraj module in AmberTools18.26 Principal component analysis (PCA) was performed on Cα atoms to examine conformational changes upon ligand binding.35 Dynamic cross-correlation matrices (DCCM) were calculated to assess correlated motions between protein atoms.36 Structural visualization was performed with Discovery studio client23 and UCSF Chimera.373. Results and discussion
3.1. PMX binds PPARγ-LBD in a partial agonist-like pose
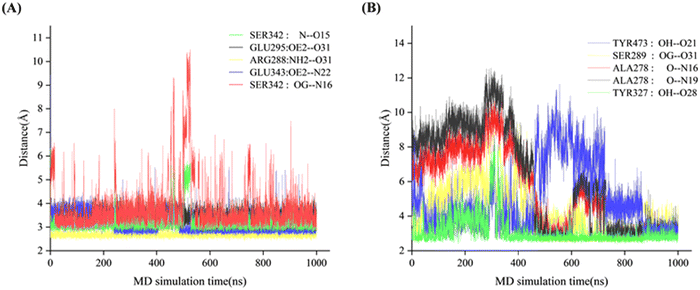
Analysis of backbone RMSD (Fig. S2) revealed greater conformational fluctuations in the apo systems (Full and Partial) than in the PMX-bound systems (PMX-Full and PMX-Partial complexes), indicating the trend of ligand-induced stabilization. Notably, the conformation of Full demonstrated higher stability than the conformation of Partial in both apo and PMX-bound states. The PMX-Partial complex exhibited a longer equilibration time and a significantly higher average backbone RMSD (3.28 ± 0.58 Å) compared to PMX-Full (2.31 ± 0.31 Å), suggesting more stable binding in the Full conformation.38 The geometric and energetic analyses are made on the average structures of 800–1001 ns MD trajectories.In the PMX-Full complex, PMX occupied the PPARγ-LBD binding pocket similarly to the full agonist rosiglitazone (Fig. 1A). Its carboxylate group (O31) formed hydrogen bonds with residues Arg288 and Glu295, with bond lengths of 3.37 Å and 2.45 Å, respectively. The O15 and N16 atoms in the nitrogenous heterocycle of PMX formed the stable hydrogen bonds with residue Ser342 (Helix 4/5), with bond lengths of 2.84 Å and 2.87 Å, respectively. Additional hydrogen bonds involved N22 and residue Glu343 (2.75 Å). The carbon skeleton of PMX occupies the portion of the large two-lobe entry of the pocket, where residue Cys285 of Helix 3, residues Ile326, Tyr327, and Leu333 from Helix 4/5 and residues Ile341, Met348 and Met364 are in the hydrophobic contacts. Residues Ser342 and Glu343 are key amino acids located within the active site responsible for the partial activation of PPARγ.15–17,39,40 Short, stable inter-residue distances (3–4 Å) were consistent with strong electrostatic contributions to binding (Table 1).
| Complexes | ΔEele | ΔEvdw | ΔGsurf | ΔGGB | ΔGbind |
|---|---|---|---|---|---|
| All values are given in kcal mol−1, and the values behind “±” are their standard deviations (SD). | |||||
| PMX-Full | −187.87 ± 23.11 | −59.43 ± 9.34 | −7.85 ± 1.14 | 192.57 ± 23.11 | −62.59 ± 9.84 |
| PMX-Partial | −26.43 ± 17.94 | −42.99± 28.24 | −5.36 ± 3.51 | 38.99 ± 25.87 | −35.79 ± 23.66 |
In the PMX-Partial complex, PMX adopted a binding mode resembling the partial agonist (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid (LRG, Fig. 1B). The carbonyl group (O31) formed a hydrogen bond with residue Ser289 (Helix 3, 2.80 Å). The carboxylate anion (O28) formed a hydrogen bond with residue Tyr327 (2.91 Å). The nitrogen heterocycle (N16 and N19) formed hydrogen bonds with residue Ala278 (Helix 3, 3.36 Å and 3.20 Å). The O21 atom formed a hydrogen bond with residue Tyr473 (2.75 Å), a key residue for the conformational change of AF-2 region and the PPARγ activation.41 The carbon skeleton engaged in hydrophobic stacking with residues from helices 3, 4/5, 7, 10, and the AF-2 region, including Cys285, Ile326, Tyr327, Leu330, Leu453, and Ile456. Unlike the PMX-Full system, larger distance fluctuations and lower hydrogen-bond stability (Fig. 2B) aligned with weaker electrostatic contributions (Table 1), supporting a partial agonist-like binding profile with higher conformational flexibility (Fig. 1B and 2B).
The stability of key hydrogen bonds was further assessed over the simulation trajectory (Fig. S3). In the PMX-Full complex, interactions with residues Ser342 and Glu343 remained stable, with bond distances fluctuating minimally around 2.8–3.0 Å. In contrast, the PMX-Partial system exhibited greater variability in hydrogen bond distances, particularly with residues Ser289 and Tyr473, where occasional breakage and reformation were observed. This transient nature of hydrogen bonding in the PMX-Partial complex underscores its dynamic binding mode and supports its classification as a partial agonist.
3.2. Binding energetics favor partial agonism
Binding free energy calculations revealed distinct energetic profiles for the two PMX-PPARγ complexes (Table 1). The PMX-Full system exhibited a significantly stronger binding affinity, with a ΔGbind of −62.59 ± 9.84 kcal mol−1, compared to −35.79 ± 23.66 kcal mol−1 for the PMX-Partial system. This difference was largely driven by more favorable electrostatic (ΔEele = −187.87 ± 23.11 kcal mol−1) and van der Waals (ΔEvdw = −59.43 ± 9.34 kcal mol−1) contributions in the PMX-Full system, relative to the PMX-Partial system (ΔEele = −26.43 ± 17.94 kcal mol−1; ΔEvdw = −42.99 ± 28.24 kcal mol−1). These results align with the structural observations in Section 3.1, where the PMX-Full complex displayed more stable and persistent interactions.The free energy landscape (FEL) was constructed using the radius of gyration (Rg) and RMSD as reaction coordinates to characterize the conformational stability and heterogeneity of PPARγ in different states (Fig. 3). The free energy for each state was estimated as G = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(p/max(p)), where R is the gas constant, T is the temperature, p is the state probability, and max(p) is the maximum probability. The Full system exhibited a broad, shallow energy basin, indicating conformational flexibility and multiple metastable states. In contrast, the Partial system displayed a more rugged landscape with several localized minima, reflecting higher structural variability. Upon PMX binding, both PMX-Full and PMX-Partial systems showed more confined and deeper energy basins, suggesting ligand-induced stabilization. Notably, the PMX-Full complex adopted a single, well-defined low-energy state, consistent with its higher structural rigidity and stronger binding affinity. The PMX-Partial system, while more compact than its apo form, retained a broader distribution of conformations, aligning with its partial agonist-like dynamic profile and weaker electrostatic contributions.
ln(p/max(p)), where R is the gas constant, T is the temperature, p is the state probability, and max(p) is the maximum probability. The Full system exhibited a broad, shallow energy basin, indicating conformational flexibility and multiple metastable states. In contrast, the Partial system displayed a more rugged landscape with several localized minima, reflecting higher structural variability. Upon PMX binding, both PMX-Full and PMX-Partial systems showed more confined and deeper energy basins, suggesting ligand-induced stabilization. Notably, the PMX-Full complex adopted a single, well-defined low-energy state, consistent with its higher structural rigidity and stronger binding affinity. The PMX-Partial system, while more compact than its apo form, retained a broader distribution of conformations, aligning with its partial agonist-like dynamic profile and weaker electrostatic contributions.
3.3. Dynamics reveal distinct conformational landscapes
Principal component analysis (PCA) and root mean square fluctuation (RMSF) were used to characterize the structural dynamics of key PPARγ regions, including the Ω loop, AF-2, and H3 Helix (Fig. 4 and 5). RMSF analysis revealed that while the overall fluctuation profiles of Full and PMX-Full were similar, PMX binding induced distinct local changes (Fig. 4). Notably, in the H2′ helix (residues 252–265), fluctuations increased in PMX-Full, suggesting PMX modulates this region to achieve synergistic movement with AF-2 (residues 467–473) and promote functional coherence. In contrast, PMX-Partial showed markedly reduced fluctuations relative to Partial, indicating enhanced global stability. Both PMX-bound systems suppressed fluctuations in the critical AF-2 region, underscoring its central regulatory role. Furthermore, the PMX-Full system exhibited greater flexibility near the active site than PMX-Partial, including the S245 loop (residues 239–251) and Ω loop (residues 266–276) regions. Despite strong ligand–protein interactions and low global RMSD (Fig. S2), PMX-Full maintained high local mobility, likely anchored by a stable hydrogen-bond network involving residues Glu295 and Ser342. By contrast, the weaker, more transient hydrogen bonds in PMX-Partial led to disorganized residual motions that lacked functional coherence, despite lower RMSF values. | ||
| Fig. 4 The time-evolution backbone-atom root-mean-square fluctuations (RMSF) for the Full (green), Partial (pink), PMX-Full (yellow) and PMX-Partial (blue) systems. | ||
PCA further highlighted differences in conformational sampling between the two systems. The first 20 eigenvectors accounted for 70–85% of backbone motions. Comparison with the apo systems revealed that PMX binding consistently induced a more compact arrangement of α-helices (blue) and increased folding in flexible regions (red), irrespective of the conformational state (Full or Partial) (Fig. 5A–D), indicating ligand-induced stabilization. Furthermore, the conformational ensembles of PMX-bound states were more concentrated in the PC space than those of the apo states (Full and Partial) (Fig. 5E–H), suggesting that PMX binding reduces conformational diversity and enhances structural stability. Notably, in the PMX-Full system, functional domains including H3 (residues 277–300) and AF-2 (residues 467–473) exhibited directed and concerted motions, as evidenced by extended eigenvectors (Fig. 5C), and tightly clustered projections in the PC1–PC2 subspace (Fig. 5G). This reflects a restricted set of dominant conformations and high thermodynamic stability, consistent with the binding energetics (Table 1). In contrast, the PMX-Partial system showed more disordered motions and broader conformational sampling (Fig. 5D and H), indicating greater dynamic heterogeneity and weaker conformational clustering.
Dynamic cross-correlation maps (DCCM) revealed distinct inter-domain coupling patterns among the systems (Fig. 6). Compared to the PMX-bound systems, the apo states exhibited largely independent and disordered motions, with fewer continuous correlated regions and more prominent anti-correlated movements. This PMX-induced dynamic coupling was particularly evident in the AF-2 region of the PMX-Partial system. Direct comparison between the two PMX-bound systems indicated higher overall coordination in PMX-Full than in PMX-Partial. In the PMX-Full complex, the H2 helix (residues 230–238) was anti-correlation with the H2′-Ω loop segment (residues 248–276), which in turn correlated positively with H4/5, β2/β3 sheets (residues 311–349), and H8 (residues 381–392). The AF-2 region served as a conformational mediator, exhibiting anti-correlation with H6 and H7 but positive correlation with H4 and residues 395–404. In the PMX-Partial system, a coordinated movement was observed among the H2 helix, β1-sheet (residues 247–249), and H2′ helix (residues 252–265), which correlated with H4, β2/β3 sheets, and H6, while being anti-correlated with H8. The AF-2 region showed positive correlation with H4/5, H6, H7, and H8, but strong anti-correlation (>60%) with the Ω loop, underscoring its central role under partial agonism.
3.4. Secondary structure stability supports functional differentiation
The α-helical conformation of the AF-2 domain (residues 467–473) is essential for coactivator recruitment in PPARγ.42 Secondary structure analysis using the Dictionary of Secondary Structure of Proteins (DSSP) method revealed that key regions, particularly AF-2, exhibited frequent fluctuations and high conformational disorder in the apo systems (Fig. S4). In contrast, PMX binding markedly enhanced global structural stability and promoted cooperative dynamics between H3/H4 and AF-2, leading to more functionally coherent and ordered motions. Specifically, in the PMX-Full system, the AF-2 domain maintained high α-helical content (>80%) throughout the simulation without significant conversion to random coil, indicating a stable active conformation.43 The Ω loop (residues 266–276) predominantly adopted bend conformations with minimal variation, thereby preserving active pocket integrity. Similarly, the H3 Helix (residues 277–300) maintained a consistent α-helical structure, supporting the overall pocket architecture.In contrast, the PMX-Partial system showed marked instability in the AF-2 domain, with α-helical content below 50% and random coil exceeding 30%. A pronounced helix-to-turn transition occurred around 500 ns, reflecting loss of the active conformation. The Ω loop also displayed frequent secondary structure transitions, further destabilizing the binding pocket. Our simulations indicated that PMX binds PPARγ similarly to known agonists, occupying the LBD in both tested conformations and contributing to receptor stabilization (Fig. 1 and 2). The carbon skeleton of the ligand is enclosed by Helices H3, H7, and H10, and engages in hydrophobic contacts with residues Cys285, Ile326, Tyr327, and Leu330 (Fig. 1).
Despite its greater stability and deeper binding (Table 1), the behavior of the PMX-Full complex aligns with a partial agonist profile. Canonical full agonists stabilize the AF-2 helix via sustained, direct interaction with Tyr473—a key residue for functional differentiation.15,41 In contrast, partial agonists avoid the direct AF-2 engagement, preferentially occupying Branches II and III, with acidic groups forming hydrogen bonds to residue Ser342 and hydrophobic contacts with residues Cys285 and Arg288.41,43 PMX fails to engage Tyr473 robustly or bind deeply within the AF-2 region. Concurrently, the dynamic heterogeneity of the PMX-Partial system destabilizes the AF-2 helix, likely impairing coactivator recruitment—a hallmark of partial agonism. Thus, although the two binding modes differ, both the lack of strong AF-2 engagement in the Full system and the inherent AF-2 instability in the Partial system converge on the same functional outcome: incomplete receptor activation. Based on these criteria, PMX exhibits characteristic features of a partial agonist, leading us to classify it as such for PPARγ.
4. Conclusions
PPARγ, a key regulator of nucleotide metabolism, has emerged as a promising target for lung cancer chemoprevention. Pemetrexed (PMX), a first-line NSCLC therapeutic that inhibits purine and pyrimidine synthesis, was investigated for its potential interaction with PPARγvia integrated docking and molecular dynamics simulations. This study provides, to our knowledge, the first atomistic view of PMX binding to PPARγ.The results showed that PMX adopts a binding mode reminiscent of known PPARγ agonists, positioning its scaffold between H3, H7, and H10 helices and engaging hydrophobic residues (Cys285, Ile326, Tyr327, and Leu330) at the pocket entrance (Fig. 1). Energetic and conformational analyses consistently demonstrated that PMX binds as a partial agonist, driven by the essential roles of its carboxylate groups and hydrophobic core, and characterized by incomplete AF-2 stabilization. Inspired by these findings, future work can focus on: (1) optimizing PMX derivatives to enhance AF-2 interaction and partial agonistic activity; (2) designing dual-target molecules that concurrently engage the PPARγ partial agonist site and folate-dependent enzyme active centers to synergize nucleotide synthesis inhibition with PPARγ activation; and (3) developing selective partial agonists by modulating PPARγ conformational dynamics, such as stabilizing the synergistic H2′-AF-2 motions. These insights may facilitate the rational development of novel chemopreventive agents targeting PPARγ.
Conflicts of interest
The authors declare no competing financial interests.Data availability
Computational instructions and data from this study are provided in the main text and supplementary information (SI), and further information and requests may be directed to and will be fulfilled by Zhiwei Yang. The supplementary information contains molecular dynamics simulation data, comprising depiction of the PPARγ-LBD starting structures, backbone RMSD evolution, key hydrogen-bond distance analyses, and secondary structure evolution profiles. See DOI: https://doi.org/10.1039/d5cp03840e.Software used: BIOVIA Discovery Studio 3.1, https://www.3ds.com/; Amber18, https://ambermd.org/.
Acknowledgements
The authors wish to thank Professor Dr Shengqing Li of Huashan Hospital, Fudan University for the useful suggestions.References
- H. Saji, M. Okada, M. Tsuboi, R. Nakajima, K. Suzuki, K. Aokage, T. Aoki, J. Okami, I. Yoshino and H. Ito, The Lancet, 2022, 399, 1607–1617 CrossRef PubMed.
- L. E. Hendriks, J. Remon, C. Faivre-Finn, M. C. Garassino, J. V. Heymach, K. M. Kerr, D. S. Tan, G. Veronesi and M. Reck, Nat. Rev. Dis. Primers, 2024, 10, 71 CrossRef PubMed.
- L. Paz-Ares and J. Corral, The Lancet, 2014, 383, 1528–1530 CrossRef PubMed.
- A. Ashrafi, Z. Akter, P. Modareszadeh, P. Modareszadeh, E. Berisha, P. S. Alemi, M. D. C. Chacon Castro, A. R. Deese and L. Zhang, Cancers, 2022, 14, 4562 CrossRef PubMed.
- L. Gandhi, D. Rodríguez-Abreu, S. Gadgeel, E. Esteban, E. Felip, F. De Angelis, M. Domine, P. Clingan, M. J. Hochmair and S. F. Powell, N. Engl. J. Med., 2018, 378, 2078–2092 CrossRef PubMed.
- T. Hang, L. Yang, X. Zhang, J. Li, F. Long, N. Zhu, Y. Li, J. Xia, Y. Zhang and P. Zhang, Am. J. Transl. Res., 2021, 13, 2296 Search PubMed.
- R. T. Gampe, Jr., V. G. Montana, M. H. Lambert, A. B. Miller, R. K. Bledsoe, M. V. Milburn, S. A. Kliewer, T. M. Willson and H. E. Xu, Mol. Cell, 2000, 5, 545–555 CrossRef PubMed.
- R. A. Nemenoff, M. Weiser-Evans and R. A. Winn, PPAR Res., 2008, 2008, 156875 CrossRef PubMed.
- F. Ondrey, Clin. Cancer Res., 2009, 15, 2–8 CrossRef PubMed.
- S. H. Sheu, T. Kaya, D. J. Waxman and S. Vajda, Biochemistry, 2005, 44, 1193–1209 CrossRef PubMed.
- N. Mitro, F. Gilardi, M. Giudici, C. Godio, E. Scotti and M. Crestani, Methods Mol. Biol., 2013, 952, 137–144 CrossRef PubMed.
- Y. Li, M. H. Lambert and H. E. Xu, Structure, 2003, 11, 741–746 CrossRef PubMed.
- L. Nagy and J. W. Schwabe, Trends Biochem. Sci., 2004, 29, 317–324 CrossRef PubMed.
- A. Farce, N. Renault and P. Chavatte, Curr. Med. Chem., 2009, 16, 1768–1789 CrossRef PubMed.
- J. S. Shang and D. J. Kojetin, Structure, 2021, 29, 940–950 CrossRef PubMed.
- M. D. Nemetchek, I. M. Chrisman, M. L. Rayl, A. H. Voss and T. S. Hughes, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2215333119 CrossRef PubMed.
- B. S. Mactavish, D. Zhu, J. S. Shang, Q. Z. Shao, Y. J. He, Z. J. Yang, T. M. Kamenecka and D. J. Kojetin, Nat. Commun., 2025, 16, 2065 CrossRef CAS PubMed.
- M. Cioce, F. Ganci, V. Canu, A. Sacconi, F. Mori, C. Canino, E. Korita, B. Casini, G. Alessandrini, A. Cambria, M. A. Carosi, R. Blandino, V. Panebianco, F. Facciolo, P. Visca, S. Volinia, P. Muti, S. Strano, C. M. Croce, H. I. Pass and G. Blandino, Oncogene, 2014, 33, 5319–5331 CrossRef CAS.
- E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Z. H. Zhang and T. Hou, Chem. Rev., 2019, 119, 9478–9508 CrossRef CAS PubMed.
- Z. W. Yang, Z. C. Zhang, Y. Z. Zhao, Q. S. Ye, X. H. Li, L. J. Meng, J. A. Long, S. L. Zhang and L. Zhang, Front. Pharmacol., 2022, 13, 935898 CrossRef CAS PubMed.
- R. T. Gampe, Jr., V. G. Montana, M. H. Lambert, A. B. Miller, R. K. Bledsoe, M. V. Milburn, S. A. Kliewer, T. M. Willson and H. E. Xu, Mol. Cell, 2000, 5, 545–555 CrossRef PubMed.
- R. Montanari, F. Saccoccia, E. Scotti, M. Crestani, C. Godio, F. Gilardi, F. Loiodice, G. Fracchiolla, A. Laghezza, P. Tortorella, A. Lavecchia, E. Novellino, F. Mazza, M. Aschi and G. Pochetti, J. Med. Chem., 2008, 51, 7768–7776 CrossRef CAS PubMed.
- Accelrys, Discovery Studio 3.1, https://accelrys.com.
- Z. Yang, Y. Zhao, D. Hao, H. Wang, S. Li, L. Jia, X. Yuan, L. Zhang, L. Meng and S. Zhang, RSC Adv., 2020, 11, 147–159 RSC.
- J. S. Zhao, Y. Z. Zhao, S. L. Zhang, L. Zhang and Z. W. Yang, Phys. Chem. Chem. Phys., 2024, 26, 28143–28154 RSC.
- I. Y. B.-S. D. A. Case, S. R. Brozell, D. S. Cerutti III, T. E. Cruzeiro, T. A. Darden, R. E. Duke, D. Ghoreishi, M. K. Gilson, H. Gohlke, A. W. Goetz, D. Greene, R. Harris, N. Homeyer, S. Izadi, A. Kovalenko, T. Kurtzman, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, D. J. Mermelstein, K. M. Merz, Y. Miao, G. Monard, C. Nguyen, H. Nguyen, I. Omelyan, A. Onufriev, F. Pan, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. L. Simmerling, J. Smith, R. Salomon-Ferrer, J. Swails, R. C. Walker, J. Wang, H. Wei, R. M. Wolf, X. Wu, L. Xiao, D. M. York and P. A. Kollman, AMBER 2018, University of California, San Francisco, 2018 Search PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- Z. K. Zeng, Z. W. Yang, C. H. Li, S. J. Liu, W. Wei, Y. Zhou, S. M. Wang, M. J. Sui, M. D. Li, S. M. Lin, Y. Y. Cheng and P. Hou, J. Med. Chem., 2024, 67, 19216–19233 CrossRef CAS PubMed.
- G. Kilroy, H. Kirk-Ballard, L. E. Carter and Z. E. Floyd, Endocrinology, 2012, 153, 1206–1218 CrossRef CAS PubMed.
- A. G. Atanasov, J. N. Wang, S. P. Gu, J. Bu, M. P. Kramer, L. Baumgartner, N. Fakhrudin, A. Ladurner, C. Malainer, A. Vuorinen, S. M. Noha, S. Schwaiger, J. M. Rollinger, D. Schuster, H. Stuppner, V. M. Dirsch and E. H. Heiss, Biochim. Biophys. Acta, 2013, 1830, 4813–4819 CrossRef CAS PubMed.
- Z. Yang, X. Fu, Y. Zhao, X. Li, J. Long and L. Zhang, Int. J. Biol. Macromol., 2023, 240, 124352 CrossRef CAS PubMed.
- J. M. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- V. Kräutler, W. F. van Gunsteren and P. H. Hünenberger, J. Comput. Chem., 2001, 22, 501–508 CrossRef.
- B. J. Grant, A. P. Rodrigues, K. M. ElSawy, J. A. McCammon and L. S. Caves, Bioinformatics, 2006, 22, 2695–2696 CrossRef CAS PubMed.
- W. F. Vangunsteren, P. H. Hunenberger, A. E. Mark, P. E. Smith and I. G. Tironi, Comput. Phys. Commun., 1995, 91, 305–319 CrossRef CAS.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- A. Grossfield and D. M. Zuckerman, Annu. Rep. Comput. Chem., 2009, 5, 23–48 CAS.
- J. S. Shang, S. A. Mosure, J. Zheng, R. Brust, J. Bass, A. Nichols, L. A. Solt, P. R. Griffin and D. J. Kojetin, Nat. Commun., 2020, 11, 956 CrossRef CAS PubMed.
- W. S. Deng, X. J. Wang, X. Y. Niu, X. J. Zhang, Y. L. Hou and M. Z. Qin, J. Med. Chem., 2025, 68, 9084–9100 CrossRef CAS PubMed.
- H. Miyachi, Int. J. Mol. Sci., 2023, 24, 3940 CrossRef CAS PubMed.
- J. B. Bruning, M. J. Chalmers, S. Prasad, S. A. Busby, T. M. Kamenecka, Y. He, K. W. Nettles and P. R. Griffin, Structure, 2007, 15, 1258–1271 CrossRef CAS PubMed.
- A. J. Kroker and J. B. Bruning, PPAR Res., 2015, 2015, 816856 CrossRef PubMed.
| This journal is © the Owner Societies 2026 |