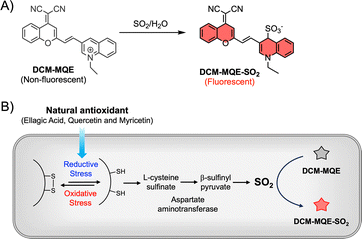
A fluorescent probe for sulfur dioxide reveals intracellular reductive stress triggered by natural antioxidants
Sijie
Luo†
ab,
Lie
Xiang†
c,
Huihong
Liu
b,
Yanyan
Luo
b,
Lei
Shi
 *a,
Guhuan
Liu
*a,
Guhuan
Liu
 *b and
Sheng
Yang
*b and
Sheng
Yang
 *bc
*bc
aGuangdong Engineering Technical Research Center for Green Household Chemicals, Guangdong Industry Polytechnic University, Guangzhou, Guangdong 510300, P. R. China. E-mail: shileinaoh@163.com
bKey Laboratory of Chemical Biology & Traditional Chinese Medicine Research, Ministry of Education, Institute of Interdisciplinary Studies, College of Chemistry and Chemical Engineering, Hunan Normal University, Changsha, Hunan 410081, China. E-mail: ghliu@hunnu.edu.cn; yangsheng@hunnu.edu.cn
cHunan Provincial Key Laboratory of Cytochemistry, School of Chemistry and Pharmaceutical Engineering, Changsha University of Science and Technology, Changsha 410114, P. R. China
First published on 24th November 2025
Abstract
Sulfur dioxide (SO2), a pivotal gasotransmitter, is intricately linked to cellular redox homeostasis. Here, we developed the red-emissive DCM-MQE probe for rapid and sensitive detection of intracellular SO2. Using this probe, we have demonstrated that natural antioxidants significantly elevate SO2 levels in both cells and tumor-bearing mice, and confirmed that antioxidants modulate SO2-mediated redox homeostasis through induction of reductive stress.
Cellular redox homeostasis, maintained through the precise balance of oxidants and reductants, is fundamental to biological function. This equilibrium is primarily governed by the glutathione (GSH/GSSG) redox couple, which orchestrates electron transfer reactions critical for signaling, metabolism, and defence against oxidative damage.1,2 Disruption of this balance—particularly an excessive reductive environment termed reductive stress—is increasingly linked to pathologies including cancer, inflammation, metabolic disorders, and neurodegenerative diseases. Unlike oxidative stress, reductive stress arises from an overabundance of reducing equivalents (e.g., elevated NAD(P)H, thiols and sulfides), leading to aberrant redox signaling and cellular dysfunction.3–6 Understanding the molecular mechanisms driving reductive stress is thus essential for unravelling redox biology and developing targeted therapies for redox-related diseases.
Among reductive sulfur species, sulfur dioxide (SO2) and its derivatives (sulfite/bisulfite, HSO3−/SO32−) occupy a unique niche. Despite sulfur's higher oxidation state and weaker reducing capacity compared to H2S or thiols, SO2 plays pivotal roles in mammalian physiology.7,8 Endogenous SO2 production in mammals occurs via a defined enzymatic cascade: L-cysteine is oxidized by cysteine dioxygenase (CDO) to L-cysteinesulfinate, transaminated to β-sulfinylpyruvate by aminotransferase, and finally decomposed to pyruvate and SO2.9–11 Critically, reductive stress induced by natural antioxidants elevates cellular thiol levels, which may perturb SO2 metabolism—yet the causal relationship remains poorly defined.12–14 This gap stems from a lack of tools capable of specifically, sensitively, and dynamically monitoring SO2 fluctuations in living systems. Existing probes often suffer from interference by reactive sulfur species (RSS) like H2S, inadequate selectivity, or limited applicability in vivo, hindering mechanistic studies of antioxidant-driven reductive stress15–21
Herein, we address this challenge by designing DCM-MQE (Scheme 1), a red-emitting fluorescent probe for SO2 with exceptional specificity and biocompatibility. DCM-MQE exhibits a >100-fold fluorescence enhancement at 660 nm upon SO2 addition, enabling high-contrast imaging with minimal background. Its response resolves SO2 from biological interferents (including H2S and cellular RSS). Leveraging this probe, we demonstrate for the first time that natural antioxidants (ellagic acid, quercetin and myricetin) from traditional Chinese medicine (TCM) induce reductive stress in tumor cells by elevating endogenous SO2 levels. By monitoring SO2 dynamics in live HeLa cells and in vivo models, we establish a direct link between antioxidant exposure, SO2 upregulation, and reductive stress in cancer (Scheme 1). This work not only provides a transformative tool for redox biology but also unveils SO2 as a key mediator in antioxidant-driven tumor modulation, opening avenues for novel redox-targeted anticancer strategies.
Dicyanomethylene-4H-pyran (DCM) derivatives represent a class of classic donor–π–acceptor (D–π–A) fluorophores exhibiting red emission and large Stokes shifts. They are a universal scaffold to construct activity-based sensing probes by regulating the intramolecular charge transfer (ICT) process between the donor and acceptor of the DCM core, triggering obvious fluorogenic signals.22,23 To enable selective detection of SO2 and its derivatives, we designed the probe DCM-MQE. It is readily synthesized via a two-step reaction (Scheme S1) and characterized by 1H NMR, 13C NMR, and MS (Fig. S1 and S2). Through a sulfite-promoted addition reaction, the quinoline moiety can be transformed into an electron-rich enamine moiety, hence resulting in a favourable D–π–A structure with a typical ICT effect.24–26
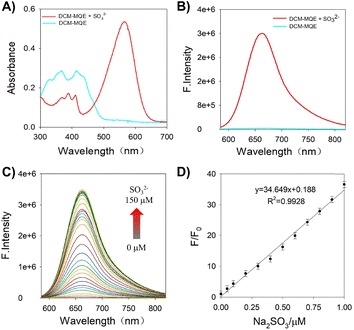
UV-vis absorption spectroscopy revealed that upon SO2 addition, the probe's maximum absorption band red-shifted from 365 nm to 561 nm, accompanied by a distinct color change from yellow to purple (Fig. 1A and Fig. S3, S4). In contrast to the non-fluorescent DCM-MQE, a bright red fluorescence was observed in the presence of Na2SO3 (Fig. 1B and Fig. S3). Notably, treatment with 10 µM Na2SO3 resulted in a 100-fold increase in emission intensity at 661 nm (λex = 561 nm), demonstrating that DCM-MQE functions as a NIR “turn-on” probe for Na2SO3 with high contrast and a significant Stokes shift. Sulfite reduces the methylquinoline moiety of DCM-MQE, converting it into a typical D–π–A structure, which likely accounts for the observed hyperchromic effect and fluorescence enhancement.
DCM-MQE exhibits an ultrafast reaction rate with SO2, reaching maximum fluorescence at 661 nm within <3 s (Fig. S5). As shown in Fig. 1C and D, the emission intensity ratio (F/F0) shows excellent linearity with Na2SO3 concentration (0–1 µM), where F and F0 represent the fluorescence intensities at 661 nm with and without Na2SO3, respectively. The limit of detection (LOD) was calculated to be 6 nM using the 3σ/k method,27 significantly below cellular SO2 thresholds.
Selectivity assays (Fig. S6) against biologically relevant species including K+, Na+, Ca2+, NO2−, CO32−, acetate anion (AcO−), ClO−, H2O2, dithiothreitol (DTT), vitamin C (VC), NADH, NADPH, cysteine (Cys), and glutathione (GSH), SO42−, S2−, SO32− revealed minimal interference. Only Na2SO3 induced a significant fluorescence enhancement and color change, while the others caused negligible fluorescence increase. Such specific recognition is actually dependent on the ultrafast fluorogenicity of DCM-MQE for SO2, thereby enabling it to be differentiated from other nucleophilic reactive species and reductants via drastic distinction in response kinetics.28,29 Competition experiments confirmed minimal interference from other species, enabling specific Na2SO3 detection in complex environments.
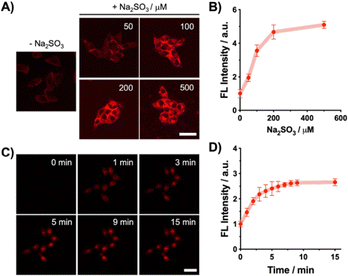
Given its robust response to Na2SO3in vitro, we evaluated DCM-MQE for imaging Na2SO3 in live cells. MTT assays confirmed negligible cytotoxicity below 10 µM (Fig. S7). Confocal imaging in HeLa cells (Fig. 2A and B) showed no fluorescence after treatment with 5 µM DCM-MQE alone, and a rapid, dose-dependent fluorescence enhancement upon incubation with Na2SO3 (10 min), intensifying with increasing concentrations (0–500 µM). It demonstrates excellent cell permeability and fast intracellular response kinetics (completed within ∼10 min), enabling real-time Na2SO3 imaging in live cells (Fig. 2C and D).
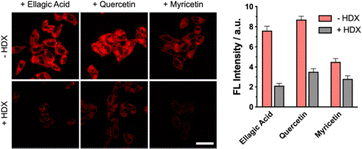
After confirming the high sensitivity and selectivity of DCM-MQE for Na2SO3, we employed this probe to monitor SO2 levels in HeLa cells during treatment with natural antioxidants (ellagic acid, quercetin, and myricetin). Notably, these antioxidants themselves exhibited negligible interference with the fluorescence signal of DCM-MQE, ensuring that the observed changes in fluorescence were solely attributable to SO2 generation (Fig. S8). Treatment with ellagic acid or quercetin (0–2 µg mL−1, 6 h) induced a concentration-dependent increase in fluorescence, which declined at 5 µg mL−1. Similarly, myricetin (0–5 µg mL−1, 6 h) enhanced fluorescence, but this effect diminished at 10 µg mL−1 (Fig. 3 and Fig. S9–S13). Further elevation of antioxidant concentrations led to decreased fluorescence, which may be attributed to the inhibition of sulfhydryl-containing amino acid oxidation processes, resulting in reduced SO2 generation (Fig. S9–S13).
To validate the mechanism, we used HDX – an inhibitor of aspartate aminotransferase30 that blocks SO2 generation from L-cysteinesulfinate – which significantly reduced the fluorescence compared to antioxidant-treated groups. Collectively, these results demonstrate that ellagic acid, quercetin, and myricetin induce reductive stress in HeLa cells, elevating intracellular thiol levels that undergo subsequent oxidation to generate SO2 (Fig. 3).
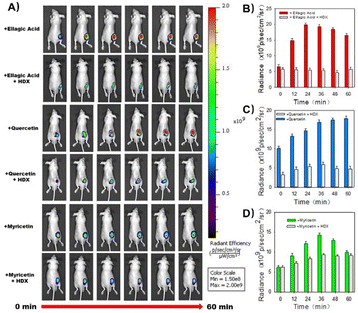
To further validate natural antioxidant-induced reductive stress in vivo, we conducted comprehensive experiments in tumor-bearing mice using DCM-MQE for SO2 imaging (Fig. S14 and S15). First, we established the probe's ability to detect both endogenous and exogenous SO2 derivatives at tumor sites. Mice were divided into three groups: (1) Control (PBS injection), (2) HDX inhibitor (HDX pre-injection), and (3) Sulfite group (Na2SO3 pre-injection). Fluorescence intensity was weaker in the HDX group than the control but stronger in the Na2SO3 group, diminishing over time—confirming DCM-MQE's ability to image SO2 derivatives in vivo.
Next, we evaluated SO2 induction by natural antioxidants. Tumor-bearing mice received subcutaneous injections of ellagic acid, quercetin, or myricetin (5 µg mL−1) for 5 h, followed by DCM-MQE administration. Fluorescence imaging revealed that all antioxidant-treated groups displayed markedly stronger signals than the HDX inhibitor group (Fig. 4). This enhancement, consistent across ellagic acid, quercetin, and myricetin treatments, directly demonstrated elevated SO2 generation in HeLa tumors during antioxidant exposure. Collectively, these in vivo results confirm that DCM-MQE effectively images SO2 dynamics and that natural antioxidants trigger SO2 production through reductive stress pathways in tumors. Notably, all HDX-supplemented control groups exhibited reduced fluorescence, indicating that in vivo, antioxidants upregulate SO2 by similarly enhancing biothiol levels, with SO2 generation resulting from their oxidation (Fig. 4).
Sulfur dioxide is an important gaseous signaling molecule closely associated with cellular redox homeostasis. In this work, we developed a red-emitting DCM-MQE probe for rapid and sensitive detection of intracellular SO2. Utilizing this probe, we demonstrated that natural antioxidants significantly elevated SO2 levels in both cells and tumor-bearing mice, and confirmed that antioxidants modulate SO2-mediated redox homeostasis by inducing reductive stress. We propose that SO2 may emerge as a novel reductive stress target, exhibiting significant concentration changes, distinct differences, and enhanced specificity compared to biological thiols under reductive stress conditions.
All authors have given approval to the final version of the manuscript. S. L.: writing – original draft and formal analysis. L. X.: conceptualization, methodology, writing – original draft, and formal analysis. H. L.: visualization, resources, and writing – original draft. Y. L.: investigation, funding acquisition. L. S.: supervision, writing – review and editing, and funding acquisition. G. L.: conceptualization, writing – review and editing. S. Y. supervision, conceptualization, writing – review and editing, and funding acquisition.
This work was supported by the National Natural Science Foundation of China (No. 22374044 and 22271089), the Feature Innovation Projects in Ordinary Universities in Guangdong Province (2022KTSCX228), and Hunan Province College Students Research Learning and Innovative Experiment Project (S202410542130) for financial support.
Conflicts of interest
There are no conflicts to declare.Data availability
All data generated or analyzed during this study are included in this published article and supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cc05915a.Notes and references
- J. B. Spinelli and M. C. Haigis, Nat. Cell Biol., 2018, 20, 745–754 CrossRef CAS
.
- S. Li, R. Liu, X. Jiang, Y. Qiu, X. Song, G. Huang, N. Fu, L. Lin, J. Song, X. Chen and H. Yang, ACS Nano, 2019, 13, 2103–2113 CAS
.
- M. Wang and R. J. Kaufman, Nature, 2016, 529, 326–335 CrossRef CAS
.
- A. Tirla and P. Rivera-Fuentes, Angew. Chem., Int. Ed., 2016, 55, 14709–14712 CrossRef CAS
.
- L. Luo, S. Chen, H. Jin, C. Tang and J. Du, Biochem. Biophys. Res. Commun., 2011, 415, 61–67 CrossRef CAS PubMed
.
- X. Pan, Y. Zhao, T. Cheng, A. Zheng, A. Ge, L. Zang, K. Xu and B. Tang, Chem. Sci., 2019, 10, 8179–8186 RSC
.
- D. Lin, T. Lu, X. Wang, X. Ye and Z. Liu, Chem. Sci., 2024, 15, 4824–4832 RSC
.
- H. Niu, Y. Zhang, F. Zhao, S. Mo, W. Cao, Y. Ye and Y. Zhao, Chem. Commun., 2019, 55, 9629–9632 RSC
.
- S. X. Du, H. F. Jin, D. F. Bu, X. Zhao, B. Geng, C. S. Tang and J. B. Du, Acta Pharmacol. Sin., 2008, 29, 923–930 CrossRef CAS PubMed
.
- B. C. Dickinson and C. J. Chang, Nat. Chem. Biol., 2011, 7, 504–511 CrossRef CAS PubMed
.
- B. Yang, J. Xu and H. L. Zhu, Free Radical Biol. Med., 2019, 145, 42–60 CrossRef CAS PubMed
.
- W. Zhang, F. Huo, Y. Yue, Y. Zhang, J. Chao, F. Cheng and C. Yin, J. Am. Chem. Soc., 2020, 142, 3262–3268 CAS
.
- W. Zhang, F. Huo, F. Cheng and C. Yin, J. Am. Chem. Soc., 2020, 142, 6324–6331 CAS
.
- V. S. Lin, W. Chen, M. Xian and C. J. Chang, Chem. Soc. Rev., 2015, 44, 4596–4618 CAS
.
- X. Yang, Y. Zhou, X. Zhang, S. Yang, Y. Chen, J. Guo and X. Li, Chem. Commun., 2016, 52, 10289–10292 RSC
.
- W. Xu, C. L. Teoh, J. Peng, D. Su, L. Yuan and Y. T. Chang, Biomaterials, 2015, 56, 1–9 CrossRef CAS PubMed
.
- Z.-Y. Lian, P.-D. Mao, K.-S. Jin, W.-N. Wu, L.-Y. Bian, Y. Wang, Y.-C. Fan, Z.-H. Xu and T. D. James, Sens. Actuators, B, 2025, 441, 137933 CrossRef CAS
.
- W.-N. Wu, Y.-F. Song, X.-L. Zhao, Y. Wang, Y.-C. Fan, Z.-H. Xu and T. D. James, Chem. Eng. J., 2023, 464, 142553 CrossRef CAS
.
- K. Li, L.-L. Li, Q. Zhou, K.-K. Yu, J. S. Kim and X.-Q. Yu, Coord. Chem. Rev., 2019, 388, 310–333 CrossRef CAS
.
- H. Wang, T. Zhu, W. Li, Z. Hu, X. Yang, Q. Zhang, P. Huang, J. Hu and Z. Feng, Anal. Chem., 2025, 97, 16439–16448 CrossRef CAS
.
- Q. Liu, Z. Dong, C. Chang, S. Chen, P. Sun, W. Shu, C. Zeng and W. Chi, Anal. Chem., 2025, 97, 4144–4150 CrossRef CAS
.
- H. Li, J. Wang, H. Kim, X. Peng and J. Yoon, Angew. Chem., Int. Ed., 2024, 63, e202311764 CrossRef CAS
.
- Q. Liu, C. Sun, R. Dai, C. Yan, Y. Zhang, W.-H. Zhu and Z. Guo, Coord. Chem. Rev., 2024, 503, 215652 Search PubMed
.
- Y. Bin, L. Huang, J. Qin, M. He, S. Zhao, Y. Huang and J. Zhao, Anal. Chem., 2025, 97, 9798–9808 CrossRef CAS PubMed
.
- L. Zeng, T. Chen, B.-Q. Chen, H.-Q. Yuan, R. Sheng and G.-M. Bao, J. Mater. Chem. B, 2020, 8, 1914–1921 RSC
.
- W. Gong, C. Zhang, X. Zhang and Y. Shen, Spectrochim. Acta, Part A, 2022, 278, 121386 CrossRef CAS PubMed
.
- J. You, H. Liu, Q. Pan, A. Sun, Z. Zhang and X. Shi, J. Anal. Test., 2023, 7, 69–78 CrossRef
.
- L. Yuan, W. Lin and Y. Yang, Chem. Commun., 2011, 47, 6275–6277 RSC
.
- J. Li, X. Yu, D. Shu, H. Liu, M. Gu, K. Zhang, G. Mao, S. Yang and R. Yang, Anal. Chem., 2024, 96, 7723–7729 CrossRef CAS PubMed
.
- S. Yang, X. Wen, X. Yang, Y. Li, C. Guo, Y. Zhou, H. Li and R. Yang, Anal. Chem., 2018, 90, 14514–14520 CrossRef CAS
.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |