Open Access Article
Open Access ArticleA 3D-bioprinted dermal-like scaffold incorporating fibroblasts and DRG neurons to investigate peripheral nerve regeneration†
Francesco
Formaggio‡
 *a,
Emanuela
Saracino‡
*a,
Emanuela
Saracino‡
 b,
Marianna
Barbalinardo
b,
Marianna
Barbalinardo
 c,
Eva
Clemente
c,
Eva
Clemente
 c,
Franco
Corticelli
d,
Sara
Buoso
c,
Franco
Corticelli
d,
Sara
Buoso
 e and
Simone
Bonetti
e and
Simone
Bonetti
 *c
*c
aDepartment of Pharmacy and Biotechnology, University of Bologna, Via San Donato 19/2, 40127 Bologna, Italy. E-mail: francesco.formaggio2@unibo.it; Tel: +39 051 20 9 5630
bInstitute for the Organic Synthesis and Photoreactivity, National Research Council of Italy, Bologna, Italy
cInstitute for the Study of Nanostructured Materials, National Research Council of Italy, Bologna, Italy. E-mail: simone.bonetti@cnr.it; Tel: +39-051639 8517
dInstitute for Microelectronics and Microsystems, National Research Council, Via P. Gobetti 101, 40129 Bologna, Italy
eKerline srl, Via Piero Gobetti 101, 40129 Bologna, Italy
First published on 19th May 2025
Abstract
Peripheral nervous system (PNS) regeneration is a rapidly advancing field with critical implications for addressing sensory impairments and neuropathic conditions. Dorsal root ganglion (DRG) neurons, essential for sensory transmission, exhibit regenerative potential through axonal regeneration. However, the mechanisms driving these processes are not yet understood. This study introduces an innovative 3D-bioprinted fibroblasts/DRG co-culture construct, specifically designed to investigate and characterize PNS regeneration and wiring mechanisms under both physiological and pathophysiological conditions. By characterizing bioink rheology and optimizing bioprinting parameters, we created a stable, biocompatible derma-like construct supporting cell adhesion and growth. Bioprinted 3T3 fibroblasts demonstrate high viability and proliferation, while DRG neurons exhibit enhanced neurite outgrowth and complex branching patterns within the co-culture system. These findings highlight the role of fibroblasts in promoting axonal regeneration and provide a robust in vitro platform for studying sensory system reinnervation. This model lays the foundation for developing personalized therapies for neuropathic pain and sensory dysfunction, advancing both fundamental neuroscience and translational medicine.
1. Introduction
Peripheral nervous system (PNS) regeneration is a vital and rapidly evolving area of research within neuroscience and regenerative medicine.1 The afferent division of the PNS, particularly the neuronal cluster located in the dorsal root ganglion (DRG), is responsible for transmitting sensory information from the peripheral tissues to the central nervous system (CNS) and is crucial for sensory function including touch, proprioception and pain.2 From an anatomical point of view, DRG cell soma originates a single process that bifurcates into a peripheral and a central branch. The peripheral branches innervate a target organ whereas the central branch passes into the spinal cord and transmits sensory information from the target organ to the CNS.3 Injuries to this component in the somatosensory pathway associated as diabetic neuropathy, lumbar spine pain, cancer pain, HIV, Herpes Zoster infections and peripheral nerve lesions can lead to severe sensory alterations, such as loss of sensory function, neuropathic pain and associated debilitating conditions.4 In case of peripheral nerve lesion, DRG neurons exhibit the potential to reinnervate the target tissue by the regeneration of the injured neuron. If an excessive gap is absent, damaged axons may grow along their original paths to reinnervate target tissues. Alternatively, if regeneration is hindered by physical obstacles or a non-conducive environment, a local mechanism of axonal branching may extend from uninjured neurons into adjacent denervated tissue (collateral sprouting) for a more rapid sensory recovery.5 Nevertheless, recurrent and persistent damages, associated with peripheral neuropathies may induce abnormal rewiring mechanisms. Particular emphasis has been recently given to these mechanisms in the skin, potentially involved in neuropathic pain conditions. For instance, the aberrant C-fibers sprouting in the skin may lead to erroneous reinnervation linked to the maintenance of chronic neuropathic pain.6 Considering that most types of neuropathic pain denote the combination of sensory loss and pain,7 artificial nerve guides could represent a promising approach to repair damaged nerves by mimicking the natural environment and promote the correct axonal regeneration.8 Several 2D systems has been developed to investigate DRG axonal regeneration and were fundamental to understand basic biological principles and initial cellular responses under investigation. Moreover, they are cost-effective methods with well-established protocols and exhibit high reproducibility. However, their inability to reproduce the in vivo environment, the cell–cell and cell-matrix interaction and the overall tissue architecture hindered more complex application and limited their translational potential.9,10 To overcome these challenges emergent studies focused their attention on regeneration processes by using engineered biomaterials and 3D scaffolds designed to mimic the natural extracellular matrix (ECM).11,12 In this scenario, the three-dimensional (3D) bioprinting represents an innovative approach at the intersection of tissue engineering and regenerative medicine.13,14 Bioprinting is a technique that enables the fabrication of complex 3D scaffolds by manipulating bioinks according to a predefined 3D-structure designed using computer-aided design (CAD) software or extrapolated from X-ray, computed tomography, or magnetic resonance imaging data.15 Microextrusion technology is particularly relevant in bioprinting due to its low development costs, broad range of printable materials, and high efficiency in preserving cell viability during and after the printing process.16 Furthermore, the bioprinting technique has been previously applied for treating tissue injuries and diseases, through both in vitro bioprinting implantation17 and in vivo bioprinting.18–20 The objective of this approach is to provide a more physiological micro-environment and provide a three-dimensional tissue framework which is lost in typical 2D cultures.21 In addition, since DRG neurons innervate target tissues to transduce sensory information, 3D bioprinting could be useful for creating multiple tissue constructs that mimic complex native tissue architectures. In this study, we apply advancements in bioprinting microextrusion technology to fabricate multilayered 3D artificial dermal constructs, to be used as an in vitro model for studying and characterizing PNS regeneration during pathophysiology. First, we characterized the rheological properties and printability of the bioink to ensure that the optimal extrusion requirements were met. Then, NIHT-3T3 fibroblasts were added to the bioink and extruded using a multitool, temperature-controlled, open-source bioprinter, to print a dermal scaffold able to mimic the extracellular environment of native tissues, providing support for cell adhesion and growth. Finally, primary murine DRG neurons were cultured on top of the construct to study the axonal regeneration. The final outcome of this study is the development of a bioprinted murine in vitro model designed to emulate innervated dermal-like tissue useful for modeling pathological processes and studying wiring mechanisms in peripheral sensory districts.2. Results
2.1. Rheological evaluation and printability characterization of bioink
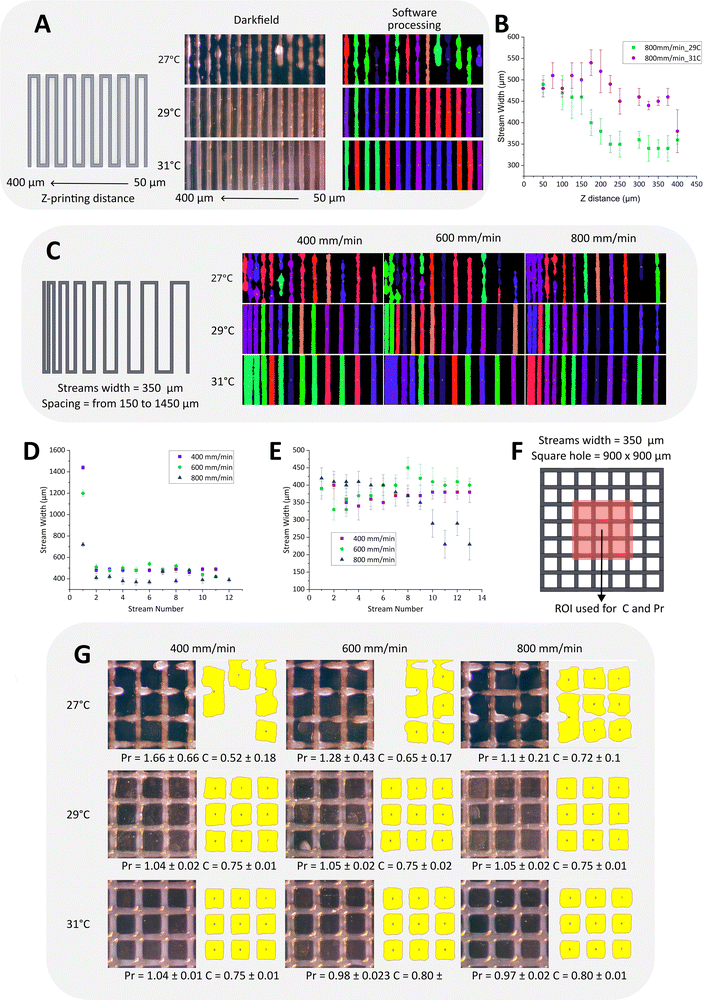
Bioink printability was assessed using various techniques such as rheology, evaluation of the printing distance (Z), filaments fusion, and the integrity of the multilayer structures. The rheological characterizations were used to extrapolate the best range of temperature usable in the printability test and to study the stress imposed on the cells during the extrusion process. As shown in Fig. 1A, the viscosities of the 2% fibrinogen solution and the 0.5% alginate solution had little to no variation when the temperature was changed, suggesting that the viscosity of both bioink components is not temperature-dependent.22 On the other hand, the viscosity of the 10% gelatin solution increased significantly at temperatures below 30 °C.22 The bioink exhibited similar viscous behavior, indicating that gelatin was the primary component responsible to the thermally mediated gelation of the mixture. As shown in Fig. 1B, the bioink exhibited different behaviors during the warming and cooling processes. The gelation temperature (Tg) during cooling was around 28 °C, while the melting temperature during warming was about 34 °C. Furthermore, the shear rate sweep test highlighted the shear-thinning properties of the bioink at different temperatures, showing a log-linear relationship between viscosity and shear rate (Fig. S2, ESI†). In the layer-by-layer approach, the distance in Z between the nozzle and the platform (Z-printing) is a critical parameter for the success of the print. If the nozzle is too close to the platform, the stream becomes wider than designed, and if it is very close to the substrate, it might lead to nozzle clogging or discontinuous printing. On the other hand, if the nozzle is too far, the layer does not adhere to the substrate, resulting in either a discontinuous or irregular strand. To check the correct Z-printing, the pattern shown in Fig. 2A was printed, changing the Z-printing for each vertical strand in steps of 50 μm, starting from 50 μm to 400 μm from right to left. The test was conducted at a standard printing speed of 800 mm min−1, within a temperature range of 27 °C to 31 °C. This range was selected to investigate printing behavior around the Tg of the bioink (28 °C), as determined by rheological analysis. It was slightly shifted toward higher temperatures to prevent nozzle clogging that may occur at temperatures below the Tg. Fig. 2A displays cropped images of the printed pattern at three different temperatures in the upper central section, while the lower central section shows processed images analyzed using the “extrusion stream width calculation” script.23 At 27 °C, the printed strands were not uniform due to the gelation of the bioink. Therefore, to analyze the optimal Z-printing, the samples printed at 29 °C and 31 °C were selected, and the average strand widths (Sw) were plotted in Fig. 2B as a function of the putative Z-printing. At the initial Z points, the Sw were comparable for both temperatures and exceeded the designed dimensions (300 μm).At 31 °C, the Sw consistently remained above the intended value due to the viscous behaviour of the bioink. Conversely, at 29 °C, a progressive decrease in Sw was observed between the Z-printing of 50 and 225 μm, with a reduction of approximately 150 μm. This resulted in a final Sw of 340 μm, more closely matching the designed specification. Notably, in the samples printed at 31 °C, the strands with smaller interspaces fused together and the individual strands had a greater width. Excluding the fused strands, the average width of the strands was constant and the standard deviations were low (±25 μm), due to the viscous behavior of the bioink printed at a temperature higher than its Tg (Fig. 2D). The printing speed did not significantly affect the printing results, except for a decrease in fusion phenomena at 800 mm min−1, considering that the strands appear to be slightly finer than the ones extruded at lower speeds. At 29 °C, the fusion of strands was absent, all printing speeds produced 13 independent strands with an average width close to the designed value (Fig. 2E). Compared to the higher temperature, the average width of the strands varied slightly within the same samples and the standard deviation of each strand was higher, because the extrusion temperature was close to the Tg and the streams became more wrinkled. The printing speed affected the quality of the sample at 800 mm min−1, producing strands with higher widths variability within the same sample, compared to lower speeds. To study the integrity of the multilayer structures, a grid was printed using different printing conditions as in the previous tests (Fig. 2F). Fig. 2G shows the cropped images of the samples for each condition and the resulting images processed with the script named ‘3-grid analysis’,23 used to calculate C and Pr.22 As in the previous experiment, at 27 °C the strands were disconnected and irregular. The holes of the grids were irregular and sometimes interconnected, showing C and Pr values that were far from the optimal ones. At higher temperatures, the samples printed at different speeds showed a good Pr, close to 1. Analysing C, the samples printed at 31 °C had slightly higher values compared to π/4, indicating that the printed construct would fuse at the cross-sites. In contrast, at 29 °C C was close to the desired value, meaning that the layers were easily distinguished. Based on these printability tests, 29 °C was chosen for printing the models for biological studies. Among the two optimal speeds, 400 and 600 mm s−1, the lower speed was selected to decrease the shear stress applied to the cells during extrusion.
2.2. Bioprinting of a dermal-like model
Once the bioprinting parameters were defined and settled, we extruded mouse 3T3 fibroblasts and monitored the 3D-bioprinted samples over time. Fig. 3A illustrates a typical 6 well plate containing 10 × 10 × 1 mm bioprinted constructs. Fluorescence microscopy was performed to monitor both the stability of the 3D scaffolds and to observe the cell morphology and distribution within the bioink matrix in which 3T3 fibroblasts were embedded. Fig. 3C shows fluorescent microscopy images captured at 3, 5, 7, and 10DIV of 3T3 fibroblasts extruded in the constructs. Collectively, the results indicate the homogeneous distribution of cells in the scaffolds (Fig. 3). Moreover, the increase in cell number over time, as shown in the fluorescence images, confirmed the trend of cell growth and proliferation. To quantify and compare the cell viability on samples over time, we performed resazurin reduction assay in triplicate after 3, 5, 7 and 10DIV on each sample (Fig. 3B). The values reported in the plot, as time-course of the cell viability of the cells in the 3D-matrix, are proportional to the cells metabolic activity and are correlated with the presence of a high number of viable cells in the single scaffold. In conclusion, data suggests that 3T3 fibroblasts proliferate into the constructs in a time-dependent manner. We suppose, by a final evaluation, that the bioink formulation provides a supportive environment for cell survival and proliferation over time. In addition, we performed SEM at 5DIV (Fig. S4A, ESI†). According to fluorescent microscopy, we observed an abundant number of 3T3 fibroblasts embedded in the superficial layers of the scaffold. Notably, cells display an elongated morphology, typical of healthy and adhering fibroblasts, indication of efficient interaction with the material blend. Moreover, to observe more in detail the intrinsic 3D structure of the matrix in which cells are dispersed, we also performed SEM on the scaffold fabricated with no cells. The image is reported in Fig. S4B (ESI†). It highlights a strong nanoscale porosity/fibrillary surface, an important feature that may improve the property nutrients uptake, useful once cells are embedded into the matrix.2.3. Cultured DRG on the bioprinted constructs and neurite outgrowth
Once the printability of mouse 3T3-fibroblasts was confirmed, we set up a protocol to generate a more complex co-culture system, by adding DRG neurons isolated from young adult mice (2–3 months age). In these experiments, DRG neurons were directly seeded onto the bioprinted constructs, 2 days after the 3T3 fibroblasts extrusion. As shown in Fig. 4A, for this experiment we designed circular structures with thick edges to confine the sensory neurons and promote their attachment (external diameter: 10 mm, height: 1 mm, edge width: 1 mm, internal heights: 0.2–0.4–0.6 mm). The selected thickness range for the internal part of the sample mimics the same thickness as the murine in vivo derma.24 The result of an extruded sample and the image of the maintained sample structure after 14 DIV are shown in Fig. 4B. After 2 days from the derma-like construct bioprint, DRG neurons were seeded both in the presence or absence of 3T3 fibroblasts to characterize the effect of the co-culture system on neuronal viability and neurite outgrowth (Fig. 4C). DRG neurons plated onto 10% gelatin/2% fibrinogen/0.5% alginate (w/v) bioink formulation show excellent viability since 2DIV, with more than 80% of DRG characterized by neurite branching (Fig. 4D, upper panels). Confocal microscopy, performed by staining DRG with PGP 9.5 (Fig. 4D, red fluorescence) suggests that DRG somas, on the samples without NT3-fibroblasts, were concentrated at the top of the surface with minimal insertion into the material (Fig. 4D upper panels and Video S1, ESI†). At 7DIV, the cell soma position was conserved over time, resembling the 2DIV condition, while neuronal sprouting was greatly increased and characterized by intricate branching in multiple directions throughout the bioprinted construct (Fig. 4D, lower panels). Neurite outgrowth was mostly localized on the surface with partial entrance in the material at both 2DIV (Video S1, ESI†) and 7DIV (Video S2, ESI†). DRG were also seeded on 3T3 fibroblasts + bioink formulation. Here, we did not observe significant difference in cell viability (Fig. 4E): DRG soma exhibited the typical round shape with extensive branching at 2DIV in the 90% of the neurons (Fig. 4E, upper panels), while 3T3 fibroblasts maintained the typical fibroblast morphology with soma elongated and primary branching. Their typical elongated morphology characterizes their distribution throughout the entire 3D matrix. Interestingly, the confocal microscopy on DRG at 2DIV, highlights that while DRG soma were almost totally enclosed into the bioprinted surface, their neurites were capable to enter inside the intrinsic 3D structure of the bioprinted material contacting the 3T3 fibroblasts (Fig. 4F left panel). At 7DIV the neurite sprouting was remarkably increased with a rather complicated neurite net, wiring the majority of the bioprinted constructs (Fig. 4E and F left panel). At the same time, also the 3T3 fibroblasts cell number increased, suggesting excellent long-term stability of the bioprinted co-culture system. At 7DIV the DRG neurites reached the entire 3D structure, even the deeper intrinsic fibroblast layers, as shown by the PGP 9.5 fluorescence staining, which co-localized with the fibroblast-derived GFP green signal in the z-stack reconstructed images (Fig. 4F). Notably, DRG neurites interacted with fibroblasts both at the level of the soma and along the elongated fibroblast processes, as shown in Fig. S5 (ESI†) (white arrows, merged image). This observation suggests that DRG neurons and fibroblasts can coexist within the same area, enabling physical interactions and potentially facilitating biological crosstalk. Consequently, a detailed analysis of DRG neurite properties was performed with the goal to demonstrate that our model of co-culture combined with 3D bioprinting increases axonal regeneration and supports the development of complex and extensive neurite outgrowth. The staining and immunofluorescence analysis of PGP 9.5 permits the visualization of primary neurite but also secondary, tertiary and quaternary levels of branching. Fig. 5A and B illustrates that in the presence of green GFP-positive 3T3 fibroblasts, DRG neurons exhibit an increase in neurite branching. When DRG were cultured alone, the total arbor length (Fig. 5C), which is the sum of all branch lengths, was 565 ± 655 μm after 2DIV and significantly increased at 7DIV (1180 ± 732 μm, Fig. 5C, left panel). When 3T3-cells were present, the total nerite length at 2DIV was three times longer (1784 ± 1254 μm), when compared to the sample where fibroblast was not present. At 7DIV a fourfold increase was measured when 3T3-fibroblasts were present in the bioprinted construct (3839 ± 1962 μm). In terms of primary process number (Fig. 5C), DRG alone shows a significant increase over time, whereas DRG + 3T3-fibroblasts exhibit a slight non-significant increase in the primary process number between 2DIV and 7DIV, indicating that DRG + 3T3 fibroblasts at 2DIV already nearly completed the first phase of neurite sprouting, in which thick primary branches are created. In contrast, fine processes falling into secondary, tertiary and quaternary sub-classes indicated that DRG + 3T3 fibroblasts are more likely to display complex branching patterns. The neurite numbers significantly increased at 7DIV, particularly at tertiary and quaternary levels (Fig. 5D, left and central panels). This increased branching capacity was also reflected in the arbor length, which differs significantly between the two groups. DRG alone demonstrated simpler branching patterns, with increased primary neurite length over time and slight differences with the DRG + 3T3 fibroblasts, confirming that the main branching structures did not vary between the different sample types (Fig. 5D, right panel). In contrast, at more complex arborization levels, the specific branch length was dramatically increased in DRG + 3T3 fibroblasts, specifically at secondary, tertiary and quaternary levels (Fig. 5E). Interestingly, DRG alone displays a decreased trend of neurite length when looking at the axonal tree from simple primary processes to more complex branches. The primary processes were the longest, whereas moving towards the finest structures the branches were progressively shorter. The opposite trend was observed in DRG + 3T3 fibroblasts where fine processes (secondary, tertiary and quaternary) mostly contributed to the total neurite length, reflecting more complex and highly branched structures.3. Discussion
The transition from 2D models of the tissue-like system to 3D approaches has been driven by recent advancements in tissue engineering technologies. Among them the nervous system represents one of the most study and complicated system to reproduce.25,26 Bioprinting has emerged as a highly promising technique for recapitulating representative tissue models, being able to produce complex 3D structures composed of heterogeneous materials.27 Herein, we present a 3D bioprinted murine in vitro dermal-like model, utilized to study wiring mechanisms driven by primary DRG neurite outgrowth, laying the groundwork for the exploration and development of a sensory model. When generating 3D bioprinted complex tissues such as the sensory system, one of the most crucial factors to consider is the investigation of biofunctional and biocompatible materials, processable using printing techniques. Various bioprinting technologies have been used to generate complex tissues but the extrusion-based method is the most employed due to its relative simplicity, affordability, and scalability.16 In this study, to emulate the dermal tissue components of the sensory system, we selected a microextruded 3D-printed scaffold-free model, previously validated using human fibroblasts.28 This model was also selected for the presence of 2% fibrinogen, which, as demonstrated in the literature, is a suitable component for DRG maintenance and neurite outgrowth, upon conversion to fibrin after thrombin treatment. Fibrin contains cell-binding domains, offering potential for live cell encapsulation and neurite extension. Additionally, it has been used in previous studies as a material for axonal guidance.29 Gelatin is the primary component of the bioink used in this study, present at the highest concentration. Processing highly concentrated gelatin bioinks using extrusion technology presents challenges due to the rheological properties of this component. To address different issues, we evaluated the printability of the selected bioink using a rheometer and the open-source microextrusion bioprinter used to bioprint the derma-like models. The printed samples were analyzed using custom-developed tool scripts for automated standard printability tests, that we published open-source for the use by bioprinting community in future studies.23 Printability was evaluated by examining the printing distance in the Z-axis, filament fusion, and the integrity of the multilayer structures, modifying temperature and printing speed. The parameters chosen to bioprint the dermal-like model was: 200 μm (Z-printing), 29 °C (printing temperature) and 400 mm min−1 (printing speed). The Z-printing value selected by the test aligns with the literature,30 corresponding to 80% of the inner tip diameter (250 μm) used in the bioprinting process. The printing temperature of 29 °C, like those reported in the literature,28 is the optimal condition for bioprinting the bioink used in this study with the proposed thermally controlled extrusion system. As demonstrated by the rheological experiments, the optimal Tg for the bioink under cooling conditions is approximately 28 °C. However, the use of a custom-made heating system requires an increment of the employed extrusion temperature by 1 °C compared to the theoretical value. The heating system covers the syringe up to 2 mm from the tip to ensure uniform thermalization up to the nozzle. Nonetheless, to maintain sterility during printing, the heating element is not in direct contact with the syringe. This design creates an air gap, a poor thermal conductor, necessitating a 1 °C increase to achieve the optimal printability temperature for the bioink. The chosen printing speed, instead, is the slowest tested because, despite yielding similar results to the highest speed, it better preserves the viability of the extruded cells by reducing shear stress.31 After setting the optimal bioprinting parameters, we utilized this extrusion-based bioprinting to generate a dermal model that replicates dermis properties. Our approach opts for a gelatin/fibrin-based bioink with a small quantity of alginate to improve the mechanical stability and the printability of the construct.28,32,33 This formulation enabled us to obtain a hydrogel with a favorable stability for up to 14DIV, creating a supportive environment for the 3T3 fibroblasts viability and cell growth. Three days after printing, 3T3 fibroblasts displayed a homogeneous cell distribution in the matrix, followed by an exponential growth between 3 days and 10 days (Fig. 3B). After 10 days, 3T3 fibroblasts populated the 3D bioprinted construct, suggesting good biocompatibility and long-term stability (Fig. S6, ESI†). In our 3D culture system, the observed time-dependent increasing cellular reduction capacity (Fig. 3B) is interpreted in the context of the well-documented biological behavior of NIH-3T3 fibroblasts, which are known for their robust proliferation in permissive hydrogel environments with comparable biochemical and mechanical properties.34–36 Moreover, fibroblast cells showed the typical elongated shape and a few polarized processes elongating from the cell body. This feature is generally present in fibroblasts when cultured in 3D scaffold.37 In addition, our observations are in line with previous studies on the same bioink preparation and cell type, where the dermal manufacture was colonized after 7 days.28 The main objective of this work was to generate an in vitro model of dermal matrix which promoted the axonal regeneration of DRG neurons. We focus on investigating the benefits and effects of bioprinted fibroblasts cells in co-colture with primary DRG and explore the specific contribution of fibroblasts within the constructs. DRG neurons cultured on top of empty bioprinted constructs showed good viability as indicated by the extensive neurite sprouting after 2DIV and after long term maintenance (7DIV). In this context, the formulated and tested bioink may mimic the ECM composition in terms of mechanical features of dermal and DRG tissues.38,39 However, the degree of complexity of the branching patterns was not remarkably increased between 2DIV and 7DIV (Fig. 5), where mostly primary neurite elongation occurred with limited development of highly branched structures. It has been recently shown that by using extrusion technology it is possible to print DRG neurons with good viability and functionality, while the neurite outgrowth is usually rather limited.40,41 To better mimic the physiological innervation of a dermal construct, we seeded the DRG on top of bioprinted dermal-like scaffolds containing 3T3 fibroblasts. Interestingly, we observed that the presence of fibroblasts increased the total neurite length up to more than 3000 μm after 7 days in culture. Moreover, we observed a higher degree of complexity of the branching patterns, suggesting that the presence of the fibroblasts can contribute directly to axon regeneration and modulation. A few previous reports indicate that DRG neurons co-cultured with skin derived fibroblasts emit multiple neurites in the absence of NGF.42 Jerregård et al. used skin-derived fibroblast and 3T3 fibroblasts to promote neurite outgrowth and this effect was mediated by the release of Neurotrophin-3 (NT-3). In another study the total neurite growth on a sciatic-derived fibroblast monolayer was calculated to be 220 ± 20 μm for small neurons (10–20 μm soma) and 508 ± 39 μm for medium neurons (20–35 μm soma) after 2DIV.43 More recently, it has been shown that dermal fibroblasts and keratinocytes produce both neurotrophic factors and chemorepellents, e.g. NGF,21,44 semaphorin 3A45 and growth factor-inducible-14 promoted neurite outgrowth.46 These factors exert opposite effects on nerve fiber growth, enabling a precise control of cutaneous innervation.47 Beside skin-derived cells, peripheral nerve-derived fibroblasts promoted neurite outgrowth in adult DRG induced by soluble factors release;48 in this study the longest neurites were in the range of 80–120 μm after 2DIV. In a recent in vivo study, a scaffold-free 3D bioprinted conduits composed entirely of fibroblast cells promoted nerve regeneration in a rat sciatic nerve model.49 In this context, our 3D-bioprinted DRG/fibroblast co-culture system stimulated a noteworthy neurite regeneration when compared to 2D DRG/fibroblast co-cultures or in 3D-bioprinted DRG.50 It is plausible to suggest that the presence of fibroblasts in combination with a 3D matrix can have a synergistic effect on neurite outgrowth and provide an excellent platform for regeneration studies. Indeed, combination of 3D bioprinted scaffold with different supportive cells may represent a promising approach to induce axonal guidance and provide orientation for DRG neurite outgrowth. To further refine the model, upcoming studies will explore how different fibroblast densities affect neurite regeneration, aiming to determine the most effective cell ratios. Besides fibroblasts, Schwann cell represent crucial players in axonal regeneration as previously reported in bioprinted scaffold44 or in hydrogel cell culture inserts.51 The integration of supportive cells with bioprinted scaffolds characterized by differential stiffness could shed light on the effect of different mechanical properties on axonal guidance.52 Moreover, by utilizing various scaffold materials and bioprinting techniques, we will conduct quantitative analyses of neurite outgrowth to offer a more comprehensive evaluation of the model's performance. Overall, these models could be useful for investigating the complex rewiring mechanisms of sensory neurons, exploring neuropathic pain mechanisms in the skin, and potentially being implemented for drug testing. In addition, these platforms could be useful to induce the full functional maturation of human induced pluripotent stem cells derived sensory neurons with greater translational potential.254. Conclusions
Recent advancements in tissue-engineering technologies represent a shift from conventional 2D cell system models to more advanced 3D culture systems. Notably, 3D bioprinting shows significant potential for creating tissue models that better replicate physiological conditions, especially when considering complex systems such as nervous tissue. Herein, we present a novel murine in vitro model, designed to emulate innervated dermal-like tissue, by combining a 3D-printed dermal construct with primary DRG neurons. Analysis of this construct highlights the model's suitability, demonstrating strong biocompatibility and bio functionality. The first is evidenced by relevant cell viability in the co-colture system and the latter by extensive DRG neurite regeneration. This study is a proof of concept to validate and characterize biomaterials and procedures needed to develop a heterogeneous in vitro sensory model. The developed construct aims to advance in the study of axonal regeneration implicated in several neuropathic disorders and pain states. Moreover, we aim to support both fundamental research and progress in personalized medicine by introducing the prospective use of patient-derived fibroblasts in the bioprinting dermal construct. In conclusion, the potential of this study could be the in vivo application of synthetic dermis with an improved re-innervation capacity. The following strategy could facilitate the appropriate innervation of damaged tissues, thereby promoting the restoration of sensory function in affected regions.5. Experimental
5.1. Bioprinting setup
The bioinks were printed using an extrusion-based 3D bioprinter like those described in literature,53 with several upgrades including a thermally controlled syringe pump tool. This system, shown in Fig. S1 (ESI†), was essential for achieving accurate printing of materials containing gelatin. Additionally, it has been inserted a support plate with an interchangeable holder adaptable to Petri dishes, microwells, microscope slides and microscope glasses. Detailed sketches of all designed parts are provided in the ESI† (Fig. S1).23 All parts were designed using SolidWorks 2021 and produced through various methods such as 3D printing and CNC milling. The printed parts were fabricated in PLA (3D Verbatim PLA) on a Prusa MK3S (Prusa, Prague, Czech Republic). All the bioprinted parts and printability test models were designed and exported in STL format using SolidWorks 2021, and processed using the slicer Simplify3D (Cincinnati, USA).5.2. Bioink preparation
The bioink was composed of a mixture of 2% (w/v) fibrinogen from bovine plasma (Sigma-Aldrich, F8630), 0.5% (w/v) sodium alginate (Sigma-Aldrich, A2158), and 10% (w/v) gelatin from porcine skin (Sigma-Aldrich, G1890), diluted in Dulbecco's modified Eagle's medium 1× (DMEM) ([+] 4.5 g L−1D-glucose, [+] L-glutamine, [−] sodium pyruvate, Gibco).28,32 Fibrinogen was dissolved in DMEM at 37 °C for 1 hour, under gentle stirring. Once a homogeneous blend was obtained, sodium alginate was added; the solution was kept under agitation for 3 hours to guarantee complete powder dissolution. Finally, gelatin was added to the blend and left under agitation for 4 hours. Afterwards, trypsinized cells were seeded into the obtained bioink for a final cell concentration of [1 × 106] mL−1, and the mixture was used for printing. Additional details are provided in the ESI.†5.3. Rheological test
The rheology measurements were performed with an Anton Paar MCR 102 Rheometer and Anton Paar software for data acquisition and analysis. A solvent reservoir was filled with water prior to every experiment to prevent the sample from drying. The viscosity (η) was measured with a cone plate CP50-1 (49.975 mm diameter, 1.001° cone angle, 101 μm truncation gap), by recording the flow curves in the range of shear rates from 0.1 to 100 s−1, at 27, 29, 31 and 33 °C. The temperature sweep oscillatory test was performed to measure the moduli (G′ and G′′) and the complex viscosity (|η*|) as a function of the temperature, through oscillatory measurements under a shear strain of 1% and frequency of 10 rad s−1, with a parallel plate PP50 (49.971 mm diameter) at a 0.5 mm gap. The warming and cooling ramps were measured between 20 and 40 °C, with a rate of 1 °C min−1.The bioink was incubated at 40 °C before testing and the temperature-controlled testing plate was also set at 40 °C as an initial temperature. The gelation (Tg) and melting (Tm) temperature were calculated as the temperature at which G′ intersected G′′ during the cooling and worming ramp, respectively.
5.4. Bioprinting of printability test samples and of bio-construct
Prior to the test, the empty syringe (Hamilton 1001TLL), the tip and the bioink were kept at 37 °C for 30 minutes. Afterwords, the syringe was filled with bioink to print the samples for the printability test, and with bioink + fibroblasts to print the bio-construct. The syringe was loaded into the syringe pump tool, pre-heated to the printing temperature and left in the tool for 20 minutes to ensure uniform temperature distribution before starting the printing test. All samples were printed using a 25G tip (925125-DHUV Metcal) sterilized with 70% ethanol and dried with N2 before printing. During the printing process, the printer plate was kept at 23 °C. Once printed, the bio-construct was incubated at 37 °C for 30 minutes in a solution of 0.1 M CaCl2 and 20 U mL−1 thrombin (Sigma-Aldrich, France, CAS No. 9002-04-4). A stereomicroscope system (KERN OBL 135C832) was used to acquire images of printability test samples.5.5. Stream and grid analysis
The images acquired from the printability test were analyzed using Python scripts specifically written for this purpose,23 after using ImageJ-Fiji54 to convert them into binary images through the “Make binary” function. The script called “Extrusion stream width calculation” was used to analyze the images to characterize the correct Z height for printing and filament fusion. This script counts the number of streams and measures their average width and standard deviation, generating a .txt file summary and an image with colored and labelled streams. To semi-quantify printability, the grid images were processed with the script called “3-grid analysis”. This script analyzed the holes formed from the connection between the upper and lower printed streams in the fabricated construct, calculating the circularity (C) and printability (Pr).22Circularity is defined as:
The bioink printability (Pr) is defined as follows, based on the square shape of the grid holes:
5.6. Cell culture preparation and maintenance
Mouse embryonic fibroblast cells (NIH-3T3/GFP, mentioned in the text as 3T3-fibroblasts) were cultured under standard conditions in the DMEM medium, supplemented with 10% (v/v) fetal bovine serum (FBS, Merck) 2 mM L-glutamine, 0.1 mM MEM Non-Essential Amino Acids (NEAA, Merck), 100 U mL penicillin and 100 U mL streptomycin. Cells were maintained in culture flasks in a humidified incubator set at 37 °C with 5% CO2. When confluence was reached, 3T3 fibroblasts were dispersed using trypsin–EDTA 0.25% and the cell suspension was extruded with the bioink directly onto 6 well plates and 19 mm (diameter) circular coverslips at a density of 1 × 106 cells per mL and maintained in the incubator. Medium was completely replaced every 2–3 days until the day of the experiments.5.7. Preparation of dorsal root ganglion neurons
Primary cultures of DRG neurons were established from adult male mice aged 8 to 12 weeks. All procedures adhered to the European Community Council Directive of 24 November 1986 (86/609/EEC) and were approved by the Ethical Committee of the University of Bologna (protocol number 141/2019PR). The preparation followed previously described protocols with minimal modifications.55,56 Briefly, prior to decapitation, the mice were anesthetized using halothane. Ganglia were dissected from each mouse and transferred to ice-cold 1× Dulbecco's phosphate-buffered saline (DPBS, Gibco). The ganglia roots were then carefully sectioned using micro dissecting scissors. After rinsing in DMEM, the ganglia were incubated in DMEM containing 5000 U mL−1 type IV collagenase (Worthington) for 45–75 minutes at 37 °C with 5% CO2. Following this, the ganglia was washed twice with FBS-containing medium and mechanically dissociated using 0.5 mm and 0.6 mm sterile needles. The resulting cells were centrifuged at low speed for 10 minutes and filtered using a 70 μm cell strainer. By using a hanging drop method, 5000 cells per samples were seeded in DMEM medium supplemented with 10% FBS (Gibco), 50 ng mL−1 nerve growth factor (NGF, Gibco), and 1.5 μg mL−1 cytosine β-D-arabinofuranoside (AraC, Sigma) and incubated for 30 minutes. Fresh media was then added to fully cover the specimen and maintained in a 37 °C incubator with 5% CO2 and media was entirely changed every 2–3 days. Neuronal samples were characterized after 3 and 7 days in vitro (DIV) by optical and confocal imaging and by scanning electron microscopy (SEM).5.8. Cell viability
Cell viability was determined by resazurin reduction assay, an oxidized form of a blue redox indicator. When incubated with viable cells, the reagent is reduced changing color from blue to red. The dermal-like constructs (square-shaped, 10 mm × 10 mm × 1 mm) were bioprinted in a 6-well plate, with each well containing one sample composed of bioink and 3T3 fibroblasts. The samples were maintained in complete cell medium. After incubation, resazurin reagent was added at 10% volume of medium contained in each sample after 3, 5 and 10 days and incubated for 4 h at 37 °C with 5% CO2. Subsequently, aliquots from each sample were transferred to a 96 multiwell plate for absorbance measurement at λ 570–630 nm (Thermo Scientific Varioskan Flash Multimode Reader). All data represented absorbance mean ± standard error of the mean (SEM). Fluorescent microscopy, from the same preparations, was performed at four different time points (3, 5, 7 and 10 days). The images were captured using a Nikon Eclipse Ti microscope, equipped with appropriate fluorescence filters, 4, 10 and 20× magnification and a Digital sight DS-U3 camera.5.9. Scanning electron microscopy
Samples were fixed with 2.5% glutaraldehyde in phosphate buffer saline (PBS) at 4 °C, for 1 hour. Each sample was then rinsed three times in PBS for 5 minutes at room temperature; three further rinsings with distilled water were then performed. Then samples were sequentially dehydrated in 50%, 75%, 95% and 99% ethanol. Dried specimens were sputter-coated (Edwards S150B) with gold (10 nm thickness) before analysis with a Zeiss LEO 1530 FEG.5.10. Immunofluorescence and confocal microscopy
Bioprinted samples were processed 2DIV and 7DIV after DRG plating. Each sample was carefully washed twice in PBS for 30 min and the cells were fixed using 4% formaldehyde at RT for 20 minutes. Following two 30-minute washes in PBS, they were blocked with a solution containing 5% bovine serum albumin (BSA) and 0.05% Triton X-100 in PBS for 1 hour at RT. Subsequently, constructs were incubated overnight at 4 °C with the primary antibody (PGP 9.5, ab108986) in 1% BSA with 0.05% Triton X-100. The next day, the preparations underwent 30-minute washes with 1× PBS. The secondary antibody Alexa 568 conjugated (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, Jackson Immuno Research), was then applied at RT for 2 hours. After another 3 × 20-minute PBS rinse, the samples were mounted with DAPI-containing Prolong Anti-Fade (Molecular Probes-Invitrogen) and optically imaged with a Nikon Ti2 microscope. Moreover, 3T3 fibroblasts were imaged and shown in green because of endogenous GFP expression (488 nm).
400, Jackson Immuno Research), was then applied at RT for 2 hours. After another 3 × 20-minute PBS rinse, the samples were mounted with DAPI-containing Prolong Anti-Fade (Molecular Probes-Invitrogen) and optically imaged with a Nikon Ti2 microscope. Moreover, 3T3 fibroblasts were imaged and shown in green because of endogenous GFP expression (488 nm).
5.11. Image acquisition
Z-Stacks of individual DRG were imaged on a confocal microscope (Nikon Ti2/A1 plus camera) with the accompanying NIS-software (Nikon). Multi-track acquisition was performed with excitation lines set at 488 nm at 0.5% and 561 nm at 0.35%. Z-Stack images of bioprinted constructs were acquired using a Plan-Apochromat 20×/0.75 air objective. Stacks were collected at a 0.4 μm slice interval, stepping through the entire scaffold. Frame size was set to 1024 × 1024 pixels. Scan averaging was set to 2 to cut down on background noise. All z-stack files were saved in (.nd2) format before export and quantitative analysis.5.12. Neurite quantification
Neuronj plugin in imageJ was used to assess neurite outgrowth and branching.57,58 Neurite tracing was manually performed by two independent operators on blinded images where single DRG neurons were clearly recognized in non-overlapping territories (soma position and DAPI staining was used to discard multiple cells in the same field of view). The obtained z-stack reconstructions (Maximum Intensity Projection) were contracted, and the background was subtracted. Subsequently, the images were transformed in 8 bits and loaded on Neuronj. Primary neurites were defined as the main branches extending directly from the cell soma, and secondary, tertiary and quaternary processes, extending from these primary branches. The neurite number and growth were quantified, according to the different arborization sub-classes.595.13. Statistical analysis
Data are reported as the mean average ± S. E. M. of the number of samples (n). Experiments were performed at least three times on independent primary DRG culture and extruded 3T3 fibroblasts from different batches. The analyses and statistics were performed with OriginPro 2023. Neurite quantification was compared by two-way ANOVA followed by Sidak's multiple comparisons test with p < 0.05 considered statistically significant.Author contributions
Francesco Formaggio designed and conducted the DRG experiments, participated in project supervision, and co-authored the manuscript. Marianna Barbalinardo and Emanuela Saracino performed the 3T3 fibroblast viability experiments, with Emanuela Saracino also assisting in manuscript writing and revision. Simone Bonetti carried out the printability tests, developed the bioprinter and open-source software, fabricated the dermal-like scaffold, supervised the project, and co-authored the manuscript. Franco Corticelli performed the SEM experiments and collected the corresponding images. Eva Clemente and Sara Buoso conducted the rheological tests, with Eva Clemente additionally contributing to manuscript editing. The study was collaboratively designed by Francesco Formaggio, Emanuela Saracino, and Simone Bonetti.Data availability
The open-source procedure for collecting printability data and the schematics of the bioprinter upgrades developed in this article are publicly available in the GitHub repository: https://github.com/simobone19?tab=repositories. The datasets related to this study will be publicly available on the Zenodo, ESI† and github platform upon the article's acceptance or upon specific request by the reviewers. While we fully support the journal's data-sharing policy, this decision has been made to ensure the protection of the work described in the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was performed with the support of Fondazione del Monte di Bologna e Ravenna AEP_C_2023_Prot. No. 1277bis/2023 and PRIN PNRR 2022-20224KF8HE. E. S. and S. B. acknowledges PNRR MUR project ECS_00000033_ECOSISTER. We are grateful to F. Bona and V. Ragona from ISMN, CNR, Bologna, for their precious technical support, to Dr. A. V. Dediu from ISMN, CNR, Bologna, that supported this work by allowing the setting up of a new laboratory dedicated to 3D-bioprinting. We also thank Dr. Davide Pasqualini from the Dept. of Pharmacy and Biotechnology at the University of Bologna for his support in cell culture preparation. We are grateful to Prof. Marco Caprini who supported this work by proving the cell culture and molecular biology laboratories and to Sveva Giovannini and Laura Cinti for their precious help in immunofluorescence experiments.Notes and references
- A. Accardo, C. Cirillo, S. Lionnet, C. Vieu and I. Loubinoux, Brain Res. Bull., 2019, 152, 202–211 CrossRef CAS PubMed.
- A. I. Nascimento, F. M. Mar and M. M. Sousa, Prog. Neurobiol., 2018, 168, 86–103 CrossRef PubMed.
- K. Jang and S. M. Garraway, Neurobiol. Pain, 2024, 15, 100151 CrossRef CAS PubMed.
- N. B. Finnerup, R. Kuner and T. S. Jensen, Physiol. Rev., 2020, 101, 259–301 CrossRef PubMed.
- A. DeFrancesco-Lisowitz, J. A. Lindborg, J. P. Niemi and R. E. Zigmond, Neuroscience, 2015, 302, 174–203 CrossRef CAS PubMed.
- V. Gangadharan, H. Zheng, F. J. Taberner, J. Landry, T. A. Nees, J. Pistolic, N. Agarwal, D. Männich, V. Benes, M. Helmstaedter, B. Ommer, S. G. Lechner, T. Kuner and R. Kuner, Nature, 2022, 606, 137–145 CrossRef CAS PubMed.
- E. Senba, K. Katanosaka, H. Yajima and K. Mizumura, Neurosci. Res., 2004, 50, 257–262 CrossRef CAS PubMed.
- J. N. Campbell and R. A. Meyer, Neuron, 2006, 52, 77–92 CrossRef CAS PubMed.
- K. Duval, H. Grover, L. H. Han, Y. Mou, A. F. Pegoraro, J. Fredberg and Z. Chen, Physiology, 2017, 266–277, DOI:10.1152/physiol.00036.2016.
- J. I. R. Labau, M. Andelic, C. G. Faber, S. G. Waxman, G. Lauria and S. D. Dib-Hajj, Exp. Neurol., 2022, 358, 114223 CrossRef CAS PubMed.
- L. Roumazeilles, N. Dokalis, E. Kaulich and V. Lelievre, Cell Adhes. Migr., 2018, 12, 87–92 CAS.
- A. C. de Luca, S. P. Lacour, W. Raffoul and P. G. di Summa, Neural Regener. Res., 2014, 9(22), 1943–1948 CrossRef PubMed.
- F. Berthiaume, T. J. Maguire and M. L. Yarmush, Annu. Rev. Chem. Biomol. Eng., 2011, 2, 403–430 CrossRef PubMed.
- A. Kantaros, Int. J. Mol. Sci., 2022, 23, 14621 CrossRef CAS PubMed.
- B. Yilmaz, A. Al Rashid, Y. A. Mou, Z. Evis and M. Koç, Bioprinting, 2021, 23, e00148 CrossRef.
- Y. S. Zhang, G. Haghiashtiani, T. Hübscher, D. J. Kelly, J. M. Lee, M. Lutolf, M. C. McAlpine, W. Y. Yeong, M. Zenobi-Wong and J. Malda, Nat. Rev. Methods Primers, 2021, 1, 75 CrossRef CAS.
- F. E. Freeman, R. Burdis and D. J. Kelly, Trends Mol. Med., 2021, 27, 700–711 CrossRef CAS PubMed.
- R. Y. Cheng, G. Eylert, J.-M. Gariepy, S. He, H. Ahmad, Y. Gao, S. Priore, N. Hakimi, M. G. Jeschke and A. Günther, Biofabrication, 2020, 12, 025002 CrossRef CAS PubMed.
- A. Urciuolo, I. Poli, L. Brandolino, P. Raffa, V. Scattolini, C. Laterza, G. G. Giobbe, E. Zambaiti, G. Selmin, M. Magnussen, L. Brigo, P. De Coppi, S. Salmaso, M. Giomo and N. Elvassore, Nat. Biomed. Eng., 2020, 4, 901–915 CrossRef CAS PubMed.
- Y. Chen, J. Zhang, X. Liu, S. Wang, J. Tao, Y. Huang, W. Wu, Y. Li, K. Zhou, X. Wei, S. Chen, X. Li, X. Xu, L. Cardon, Z. Qian and M. Gou, Sci. Adv., 2024, 6, eaba7406 CrossRef PubMed.
- E. Di Marco, P. Carlo MarchisioS, S. Bondanza, A. Tito FranziP, R. Cancedda and M. De Lucall, J. Biol. Chem., 1991, 266, 21718–21722 CrossRef CAS PubMed.
- L. Ouyang, R. Yao, Y. Zhao and W. Sun, Biofabrication, 2016, 8, 035020 CrossRef PubMed.
- S. Bonetti, github-Bioprinting, https://github.com/simobone19, (accessed 17 December 2024) Search PubMed.
- Y. Wang, K. L. Marshall, Y. Baba, G. J. Gerling and E. A. Lumpkin, PLoS One, 2013, 8(6), e67439 CrossRef CAS PubMed.
- M. Clair-Glover, R. K. Finol-Urdaneta, M. Maddock, E. Wallace, S. Miellet, G. Wallace, Z. Yue and M. Dottori, Biofabrication, 2024, 16, 045022 CrossRef PubMed.
- Y. Yan, X. Li, Y. Gao, S. Mathivanan, L. Kong, Y. Tao, Y. Dong, X. Li, A. Bhattacharyya, X. Zhao and S.-C. Zhang, Cell Stem Cell, 2024, 31, 260–274.e7 CrossRef CAS PubMed.
- W. S. Harley, C. C. Li, J. Toombs, C. D. O’Connell, H. K. Taylor, D. E. Heath and D. J. Collins, Bioprinting, 2021, 23, e00147 CrossRef.
- L. J. Pourchet, A. Thepot, M. Albouy, E. J. Courtial, A. Boher, L. J. Blum and C. A. Marquette, Adv. Healthcare Mater., 2017, 6, 1601101 CrossRef PubMed.
- S. England, A. Rajaram, D. J. Schreyer and X. Chen, Bioprinting, 2017, 5, 1–9 CrossRef.
- E. Sodupe-Ortega, A. Sanz-Garcia, A. Pernia-Espinoza and C. Escobedo-Lucea, Materials, 2018, 11, 1402 CrossRef PubMed.
- L. Lemarié, A. Anandan, E. Petiot, C. Marquette and E.-J. Courtial, Bioprinting, 2021, 21, e00119 CrossRef.
- Y. Zhao, R. Yao, L. Ouyang, H. Ding, T. Zhang, K. Zhang, S. Cheng and W. Sun, Biofabrication, 2014, 6, 035001 CrossRef CAS PubMed.
- T. A. M. Andrade, V. A. da Silva, K. Scheck, T. Garay, R. Sharma and S. M. Willerth, J. Biomed. Mater. Res., Part A, 2025, 113, e37831 CrossRef CAS PubMed.
- J. I. R. Labau, M. Andelic, C. G. Faber, S. G. Waxman, G. Lauria and S. D. Dib-Hajj, Exp. Neurol., 2022, 358, 114223 CrossRef CAS PubMed.
- H. Aubin, J. W. Nichol, C. B. Hutson, H. Bae, A. L. Sieminski, D. M. Cropek, P. Akhyari and A. Khademhosseini, Biomaterials, 2010, 31, 6941–6951 CrossRef CAS PubMed.
- S. R. Shin, H. Bae, J. M. Cha, J. Y. Mun, Y. C. Chen, H. Tekin, H. Shin, S. Farshchi, M. R. Dokmeci, S. Tang and A. Khademhosseini, ACS Nano, 2012, 6, 362–372 CrossRef CAS PubMed.
- K. M. Hakkinen, J. S. Harunaga, A. D. Doyle and K. M. Yamada, Tissue Eng., Part A, 2011, 17, 713–724 CrossRef CAS PubMed.
- C. F. Guimarães, L. Gasperini, A. P. Marques and R. L. Reis, Nat. Rev. Mater., 2020, 5, 351–370 CrossRef.
- L. Zhang, Q. Han, S. Chen, D. Suo, L. Zhang, G. Li, X. Zhao and Y. Yang, Colloids Surf., B, 2021, 199, 111503 CrossRef CAS PubMed.
- K. Roversi, H. Ebrahimi Orimi, M. Falchetti, E. da Rocha, S. Talbot and C. Boutopoulos, Micromachines, 2021, 12, 865 CrossRef PubMed.
- H. Oliveira, C. Médina, M.-L. Stachowicz, B. Paiva dos Santos, L. Chagot, N. Dusserre and J.-C. Fricain, Bioprinting, 2021, 22, e00134 CrossRef.
- H. Jerregård, P. Åkerud, E. Arenas and C. Hildebrand, J. Neurocytol., 2000, 29, 653–663 CrossRef PubMed.
- B. Seilheimer and M. Schachner, J. Cell Biol., 1988, 107, 341–351 CrossRef CAS PubMed.
- V. A. Botchkarev, M. Yaar, E. M. J. Peters, S. P. Raychaudhuri, N. V. Botchkareva, A. Marconi, S. K. Raychaudhuri, R. Paus and C. Pincelli, J. Invest. Dermatol., 2006, 126, 1719–1727 CrossRef CAS PubMed.
- M. Tominaga, H. Ogawa and K. Takamori, Br. J. Dermatol., 2008, 842, DOI:10.1111/j.1365-2133.2007.08410.x.
- K. Tanabe, I. Bonilla, J. A. Winkles and S. M. Strittmatter, J. Neurosci., 2003, 23, 9675–9686 CrossRef CAS PubMed.
- V. D. Dontchev and P. C. Letourneau, Nerve Growth Factor and Semaphorin 3A Signaling Pathways Interact in Regulating Sensory Neuronal Growth Cone Motility, J. Neurosci., 2002, 6659–6669 CrossRef CAS PubMed.
- M. Hara, K. Kadoya, T. Endo and N. Iwasaki, Exp. Physiol., 2023, 108, 621–635 CrossRef CAS PubMed.
- H. Yurie, R. Ikeguchi, T. Aoyama, Y. Kaizawa, J. Tajino, A. Ito, S. Ohta, H. Oda, H. Takeuchi, S. Akieda, M. Tsuji, K. Nakayama and S. Matsuda, PLoS One, 2017, 12, e0171448 CrossRef PubMed.
- L. Ning, N. Zhu, F. Mohabatpour, M. D. Sarker, D. J. Schreyer and X. Chen, J. Mater. Chem. B, 2019, 7, 4538–4551 RSC.
- P. Khoshakhlagh, A. Sivakumar, L. A. Pace, D. W. Sazer and M. J. Moore, J. Neural. Eng., 2018, 15, 064001 CrossRef PubMed.
- L. Honkamäki, O. Kulta, P. Puistola, K. Hopia, P. Emeh, L. Isosaari, A. Mörö and S. Narkilahti, Adv. Healthcare Mater., 2024, 2402504 Search PubMed.
- A. Engberg, C. Stelzl, O. Eriksson, P. O’Callaghan and J. Kreuger, Sci. Rep., 2021, 11, 21547 CrossRef CAS PubMed.
- imagej_fiji, https://imagej.net/software/fiji/, (accessed 17 December 2024).
- A. Pistone, A. Sagnella, C. Chieco, G. Bertazza, G. Varchi, F. Formaggio, T. Posati, E. Saracino, M. Caprini, S. Bonetti, S. Toffanin, N. Di Virgilio, M. Muccini, F. Rossi, G. Ruani, R. Zamboni and V. Benfenati, Biopolymers, 2016, 105, 287–299 CrossRef CAS PubMed.
- F. Formaggio, R. Rimondini, C. Delprete, L. Scalia, E. Merlo Pich, R. Liguori, F. Nicoletti and M. Caprini, Mol. Pain, 2022, 18, 17448069221087032 CrossRef PubMed.
- J. Popko, A. Fernandes, D. Brites and L. M. Lanier, Cytometry, Part A, 2009, 75A, 371–376 CrossRef PubMed.
- S.-Y. Ho, C.-Y. Chao, H.-L. Huang, T.-W. Chiu, P. Charoenkwan and E. Hwang, BMC Bioinf., 2011, 12, 230 CrossRef PubMed.
- P. Vallotton, R. Lagerstrom, C. Sun, M. Buckley, D. Wang, M. De Silva, S.-S. Tan and J. M. Gunnersen, Cytometry, Part A, 2007, 71A, 889–895 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb02823f |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |