Decoding dissociation pathways of ligands in prolyl oligopeptidase
Katarzyna
Walczewska-Szewc
 * and
Jakub
Rydzewski
* and
Jakub
Rydzewski
Institute of Physics, Faculty of Physics, Astronomy and Informatics, Nicolaus Copernicus University, Grudziadzka 5, 87-100 Toruń, Poland. E-mail: kszewc@umk.pl
First published on 4th December 2025
Abstract
Neurodegenerative diseases, such as Alzheimer's and Parkinson's, pose a growing global health burden. Prolyl oligopeptidase (PREP) has emerged as a potential therapeutic target in these diseases. Recent studies have shown that direct interaction between PREP and pathological proteins, such as α-synuclein and Tau, influences protein aggregation and neuronal function. While most known PREP inhibitors primarily target its enzymatic functions, a new class of ligands, known as HUPs, specifically modulate protein–protein interactions (PPIs), which are crucial in the pathology of neurodegenerative diseases. These structurally distinct ligands exhibit diverse binding behaviors, highlighting the importance of understanding their binding pathways. In this study, we analyzed the binding pathways and stability of structurally diverse ligands using molecular dynamics simulations and enhanced sampling techniques. Traditional inhibitors, such as KYP-2047, target the active site between the catalytic domains of PREP and the β-propeller domain, while HUP ligands bind to alternative regions, such as the hinge site, potentially disrupting non-enzymatic PPIs. Using a PLUMED module called maze, we demonstrated that structural variations among ligands lead to distinct binding and unbinding pathways. Free-energy profiles from umbrella sampling revealed key kinetic bottlenecks and differences in pathway selection. For example, HUP-55 exhibits pathway hopping, characterized by diffuse exploration of binding regions before selecting an exit, while KYP-2047 strongly prefers the central tunnel of the β-propeller domain even under perturbations. These results suggest that the dynamic interaction between ligands and PREP plays a critical role in their mechanism of action. The ability of HUPs to interact with multiple binding sites and adapt to PREP's conformational changes may be essential for their PPI-targeting effects. This work highlights the need to consider both binding pathways and ligand dynamics in the design of next-generation ligands for PREP and related targets.
Introduction
Neurodegenerative diseases, such as Alzheimer's disease (AD) and Parkinson's disease (PD), pose significant challenges to our aging society. With the increasing prevalence of these conditions, there is an urgent need to develop effective therapeutic interventions.1,2 Prolyl oligopeptidase (PREP), an enzyme implicated in various neurodegenerative disorders, has emerged as a promising target for this reason.3,4 PREP, initially identified as a serine protease involved in neuropeptide metabolism, has been associated with neurodegenerative diseases such as AD, PD, and others. Previous research has shown that the abnormal activity of PREP is associated with cognitive decline and dementia in animal models, making it an attractive target for therapy.5,6 Currently, the primary focus is on non-enzymatic protein–protein interactions (PPI) related to PREP functions. Recent studies have revealed that direct interaction between PREP, α-synuclein (α Syn), and Tau results in increased aggregation of such proteins.7–12 Anomalous processing and aggregation of α Syn and Tau are considered the main factors in cellular toxicity in AD and PD. Furthermore, PREP interactions with protein phosphatase 2A (PP2A) decrease PP2A activity and autophagy, further advancing these neurodegenerative diseases.13Most known PREP ligands have been designed to inhibit its enzymatic functions, targeting the well-characterized active site located at the interface of the two primary domains of PREP – an α/β-catalytic domain and a β-propeller domain composed of seven β-sheet blades. This active site contains a catalytic triad of S554, H680, and D641. In some cases, inhibitors also modulate PPI-related effects, although this is often an unintended side effect rather than the primary goal.14 Recently developed HUP ligands have been optimized to selectively regulate the nonenzymatic functions of PREP.15 Such ligands bind deeper within the hinge region, connecting the two main domains of PREP.16 However, their broader impact on the protein, particularly the mechanisms by which they modulate PPI properties, remains unclear.
The emergence of new drugs targeting alternative binding sites, in general, raises questions about their ligand-binding properties.17–21 Particularly important in this process are diverse binding pathways, which have recently been shown to influence rate-limiting processes during ligand–protein dissociation.22–24 Recently, identifying binding pathways has been addressed in many studies, underlining its value in studying complex protein–ligand systems.25–38 The differences in the effectiveness of these two structurally distinct groups of ligands may arise from their interaction sites with PREP and how they associate with the protein along multiple binding pathways.39 A recent hypothesis suggests that known ligands enter the cavity between the two subunits via flexible loops referred to as loop A (residues 189–209), loop B (residues 577–608), loop C (residues 636–646), and the His loop (residues 676–685).40,41 The access to the interior of the protein depends on the local conformations of its loops, which may be open or closed at any given time. An earlier idea proposed that ligands and substrates might enter through the β-propeller pore.42,43 Understanding how ligands bind and unbind from the protein could be key to achieving favorable inhibitory/regulatory characteristics, such as low IC50 values.44
Previous studies on PREP ligand binding pathways have focused primarily on peptide-mimicking inhibitors that target the active site. Multiple binding pathways have been explored in one such study, with particular emphasis on the β-propeller domain and the flexible loop interface among loops A, B, C, and His.45 This suggests that the flexible loop pathway is the most probable route, although it would require large-scale domain reorientation.40,45–47 However, the transition of human PREP from the closed to open conformation appears to occur frequently enough to allow the ligand access to the protein.40,48,49 Another approach has also identified two possible ligand entry routes: one via the β-propeller pore and another through the loop region.50
To gain insight into the ligand-binding pathways in PREP, we comprehensively compared various types of compounds. First, we identified their binding sites and assessed their stability within these regions. Next, we demonstrated that slight differences in the structure of ligands and their chemical properties lead to completely different binding pathways in PREP. Lastly, the binding pathways were examined using umbrella sampling (US) to quantify the corresponding free-energy profiles. This approach enabled us to identify the key kinetically limiting steps involved in these binding transitions.
Methods
Models
The structure of human PREP51 (PDB ID: 3DDU) was processed using Maestro within Schrodinger,52 employing the OPLS3e force field53 and PROPKA at pH 7.4. The list of amino acids with non-standard protonation states can be found in SI. We also prepared four mutated systems to investigate the potential effect of mutations on the identified binding pathways and assess the stability of the complexes. The structures of three ligands, KYP-2047, HUP-46, and HUP-55, underwent geometry optimization through quantum chemical calculations using the ORCA 5.0.3 program.54 These calculations employed density functional theory (DFT) with the B3LYP functional, the def2-SVP basis set, and included Grimme's D3 empirical dispersion correction with Becke–Johnson damping (DFT-D3(BJ)). The Resolution-of-Identity (RI) and Chain-of-Spheres (COSX) approximations were used. CHARMM36m parameters55 for ligands were assigned via the Swissparam server.56,57 Flexible docking of the ligands was performed using Glide (Schrodinger58) in the Standard Precision (SP) mode. The docking grid was positioned on the geometric center of the entire binding cavity with an inner box of 10 Å and an outer box of 46 Å. The top-ranked pose for KYP-2047 was selected as it reproduced the binding mode from the PDB 4AN0 structure,46 while the poses for HUP-46 and HUP-55 corresponded to those reported in the literature for analogous compounds.16 These validated poses were subsequently used as starting structures for MD simulations.After selecting the optimal conformation, each protein–ligand complex was immersed in a water box with dimensions of 10 nm in each direction and neutralized by adding Na+ and Cl− ions (0.15 M concentration). These systems comprised approximately 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms. This protocol resulted in the following seven complexes: PREP with the KYP-2047, HUP-46, and HUP-55 ligands; mutated PREPs with HUP-46 (L94 and I690C, T68C and T686C); and mutated PREPs with KYP-2047 (Q397C, Q397C and C255T). Notably, KYP-2047's covalent bond with S554 was omitted in our simulations to isolate non-covalent dissociation pathways. While this simplification excludes the bond's energetic contribution, it allows direct comparison with HUP ligands.
000 atoms. This protocol resulted in the following seven complexes: PREP with the KYP-2047, HUP-46, and HUP-55 ligands; mutated PREPs with HUP-46 (L94 and I690C, T68C and T686C); and mutated PREPs with KYP-2047 (Q397C, Q397C and C255T). Notably, KYP-2047's covalent bond with S554 was omitted in our simulations to isolate non-covalent dissociation pathways. While this simplification excludes the bond's energetic contribution, it allows direct comparison with HUP ligands.
Molecular dynamics simulations
All MD simulations were run using the Gromacs 2021.3 software59 patched with PLUMED 2.8.60–62 The CHARMM36m force field was used in all simulation systems for its accuracy in generating backbone conformational ensembles of proteins.55 Each protein–ligand complex was initially fixed while water and ions underwent equilibration for 0.5 ns. Following this, three MD simulations of 0.5 ns each were conducted in the NVT ensemble at 310 K, applying position restraints of varying magnitudes (1000, 500, and 100 kJ mol−1 nm−2) to the protein Cα atoms and the ligand. Subsequently, a fourth MD simulation with a duration of 1 ns was performed in the NPT ensemble, applying a harmonic restraint of 10 kJ mol−1 nm−2 to the protein Cα atoms and ligand while maintaining a constant pressure of 1 atm using the Berendsen barostat. Finally, an unrestrained equilibration MD simulation lasting 50 ns was conducted in the NPT ensemble, with constant pressure of 1 atm maintained using the Parrinello–Rahman barostat. The further production runs were simulated through 500 ns with the same parameters as the last step of equilibration. The temperature was held constant at 310 K by applying the velocity rescaling thermostat.63 A time step of 2 fs was used. Bonds involving hydrogen atoms were constrained using LINCS.64 We refer to SI for a detailed list of all MD simulations performed in this study (Table S1).To identify binding pathways, we used an adaptive biasing method implemented in the maze module29,65 of PLUMED.60–62 Following our previous protocol,62 for each complex (HUP-46, HUP-55, KYP-2047-PREP, HUP-46-KYP-2047, and the mutated PREPs), we ran 50 MD simulations in the NVT ensemble at 310 K. During these simulations, the ligands were pulled from PREP with a constant velocity of 0.00035 nm ps−1. The pulling direction was determined by minimizing a loss function that described the interactions between the ligands and proteins using simulated annealing. The loss function was 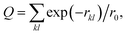 where rkl is the distance between the k-th atom of PREP and the l-th atom of the ligand, with a cutoff r0 set to 0.7 nm. By minimizing Q in the local neighborhood of the ligand, the maze algorithm identified an optimal configuration that corresponded to the minimum of the loss function. The ligand was then pulled in the direction of this identified minimum, resulting in a pathway where the native contacts decreased. This minimization process was relaunched every 10 ns to adjust the pulling direction in response to protein conformational fluctuations. The simulations were terminated either when the ligands dissociated from PREP or when the simulation exceeded 200 ns. For a detailed description of the maze algorithm, we refer to SI.
where rkl is the distance between the k-th atom of PREP and the l-th atom of the ligand, with a cutoff r0 set to 0.7 nm. By minimizing Q in the local neighborhood of the ligand, the maze algorithm identified an optimal configuration that corresponded to the minimum of the loss function. The ligand was then pulled in the direction of this identified minimum, resulting in a pathway where the native contacts decreased. This minimization process was relaunched every 10 ns to adjust the pulling direction in response to protein conformational fluctuations. The simulations were terminated either when the ligands dissociated from PREP or when the simulation exceeded 200 ns. For a detailed description of the maze algorithm, we refer to SI.
We selected representative binding pathways and used them to define collective variables (CVs) for US free-energy simulations.66,67 The pathways were clustered based on their exit configurations using the DBSCAN algorithm68,69 implemented in scikit-learn70 (see SI for details about the employed parameters). For each cluster, we identified the most representative pathway as the one closest to the cluster centroid (see Fig. S4 in SI). These representatives were then visually inspected to ensure they were reasonable compared to other pathways, and a subset of pathways was chosen based on this evaluation. Each binding pathway was divided into frames, with the center of ligand mass traveling approximately 0.2 nm between consecutive frames. Depending on the length of the pathway, this resulted in 18 to 24 frames per CV, defined as the center-of-mass distance between the bound ligand and the dissociating ligand. We ran a separate 10-ns MD simulation for each window, restraining the ligand with a harmonic potential of 500 kJ mol−1 nm−2.
The free-energy profiles were calculated using the weighted histogram analysis method71 implemented in Gromacs. Error bars were determined with bootstrapping. For each case, the free-energy difference between the bound (B) and unbound (U) states was estimated as 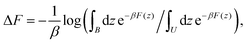 where β = 1/kBT is the inverse temperature with temperature T and Boltzmann's constant kB, and z denotes a CV. The bound state (B) corresponds to the free-energy minimum of the initial ligand position. The unbound state (U) represents the dissociated state obtained from maze simulations, where the loss function reached approximately zero (see SI). This practical definition of (U) captures the complete dissociation from the protein matrix while remaining computationally tractable.
where β = 1/kBT is the inverse temperature with temperature T and Boltzmann's constant kB, and z denotes a CV. The bound state (B) corresponds to the free-energy minimum of the initial ligand position. The unbound state (U) represents the dissociated state obtained from maze simulations, where the loss function reached approximately zero (see SI). This practical definition of (U) captures the complete dissociation from the protein matrix while remaining computationally tractable.
We used the MDAnalysis Python library for MD analysis.72 In the analysis of close-contact frequency, a cut-off distance of 3.5 Å between atoms was applied. Molecular visualizations were created using VMD.73 All the data and PLUMED input files required to reproduce the results reported in this paper are available in a RepOD repository (https://doi.org/10.18150/ZUISNK). The implementation of the maze module for PLUMED is available in a git repository (https://github.com/jakryd/plumed2-maze).
Results
Binding sites
Our results demonstrate different binding site preferences among the selected ligands. The classical peptide-like inhibitor KYP-2047, which affects catalytic functions and PPIs, binds predominantly to the well-known and characterized binding site. Previous studies have shown that KYP-2047 can form a covalent bond with S554 in the active center of the enzyme45 (see Fig. 1b). Although this covalent bond was not included in our model, the ligand maintains significant stability, occupying the same binding site as its covalently bound counterpart.45,48 The positioning of KYP-2047 within the binding cavity is regulated by interactions involving two carbonyl groups with R643 and W595, demonstrating a near 100% contact frequency throughout the eight sets of 500 ns simulations (see Table S1 in SI). The part of the ligand containing the CN functional group exhibits frequent interactions with residues S554, F476, W595, and Y473. Furthermore, the octahydroindole moiety of the ligand frequently interacts with C255, F476, W595, and R643. The aromatic benzene ring engages in a series of hydrophobic interactions with F173, M235, and I591, further enhancing the binding of the ligand.Two additional ligands, belonging to the newly developed HUP group, have been confirmed by recent studies15 through MD simulations and mutagenesis to bind at an alternative site located approximately 1.8 nm away from the active site toward the hinge region. This alternative binding site of PREP appears to be more significant than the primary active site for modulating its PPI-mediated functions. For the 5-aminooxazole-based HUP-55, after docking to a specific binding site similar to that reported by Pätsi et al. (see Fig. 1b and Fig. S1 in SI), the ligand remained stable for nearly 500 ns of MD simulation. The stability of HUP-55 binding is mainly attributed to interactions between the oxazole ring and residues Y471 and R488. Additionally, our results suggest that the CN functional group engages in hydrogen bonding with S485 and N483, while hydrophobic interactions with V455, L499, and F401 help stabilize the benzene ring. However, in three out of five simulations, the ligand diffuses out of the binding site around 500 ns, suggesting that this binding is not as stable as KYP-2047 in the active site.
The 5-aminothiazole-based HUP-46, despite its clear preference for the alternative binding site, resides there for approximately 200 ns before diffusing into the internal cavity of PREP (see Fig. S1 in SI). Interestingly, the ligand frequently returns to the alternative binding site, suggesting a dynamic and transient interaction rather than stable binding. Analysis of the simulations reveals that while HUP-46 forms similar initial contacts to HUP-55 (e.g., with Y471 and R488), its binding in the alternative site is characterized by lower contact frequency and shorter persistence of key interactions. Specifically, the thiazole ring of HUP-46 engages in weaker polar interactions compared to the oxazole of HUP-55, and its overall binding pose is less restrained, facilitating its dissociation. This dynamic behavior of HUP-46, contrasted with the more stable HUP-55, presents a compelling puzzle regarding the molecular determinants of their functional potency. The apparent inverse correlation between the stability in the alternative binding site and the experimental inhibitory efficacy (as reflected in IC50 values) suggests a complex mechanism of action that may involve binding to multiple sites or allosteric communication between sites. Understanding the precise reason why weaker binding of HUP-46 to the characterized alternative site correlates with its higher functional potency remains a critical and open question.
These observations highlight a potential mechanism of action for HUP ligands, characterized by greater flexibility in the selection of binding sites compared to standard inhibitors like KYP-2047. However, it is unclear whether this is relevant to their role in modulating PPIs. The ability to interact dynamically with multiple binding sites may be due to the adaptation to conformational changes in PREP, which could be critical for their PPI-targeting mechanism. Unlike conventional inhibitors like KYP-2047, HUP-46 and HUP-55 were specifically developed as modulators of protein–protein interactions (PPIs). Their capacity for pathway hopping and binding at alternative sites may directly interfere with PREP's interactions with pathological partners such as α-synuclein and Tau. We propose that this structural adaptability allows HUP ligands to influence transient protein complexes in ways that rigid, active-site inhibitors cannot achieve.
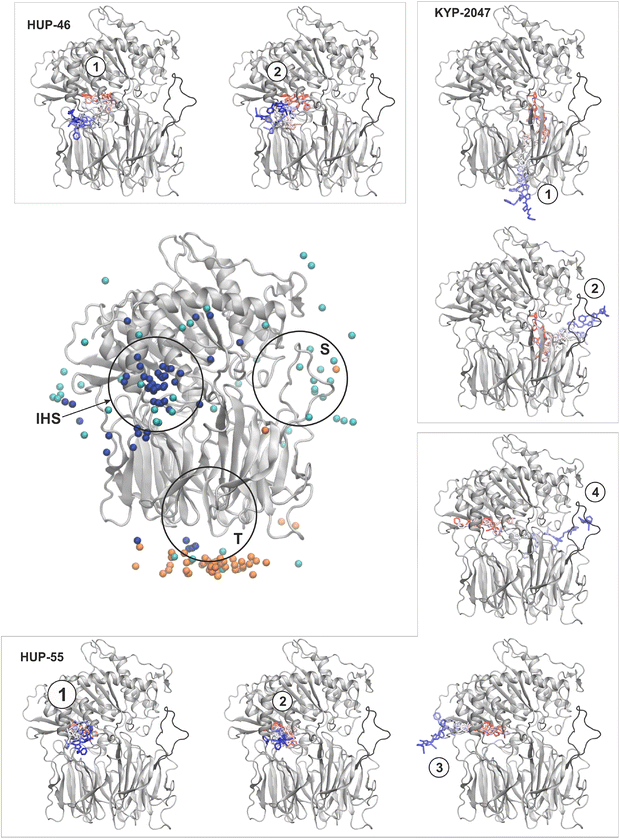
Unbinding reaction pathways
Starting from the previously identified binding site (the active site in the case of KYP-2047 and alternative site for HUP-46 and HUP-55, see Fig. 1b), we conducted independent ligand unbinding simulations using the maze module implemented in PLUMED. Fig. 2a shows the final positions of the ligands after leaving the central pocket (orange spheres represent KYP-2047, cyan HUP-55, and dark blue HUP-46). Interestingly, we found that the three ligands exhibit distinct exit patterns, with the full list summarized in Table 1. The HUP-55 ligand jumps between various possible pathways, exiting through different protein regions as it searches for transient tunnels. In contrast, HUP-46 consistently follows an exit pathway between helices 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 71, 685
71, 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 705, and Blade1 (residues 92
705, and Blade1 (residues 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 97), an area we designated “inter-helical site” (marked as IHS in Fig. 2). Although HUP-55 occasionally populates this route, its dissociation events are more uniformly distributed as it explores alternative options when the primary binding pathway is temporarily closed. These results suggest that HUP-55 can show a behavior known as pathway hopping, where ligands migrate more diffusively in proteins due to large fluctuations in binding regions before selecting an exit.23 As recently shown, pathway jumping occurs in the acetylcholinesterase-Huperzine A complex, which is also considered important for neurological disorders.22
97), an area we designated “inter-helical site” (marked as IHS in Fig. 2). Although HUP-55 occasionally populates this route, its dissociation events are more uniformly distributed as it explores alternative options when the primary binding pathway is temporarily closed. These results suggest that HUP-55 can show a behavior known as pathway hopping, where ligands migrate more diffusively in proteins due to large fluctuations in binding regions before selecting an exit.23 As recently shown, pathway jumping occurs in the acetylcholinesterase-Huperzine A complex, which is also considered important for neurological disorders.22
| Protein | Ligand | Pathway | Description |
|---|---|---|---|
| PREP | KYP-2047 | 1 | Tunnel in the β-propeller domain |
| 2 | Side opening through flexible loops: A, B and His | ||
| PREP | HUP-46 | 1 | Inter-helical site, the benzene ring toward solvent |
| 2 | Inter-helical site, the pyrrolidine ring toward solvent | ||
| PREP | HUP-55 | 1 | Inter-helical site, the pyrrolidine ring toward solvent |
| 2 | inter-helical site, benzene ring toward solvent | ||
| 3 | Hinge region | ||
| 4 | Side opening, through flexible loops: A, B and His |
The dissociation of HUP-46 through the inter-helical site occurs more quickly, primarily within the first 20 ns of simulation (see Fig. S2 in SI for the time distribution of dissociation events). In comparison, HUP-55 exits between 20 to 40 ns, although there can be occasional delays of up to 200 ns. The fastest exits for HUP-55 tend to occur near the inter-helical site. Consequently, attempts to use alternative pathways often prolong the process, as the ligand must wait for conformational changes to open these routes, such as the side opening located next to loops A, B, and the His-loop (marked as S in Fig. 2).
Interestingly, KYP-2047 prefers a different exit route than suggested in earlier findings.39,44 The average exit times for KYP-2047 are relatively short, generally occurring between 20 and 40 ns (see Fig. S2 in SI). However, the inhibitor predominantly exits via a tunnel in the β-propeller domain (marked as T in Fig. 2). This exit route has been previously considered less favorable based on free-energy calculations by St-Pierre et al.,44 who have concluded that the corresponding tunnel is too narrow and has insufficient fluctuations to enable dissociation for such inhibitors effectively. However, Kotev et al.49 have suggested that dissociation through this pathway is possible and can be biologically relevant. The alternative, more challenging exit through the side opening next to loops A, B, and the His-loop generally takes longer, often requiring over 100 ns.
The representative exit pathways for each ligand are shown as colored trajectories in the inset of Fig. 2, with positions shaded from red (trajectory start) to blue (trajectory end). For HUP-46, two exits occur through the inter-helical site with slight variations in the ligand's initial orientation. KYP-2047 shows one exit through the tunnel in the β-propeller domain and another via the flexible loop region. HUP-55 exhibits two exits through the inter-helical site (varying in ligand's orientation), another through a temporary tunnel near the hinge region, and one through the loop region (A, B, and His-loop). Although we used a single representative binding pose per ligand, these were selected based on stable conformations observed in prior MD simulations and are consistent with previous studies (e.g., Patsi et al.15). Since our focus was on dissociation pathways rather than binding affinities, this approach was sufficient for kinetic analysis. Notably, we observed that ligands could exit via the same pathway in different orientations, suggesting that moderate changes in the initial pose are unlikely to affect the main conclusions regarding exit route selection. The dissociation pathways we identified, particularly through the β-propeller tunnel and the inter-helical site, likely represent natural access routes for endogenous substrates and modulators. This interpretation aligns with previous structural studies indicating that the β-propeller domain and flexible loops regulate substrate entry to the active site, supporting the physiological relevance of these pathways.40,41 From a therapeutic perspective, targeting specific exit routes could enable rational design of ligands with optimized binding kinetics and functional selectivity.
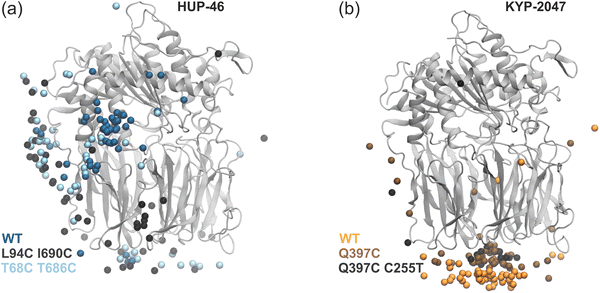
Blocking the preferred exit route of HUP-46
To further investigate the role of specific exit pathways, we performed additional simulations of ligand unbinding, with a focus on two specific cases. We introduced two sets of mutations in PREP to block the inter-helical exit pathway preferred by HUP-46. In the first system, L94 and I690 were mutated to cysteines, while in the second, cysteines were introduced at T68 and T686 to form a disulfide bridge obstructing the transient tunnel.The dynamics of the rest of the protein was minimally affected (see Fig. S3 in SI). Our simulations of the mutated systems reveal that the preferred exit route through the inter-helical site is no longer accessible, which forces the ligand to seek alternative pathways. In Fig. 3a, the locations of exits for the mutated systems (light blue and black spheres) are more evenly spread than the wild-type PREP (dark blue spheres). Additionally, the time needed to find an exit increased, with average exit times for the mutated systems comparable to those observed for HUP-55 and KYP-2047 (20–40 ns). These findings indicate that such mutations may alter ligand behavior, which could be further verified experimentally.
Preference of KYP-2047 for the β-propeller exit tunnel
Given the structural arrangement of the β-propeller domain, where the tunnel is formed by seven “blades”, we introduced the Q397C mutation to form a disulfide bridge with the native C78. This mutation reduces the flexibility of the blades, limiting their ability to undergo “breathing” fluctuations that could otherwise widen the tunnel. To prevent unintended interactions with the native C255, this residue was mutated to threonine in an additional mutated system.Despite these mutations, KYP-2047 continues to exhibit a strong preference for exiting through the tunnel in the β-propeller domain (see Fig. 3b, with brown and black spheres representing mutated systems, and orange spheres for the wild-type PREP). Neither the exit preference nor the average exit times are significantly altered by these mutations. These results suggest that the ligand not only fits well through the tunnel but also retains sufficient flexibility to overcome additional steric hindrance introduced by the mutations.
Diffusion in binding sites for nonpeptidic HUP ligands
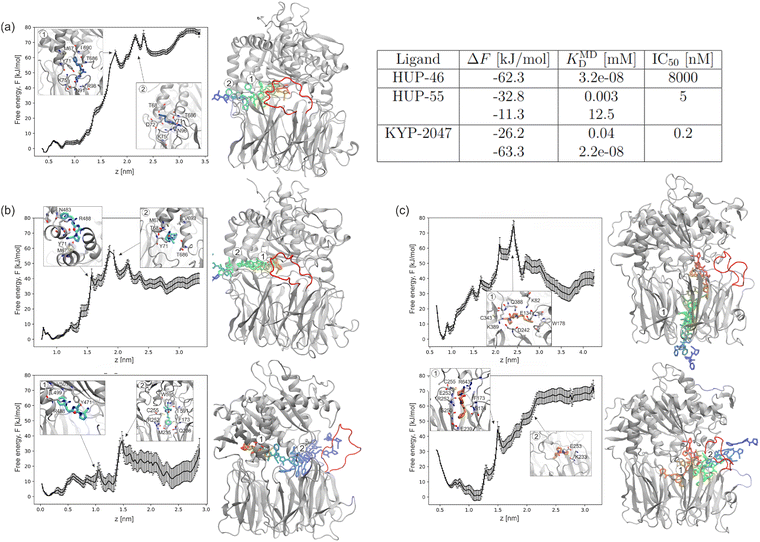
To analyze possible reaction pathways, we calculated the free energy required for ligand dissociation along the representative trajectories in each complex. To achieve this, we used US, a method successfully used in previous studies to investigate ligand dissociation from PREP.44 We note that the estimated free-energy profiles are calculated along specific unbinding pathways (i.e., different reaction coordinates), and thus, they may not exactly agree with experimental estimates. Although there are methods that can combine multiple free-energy profiles along many unbinding pathways,31,37 we instead decided to analyze each free-energy profile on the microscopic level individually, as each unbinding pathway characterizes not only the ligand but also the fluctuating protein.For HUP-46, which favors the exit through the inter-helical site, the free-energy difference between the bound and unbound states is around −62 kJ mol−1 (Fig. 4a). The main kinetic bottleneck during dissociation occurs after the ligand exits its binding site, specifically when detaching from the well-adapted pocket formed along the inner wall of the protein cavity. Here, the benzene ring of HUP-46 resides in a hydrophobic pocket formed by M67, I690, and Y71, while the nitrile group engages with polar and positively charged residues K75, N91, R98, and T686. Upon release of the benzene ring, the ligand is also restrained by interactions with T68 and D72.
For HUP-55, we investigated two possible dissociation pathways in more detail. The first pathway we consider is through the inter-helical site (Fig. 4b, top), similar to where HUP-46 exits PREP. We can see that the free-energy difference of around −33 kJ mol−1 is primarily caused by leaving the binding site of HUP-55 and traversing the sequence of residues that form the “wall” of the protein. Notably, the free-energy difference for HUP-55 is lower than that of HUP-46. This raises questions about whether this change is attributed to the structural differences between aminooxazole and aminothiazole or whether it results from fluctuations in the side chains of PREP during the simulation.
The second dissociation pathway for HUP-55 is longer, passing through the internal cavity of the protein to the side exit near the flexible loops (Fig. 4b, bottom). This pathway is particularly interesting as it demonstrates that the most challenging steps are associated with leaving two previously characterized binding sites within PREP. The first site, located deep in the hinge region, is typical for HUP ligands, while the second is closer to the active center of PREP. Transitioning from the first binding site to the second proves relatively easy, with an energy difference of just around −11 kJ mol−1. This suggests that movement between these two sites is somewhat straightforward and may occur frequently, possibly as part of the natural processing of physiological substrates in PREP. Exiting from the second binding site through the flexible loop region required overcoming an additional energy barrier of approximately 30 kJ mol−1.
While the pathway for HUP-46 is dominant in the maze simulations, the US calculations revealed relatively high free-energy barriers along the same path. This discrepancy likely arises from the fundamental differences between these two computational approaches. maze primarily explores sterically accessible routes that do not necessarily correspond to the most thermodynamically favorable pathways. Consequently, maze simulations may overrepresent pathways that are structurally easier to traverse but energetically less favorable. This difference stems primarily from the definition of the native-contact-like loss function in maze (see Methods), which enables computation of many dissociation simulations within a feasible computational time, unlike computationally demanding methods based on free-energy estimation.
Another plausible explanation is that other potential pathways were temporarily inaccessible because of protein conformational fluctuations during the simulations. Dynamic elements, such as loops or domains, could sporadically obstruct access to alternative, potentially more favorable routes, like the side opening. This transient occlusion might explain why the inter-helical pathway appeared more frequently in maze, even though it exhibits higher free-energy barriers in US.
Furthermore, the unique interactions of HUP-46 with the protein may favor this specific pathway. The aminooxazole group of the ligand, for example, can form stabilizing hydrogen bonds or electrostatic interactions that restrict its mobility in other directions. This could contrast with ligands like HUP-55, which feature an aminothiazole group, potentially leading to different binding dynamics and preferred pathways.
KYP-2047 unbinding
The final two dissociation pathways analyzed involve the peptidic ligand KYP-2047, which binds near the active center of PREP (Fig. 4c). One of the most commonly chosen and time-efficient exit routes for KYP-2047 is through the tunnel in the β-propeller domain (Fig. 4c, top). The most challenging step along this route is passing through the cluster of charged residues at the entrance of the tunnel (inset 1, the peak around z = 2.5), which creates an energy barrier of approximately 80 kJ mol−1 (measured from the baseline). However, the estimated free-energy difference between the bound and unbound states is about −26 kJ mol−1, which is relatively small and suggests that, despite the high transition barrier, this pathway may still be the preferred dissociation route.In the study by St-Pierre et al. on ZPP dissociation pathways in PREP,44 this entry route was associated with a transition barrier of approximately −101 kJ mol−1, leading to estimates of inhibition constants that were several orders of magnitude lower than the values observed experimentally. As a result, this pathway was deemed nonphysical. However, it is important to note that we have two different ligands and two distinct approaches to select trajectory points for US: in our case, maze and, in theirs, SMD. Comparison of ZPP with KYP-2047 requires caution, as the P3 part of the inhibitor differs significantly. Specifically, ZPP features a carbamate group instead of an amide bond, which could be considered to be more bulky and less flexible than the corresponding moiety in KYP-2047.
The second pathway for KYP-2047 involves exiting through the flexible loop region (Fig. 4c, bottom). All observed exit trajectories for this pathway require a slight downward shift toward the β-propeller domain. After overcoming the interactions that anchor the ligand just below the binding site (inset 1), the ligand moves through the flexible loop region, utilizing the flexibility of the loops between blades 3 and 4 of the β-propeller domain (inset 2). The free-energy difference for exiting along this pathway is approximately −63 kJ mol−1.
In the case of StPierre's work with ZPP,44 the free-energy difference between the bound and unbound states was much smaller, around −18 kJ mol−1, with a transition barrier of approximately 25 kJ mol−1. In our study, both the barrier and the energy differences are significantly higher, primarily because of the challenge of ‘pushing’ the ligand through the unfavorably positioned loops, which likely hinders dissociation.
A notable difference can be observed in the initial profiles of the free-energy curves near the starting positions of the ligands. For HUP ligands, the curve is relatively flat, while for KYP-2047, it forms a distinct deep minimum. This could explain the differences in ligand behavior observed during the dynamics of the system: KYP-2047 binds strongly and stably, whereas HUPs exhibit a more dynamic and transient binding mode, reflecting their ability to adapt to conformational changes in PREP and interact with multiple binding sites.
Estimated dissociation constants
We aimed to compare our results with available experimental data on ligand binding affinities. For KYP-2047, experimental inhibition constant (Ki) data is available, quantifying the inhibitor concentration required to occupy half of the binding sites in the enzyme or receptor. Using the formula KD = [1 M]e−βΔF, we estimated the dissociation constant (KD) from the free-energy difference between the bound and unbound states.44 We emphasize that these estimated KD values are highly approximate, as they are derived from specific modeled dissociation pathways and are subject to methodological limitations, including force-field inaccuracies and sampling constraints. For the pathways analyzed, KD values ranged from 0.02 nM for pathway 2 (side exit) to approximately 0.4 mM for pathway 1 (exit via the β-propeller domain). The experimental value for KYP-2047 is 0.02 nM, which falls at the very lower end of this range. This suggests that while our simulations can capture the existence of high- and low-affinity pathways, the absolute values of the calculated KD should be interpreted with caution. The direct comparison between KD and Ki should be interpreted with caution due to differences in their mechanistic basis.For HUP-46 and HUP-55, experimental Ki values are not available; only IC50 values have been reported (8 µM and 5 nM, respectively), along with EC50 values of 100 nM for HUP-46 and 275 nM for HUP-55.15 This presents a significant challenge in interpretation, as IC50 values, which represent the concentration required to inhibit a specific biological activity (typically enzymatic function), are highly dependent on the available enzyme concentration and experimental conditions. Consequently, while HUP-55 can be classified as an inhibitor due to its low IC50 value (5 nM), HUP-46, with an IC50 of 8 µM, is more likely to function as a ligand rather than a potent inhibitor. However, EC50 values, which quantify the concentration needed to induce a specific biological effect (in this case, modulating the function derived from PPI), may provide a more relevant measure for these compounds, as both HUP-46 and HUP-55 were specifically designed to modulate PPIs.
The estimated KD values from our simulations for HUP-46 (0.03 nM) and HUP-55 (ranging from 0.003 nM to 12.5 mM) exhibit large discrepancies with the experimental IC50 data, differing by several orders of magnitude. This is particularly striking for HUP-46, where the calculated KD (0.03 nM) and experimental IC50 (8 µM) differ by approximately 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fold. Such a large deviation from the experimental values most likely arises from limitations inherent to our approach, such as the choice of reaction coordinate, insufficient sampling, or force field inaccuracies. Therefore, we focus on comparing the relative favorability of different dissociation pathways, rather than absolute estimates.
000 fold. Such a large deviation from the experimental values most likely arises from limitations inherent to our approach, such as the choice of reaction coordinate, insufficient sampling, or force field inaccuracies. Therefore, we focus on comparing the relative favorability of different dissociation pathways, rather than absolute estimates.
The results indicate that for HUP-55, the free-energy barriers for dissociation are generally lower and the estimated KDs are weaker (spanning µM to mM) compared to the sub-nanomolar KD estimated for the most favorable pathways of KYP-2047 and HUP-46. This aligns with the observed hierarchy of functional potency from experimental IC50 values, where HUP-55 is a less potent inhibitor than KYP-2047. This comparison is qualitative, however, as IC50, while often correlating with binding affinity, is not a direct measure of binding affinity. The case of HUP-46 highlights a significant challenge: our method predicts a very stable bound state and high energy barriers for exit, resulting in an extremely low calculated KD that is inconsistent with the much weaker activity observed in functional assays. This underscores the difficulty in using such calculations to predict absolute binding affinities for certain ligand–receptor systems.
In conclusion, while the relative ranking of pathways may provide mechanistic insights, the discrepancies with experimental data confirm that these estimated KD cannot be treated as definitive physical constants and highlight the need for further methodological refinement and experimental validation.
Conclusions
This work investigates inhibitor binding and unbinding pathways in prolyl oligopeptidase (PREP), demonstrating how slight structural differences between ligands can lead to distinct binding behaviors and exit routes. We have examined these pathways by combining extensive MD simulations and enhanced sampling techniques and quantified the corresponding free-energy profiles.The analysis revealed that HUP-55 diffuses out of the binding site after approximately 500 ns, suggesting that its binding is less stable than KYP-2047. In contrast, HUP-46 shows greater flexibility in selecting binding sites, which is unusual for standard inhibitors such as KYP-2047. In particular, the initial profiles of the free-energy curves also reflect these differences: for the HUP ligands, the curve is relatively flat near the starting position, while for KYP-2047, it forms a distinct deep minimum. This observation further supports the idea that KYP-2047 binds strongly and stably. In contrast, HUPs exhibit a more dynamic and transient binding mode, likely due to their ability to adapt to conformational changes in PREP and interact with multiple binding sites.
The unbinding pathways differ significantly among the inhibitors: KYP-2047 retains a preference for exiting through the tunnel in the β-propeller domain, even when mutations narrow the pathway, whereas HUP-46 strongly favors the inter-helical site, a behavior not observed for HUP-55. This distinction may arise from structural differences in the aminooxazole and aminothiazole moieties of the HUP ligands or transient fluctuations in PREP's side chains during the simulations.
A key observation was the phenomenon of “pathway hopping,” particularly evident in HUP-55, where the ligand diffusively explored multiple binding regions before selecting its exit. The US results also showed that transitioning between specific binding sites, such as from an alternative site to the active site for HUP-55, involves a relatively low free-energy difference of about 11 kJ mol−1. This suggests that such movements could be a natural part of the physiological substrate processing of PREP. The distinct binding behaviors of the HUP ligands, compared to KYP-2047, raise questions about their potential role in modulating PPIs. The ability of the HUP ligands to interact dynamically with multiple binding sites might reflect their adaptation to PREP's conformational flexibility, which could be integral to their mechanism for targeting PPIs. Our findings also highlight the complexity introduced by pathway hopping in systems with multiple unbinding mechanisms. When transition states for alternative unbinding pathways are energetically comparable, even minor changes to drug–protein interactions can switch the dominant pathway, complicating the design of inhibitors targeting specific kinetic properties. This highlights the importance of computational methods that capture the full range of binding and unbinding pathways, especially in drug discovery.
These insights suggest possible approaches for future drug design. First, ligands intended to modulate PPIs should be designed with the capacity to populate multiple binding and dissociation pathways, as this adaptability may be crucial for effectively interfering with dynamic protein interactions. Second, strategically blocking specific dissociation routes, for instance, through mutations at the inter-helical site, could be a viable strategy to engineer ligands with prolonged residence time and enhanced functional selectivity. The physiological relevance of the identified pathways could be tested experimentally by combining targeted mutagenesis of key residues lining these exits with cellular assays measuring their impact on α-synuclein aggregation.
While our pathway analysis provides solid mechanistic insights, full quantitative validation of the binding dynamics, especially for the HUP ligands, requires additional experimental work. Although our free-energy estimates for KYP-2047 show correlation with experimental inhibition constants, the corresponding values for HUP ligands highlight the limitations of current predictions. Direct measurement of binding affinities and residence times for these ligands is necessary to link our results to biological activity and to provide insights into their mechanistic roles and potential to modulate PREP-mediated processes.
Conflicts of interest
There are no conflicts to declare.Data availability
All the data and PLUMED input files required to reproduce the results reported in this paper are available in a RepOD repository (https://doi.org/10.18150/ZUISNK). The implementation of the maze module for PLUMED is available in a git repository (https://github.com/jakryd/plumed2-maze).The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cp02804c.
Acknowledgements
We thank Timo Myöhänen and Erik Wallén for the insightful discussions that shaped this work. J. R. acknowledges funding the Ministry of Science and Higher Education in Poland. We acknowledge Polish high-performance computing infrastructure PLGrid for awarding this project access to the LUMI supercomputer, owned by the EuroHPC Joint Undertaking, hosted by CSC (Finland) and the LUMI consortium through PLL/2024/07/017528.References
- M. S. Forman, J. Q. Trojanowski and V. M. Lee, Neurodegenerative Diseases: A Decade of Discoveries Paves the Way for Therapeutic Breakthroughs, Nat. Med., 2004, 10(10), 1055–1063 CrossRef CAS PubMed.
- D. M. Skovronsky, V. M. Y. Lee and J. Q. Trojanowski, Neurodegenerative Diseases: New Concepts of Pathogenesis and their Therapeutic Implications, Annu. Rev. Pathol.: Mech. Dis., 2006, 1(1), 151–170 CrossRef CAS PubMed.
- T. Myöhänen, M. Hannula, R. Van Elzen, M. Gerard, P. Van Der Veken and J. Garca-Horsman, et al., A Prolyl Oligopeptidase Inhibitor, KYP-2047, Reduces α-Synuclein Protein Levels and Aggregates in Cellular and Animal Models of Parkinson's Disease, Br. J. Pharmacol., 2012, 166(3), 1097–1113 CrossRef PubMed.
- P. Van der Veken, V. Fulop, D. Rea, M. Gerard, R. Van Elzen and J. Joossens, et al., P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy, J. Med. Chem., 2012, 55(22), 9856–9867 CrossRef CAS PubMed.
- I. Brandt, S. Scharpé and A. M. Lambeir, Suggested Functions for Prolyl Oligopeptidase: A Puzzling Paradox, Clin. Chim. Acta, 2007, 377(1–2), 50–61 CrossRef CAS.
- R. S. Williams, L. Cheng, A. W. Mudge and A. J. Harwood, A Common Mechanism of Action for Three Mood-Stabilizing Drugs, Nature, 2002, 417(6886), 292–295 CrossRef CAS PubMed.
- I. Brandt, K. De Vriendt, B. Devreese, J. Van Beeumen, W. Van Dongen and K. Augustyns, et al., Search for Substrates for Prolyl Oligopeptidase in Porcine Brain, Peptides, 2005, 26(12), 2536–2546 CrossRef CAS PubMed.
- I. Brandt, M. Gérard, K. Sergeant, B. Devreese, V. Baekelandt and K. Augustyns, et al., Prolyl Oligopeptidase Stimulates the Aggregation of α-Synuclein, Peptides, 2008, 29(9), 1472–1478 CrossRef CAS PubMed.
- M. H. Savolainen, X. Yan, T. T. Myöhänen and H. J. Huttunen, Prolyl Oligopeptidase Enhances α-Synuclein Dimerization via Direct Protein-Protein Interaction, J. Biol. Chem., 2015, 290(8), 5117–5126 CrossRef CAS.
- R. Svarcbahs, U. Julku, T. Kilpeläinen, M. Kyyrö, M. Jäntti and T. T. Myöhänen, New Tricks of Prolyl Oligopeptidase Inhibitors-A Common Drug Therapy for Several Neurodegenerative Diseases, Biochem. Pharmacol., 2019, 161, 113–120 CrossRef CAS PubMed.
- T. S. Eteläinen, M. C. Silva, J. K. Uhari-Väänänen, F. De Lorenzo, M. H. Jäntti and H. Cui, et al., A Prolyl Oligopeptidase Inhibitor Reduces Tau Pathology in Cellular Models and in Mice with Tauopathy, Sci. Trans. Med., 2023, 15(691), eabq2915 CrossRef PubMed.
- R. Van Elzen, Y. Waumans, S. Nath, P. Van der Veken, S. Kerckhoff and E. Van Dijk, et al., The Prolyl Oligopeptidase and α-Synuclein Connection Revisited, Biochimie, 2025, 233, 1–13 CrossRef CAS PubMed.
- T. Eteläinen, V. Kulmala, R. Svarcbahs, M. Jäntti and T. Myöhänen, Prolyl Oligopeptidase Inhibition Reduces Oxidative Stress via Reducing NADPH Oxidase Activity by Activating Protein Phosphatase 2A, Free Radical Biol. Med., 2021, 169, 14–23 CrossRef PubMed.
- T. P. Kilpeläinen, L. Hellinen, J. Vrijdag, X. Yan, R. Svarcbahs and K. S. Vellonen, et al., The Effect of Prolyl Oligopeptidase Inhibitors on Alpha-Synuclein Aggregation and Autophagy Cannot be Predicted by their Inhibitory Efficacy, Biomed. Pharmacother., 2020, 128, 110253 CrossRef PubMed.
- T. P. Kilpelainen, H. T. Patsi, R. Svarcbahs, U. H. Julku, T. S. Etelainen, H. Cui, S. Auno, N. Sipari, S. Norrbacka and T. O. Leino, et al., Nonpeptidic Oxazole-Based Prolyl Oligopeptidase Ligands with Disease-Modifying Effects on α-Synuclein Mouse Models of Parkinson's Disease, J. Med. Chem., 2023, 66, 7475–7496 CrossRef CAS PubMed.
- H. T. Pätsi, T. P. Kilpelainen, M. Jumppanen, J. Uhari-Vaananen, P. V. Wielendaele and F. De Lorenzo, et al., 5-Aminothiazoles Reveal a New Ligand-Binding Site on Prolyl Oligopeptidase Which is Important for Modulation of Its Protein-Protein Interaction-Derived Functions, J. Med. Chem., 2024, 67(7), 5421–5436 CrossRef PubMed.
- M. J. Carroll, R. V. Mauldin, A. V. Gromova, S. F. Singleton, E. J. Collins and A. L. Lee, Evidence for Dynamics in Proteins as a Mechanism for Ligand Dissociation, Nat. Chem. Biol., 2012, 8(3), 246–252 CrossRef CAS PubMed.
- R. Baron and J. A. McCammon, Molecular Recognition and Ligand Association, Annu. Rev. Phys. Chem., 2013, 64, 151–175 CrossRef CAS PubMed.
- J. Rydzewski and W. Nowak, Ligand Diffusion in Proteins via Enhanced Sampling in Molecular Dynamics, Phys. Life Rev., 2017, 22, 58–74 CrossRef PubMed.
- J. M. L. Ribeiro, S. T. Tsai, D. Pramanik, Y. Wang and P. Tiwary, Kinetics of Ligand-Protein Dissociation from All-Atom Simulations: Are We There Yet?, Biochemistry, 2018, 58(3), 156–165 CrossRef PubMed.
- M. Bernetti, M. Masetti, W. Rocchia and A. Cavalli, Kinetics of Drug Binding and Residence Time, Annu. Rev. Phys. Chem., 2019, 70, 143–171 CrossRef CAS PubMed.
- P. Tiwary, V. Limongelli, M. Salvalaglio and M. Parrinello, Kinetics of Protein-Ligand Unbinding: Predicting Pathways, Rates, and Rate-Limiting Steps, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(5), E386–E391 CrossRef CAS PubMed.
- J. Rydzewski, R. Jakubowski, W. Nowak and H. Grubmuller, Kinetics of Huperzine A Dissociation from Acetylcholinesterase via Multiple Unbinding Pathways, J. Chem. Theory Comput., 2018, 14(6), 2843–2851 CrossRef CAS PubMed.
- S. D. Lotz and A. Dickson, Unbiased Molecular Dynamics of 11 min Timescale Drug Unbinding Reveals Transition State Stabilizing Interactions, J. Am. Chem. Soc., 2018, 140(2), 618–628 CrossRef CAS PubMed.
- J. Rydzewski and W. Nowak, Machine Learning Based Dimensionality Reduction Facilitates Ligand Diffusion Paths Assessment: A Case of Cytochrome P450cam, J. Chem. Theory Comput., 2016, 12(4), 2110–2120 CrossRef CAS PubMed.
- J. Rydzewski and W. Nowak, Thermodynamics of Camphor Migration in Cytochrome P450cam by Atomistic Simulations, Sci. Rep., 2017, 7(1), 7736 CrossRef CAS PubMed.
- A. Nunes-Alves, D. M. Zuckerman and G. M. Arantes, Escape of a Small Molecule from Inside T4 Lysozyme by Multiple Pathways, Biophys. J., 2018, 114(5), 1058–1066 CrossRef CAS PubMed.
- R. Capelli, P. Carloni and M. Parrinello, Exhaustive Search of Ligand Binding Pathways via Volume-Based Metadynamics, J. Phys. Chem. Lett., 2019, 10(12), 3495–3499 CrossRef CAS PubMed.
- J. Rydzewski and O. Valsson, Finding Multiple Reaction Pathways of Ligand Unbinding, J. Chem. Phys., 2019, 150(22), 221101 CrossRef PubMed.
- V. Rizzi, L. Bonati, N. Ansari and M. Parrinello, The Role of Water in Host-Guest Interaction, Nat. Commun., 2021, 12(1), 93 CrossRef CAS PubMed.
- H. Fu, H. Chen, M. Blazhynska, E. Goulard Coderc de Lacam, F. Szczepaniak and A. Pavlova, et al., Accurate Determination of Protein: Ligand Standard Binding Free Energies from Molecular Dynamics Simulations, Nat. Protocols, 2022, 17(4), 1114–1141 CAS.
- S. Wolf, M. Post and G. Stock, Path Separation of Dissipation-Corrected Targeted Molecular Dynamics Simulations of Protein-Ligand Unbinding, J. Chem. Phys., 2023, 158(12), 124106 CrossRef CAS PubMed.
- D. Ray and M. Parrinello, Kinetics from Metadynamics: Principles, Applications, and Outlook, J. Chem. Theory Comput., 2023, 19(17), 5649–5670 CrossRef CAS PubMed.
- M. Karrenbrock, A. Borsatto, V. Rizzi, D. Lukauskis, S. Aureli and F. Luigi Gervasio, Absolute Binding Free Energies with OneOPES, J. Phys. Chem. Lett., 2024, 15(39), 9871–9880 CrossRef CAS PubMed.
- V. Tänzel, M. Jäger and S. Wolf, Learning Protein-Ligand Unbinding Pathways via Single-Parameter Community Detection, J. Chem. Theory Comput., 2024, 20(12), 5058–5067 CrossRef PubMed.
- S. R. Zia, A. Coricello and G. Bottegoni, Increased Throughput in Methods for Simulating Protein Ligand Binding and Unbinding, Curr. Opin. Struct. Biol., 2024, 87, 102871 CrossRef CAS PubMed.
- D. Ray and M. Parrinello, Data-Driven Classification of Ligand Unbinding Pathways, Proc. Natl. Acad. Sci. U. S. A., 2024, 121(10), e2313542121 CrossRef CAS PubMed.
- N. Aho, G. Groenhof and P. Buslaev, Do All Paths Lead to Rome? How Reliable is Umbrella Sampling along a Single Path?, J. Chem. Theory Comput., 2024, 20(15), 6674–6686 CrossRef CAS PubMed.
- K. Walczewska-Szewc and J. Rydzewski, Structural Visualization of Inhibitor Binding in Prolyl Oligopeptidase, Biophys. Rev., 2024, 5(3), 032105 CrossRef CAS.
- A. Tsirigotaki, R. V. Elzen, P. V. D. Veken, A. M. Lambeir and A. Economou, Dynamics and Ligand-Induced Conformational Changes in Human Prolyl Oligopeptidase Analyzed by Hydrogen/Deuterium Exchange Mass Spectrometry, Sci. Rep., 2017, 7(1), 2456 CrossRef PubMed.
- Z. Szeltner, D. Rea, T. Juhász, V. Renner, V. Fülöp and L. Polgár, Concerted Structural Changes in the Peptidase and the Propeller Domains of Prolyl Oligopeptidase are Required for Substrate Binding, J. Mol. Biol., 2004, 340(3), 627–637 CrossRef CAS PubMed.
- V. Fülöp, Z. Böcskei and L. Polgár, Prolyl Oligopeptidase: An Unusual β-Propeller Domain Regulates Proteolysis, Cell, 1998, 94(2), 161–170 CrossRef PubMed.
- V. Fülöp, Z. Szeltner and L. Polgár, Catalysis of Serine Oligopeptidases is Controlled by a Gating Filter Mechanism, EMBO Rep., 2000, 1(3), 277–281 CrossRef PubMed.
- P. J. Tummino and R. A. Copeland, Residence Time of Receptor-Ligand Complexes and its Effect on Biological Function, Biochemistry, 2008, 47(20), 5481–5492 CrossRef CAS PubMed.
- J. F. St-Pierre, M. Karttunen, N. Mousseau, T. Róg and A. Bunker, Use of Umbrella Sampling to Calculate the Entrance/Exit Pathway for Z-Pro-Prolinal Inhibitor in Prolyl Oligopeptidase, J. Chem. Theory Comput., 2011, 7(6), 1583–1594 CrossRef CAS PubMed . PMID: 26596426.
- K. Kaszuba, T. Róg, R. Danne, P. Canning, V. Fülöp and T. Juhász, et al., Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyloligopeptidase, Biochimie, 2012, 94(6), 1398–1411 CrossRef CAS PubMed.
- Z. Szeltner, T. Juhász, I. Szamosi, D. Rea, V. Fülöp and K. Módos, et al., The Loops Facing the Active Site of Prolyl Oligopeptidase are Crucial Components in Substrate Gating and Specificity, Biochim. Biophys. Acta, Proteins Proteomics, 2013, 1834(1), 98–111 CrossRef CAS PubMed.
- K. Ellis-Guardiola, H. Rui, R. L. Beckner, P. Srivastava, N. Sukumar and B. Roux, et al., Crystal Structure and Conformational Dynamics of Pyrococcus Furiosus Prolyl Oligopeptidase, Biochemistry, 2019, 58(12), 1616–1626 CrossRef CAS PubMed.
- K. Walczewska-Szewc, J. Rydzewski and A. Lewkowicz, Inhibition-Mediated Changes in Prolyl Oligopeptidase Dynamics Possibly Related to α-Synuclein Aggregation, Phys. Chem. Chem. Phys., 2022, 24(7), 4366–4373 RSC.
- M. Kotev, D. Lecina, T. Tarragó, E. Giralt and V. Guallar, Unveiling Prolyl Oligopeptidase Ligand Migration by Comprehensive Computational Techniques, Biophys. J., 2015, 108(1), 116–125 CrossRef CAS PubMed.
- C. D. Haffner, C. J. Diaz, A. B. Miller, R. A. Reid, K. P. Madauss and A. Hassell, et al., Pyrrolidinyl Pyridone and Pyrazinone Analogues as Potent Inhibitors of Prolyl Oligopeptidase (POP), Bioorg. Med. Chem. Lett., 2008, 18(15), 4360–4363 CrossRef CAS PubMed.
- G. Madhavi Sastry, M. Adzhigirey, T. Day, R. Annabhimoju and W. Sherman, Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments, J. Comput-Aided Mol. Des., 2013, 27, 221–234 CrossRef CAS.
- K. Roos, C. Wu, W. Damm, M. Reboul, J. M. Stevenson and C. Lu, et al., OPLS3e: Extending Force Field Coverage for Drug-like Small Molecules, J. Chem. Theory Comput., 2019, 15(3), 1863–1874 CrossRef CAS PubMed.
- F. Neese, F. Wennmohs, U. Becker and C. Riplinger, The ORCA Quantum Chemistry Program Package, J. Chem. Phys., 2020, 152(22), 224108 CrossRef CAS PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig and B. L. De Groot, et al., CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins, Nat. Methods, 2017, 14(1), 71–73 CrossRef CAS PubMed.
- V. Zoete, M. A. Cuendet, A. Grosdidier and O. Michielin, SwissParam: A Fast Force Field Generation Tool for Small Organic Molecules, J. Comput. Chem., 2011, 32(11), 2359–2368 CrossRef CAS PubMed.
- M. Bugnon, M. Goullieux, U. F. Röhrig, M. A. Perez, A. Daina and O. Michielin, et al., SwissParam 2023: A Modern Web-based Tool for Efficient Small Molecule Parametrization, J. Chem. Inf. Model., 2023, 63(21), 6469–6475 CrossRef CAS PubMed.
- R. A. Friesner, R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood and T. A. Halgren, et al., Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes, J. Med. Chem., 2006, 49(21), 6177–6196 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith and B. Hess, et al., Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- G. A. Tribello, M. Bonomi, D. Branduardi, C. Camilloni and G. Bussi, plumed 2: New Feathers for an Old Bird, Comput. Phys. Commun., 2014, 185(2), 604–613 CrossRef CAS.
- The PLUMED consortium, Promoting transparency and reproducibility in enhanced molecular simulations, Nat. Methods, 2019, 16, 670–673.
- G. A. Tribello, M. Bonomi, G. Bussi and C. Camilloni, et al., PLUMED Tutorials: A Collaborative, Community-Driven Learning Ecosystem, J. Chem. Phys., 2025, 162(9), 092501 CrossRef CAS PubMed.
- G. Bussi, D. Donadio and M. Parrinello, Canonical Sampling through Velocity Rescaling, J. Chem. Phys., 2007, 126(1), 014101 CrossRef PubMed.
- B. Hess, P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation, J. Chem. Theory Comput., 2008, 4(1), 116–122 CrossRef CAS PubMed.
- J. Rydzewski, maze: Heterogeneous Ligand Unbinding along Transient Protein Tunnels, Comput. Phys. Commun., 2020, 247, 106865 CrossRef CAS.
- G. M. Torrie and J. P. Valleau, Nonphysical Sampling Distributions in Monte Carlo Free-Energy Estimation: Umbrella Sampling, J. Comput. Phys., 1977, 23(2), 187–199 CrossRef.
- J. Kästner, Umbrella Sampling, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1(6), 932–942 Search PubMed.
- M. Ester, H. P. Kriegel, J. Sander and X. Xu, et al., A density-based algorithm for discovering clusters in large spatial databases with noise, Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining, 1996, vol. 96, pp. 226–231 Search PubMed.
- E. Schubert, J. Sander, M. Ester, H. P. Kriegel and X. Xu, DBSCAN revisited, revisited: why and how you should (still) use DBSCAN, ACM Trans. Database Systems, 2017, 42(3), 1–21 CrossRef.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion and O. Grisel, et al., Scikit-learn: Machine Learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- J. S. Hub, B. L. De Groot and D. Van Der Spoel, g_wham-A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates, J. Chem. Theory Comput., 2010, 6(12), 3713–3720 CrossRef CAS.
- N. Michaud-Agrawal, E. J. Denning, T. B. Woolf and O. Beckstein, MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations, J. Comput. Chem., 2011, 32(10), 2319–2327 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD – Visual Molecular Dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
| This journal is © the Owner Societies 2026 |