Open Access Article
Open Access ArticleQuercetin-doped sol–gel coatings on titanium implants: a promising approach for enhanced immune response and cell adhesion†
C.
Arias-Mainer
 a,
F.
Romero-Gavilán
a,
F.
Romero-Gavilán
 *a,
A.
Cerqueira
*a,
A.
Cerqueira
 a,
D.
Peñarocha-Oltra
b,
I.
García-Arnáez
c,
O.
Amorrotu
c,
M.
Azkargorta
d,
F.
Elortza
d,
M.
Gurruchaga
a,
D.
Peñarocha-Oltra
b,
I.
García-Arnáez
c,
O.
Amorrotu
c,
M.
Azkargorta
d,
F.
Elortza
d,
M.
Gurruchaga
 c,
I.
Goñi
c and
J.
Suay
a
c,
I.
Goñi
c and
J.
Suay
a
aDepartment of Industrial Systems Engineering and Design, Universitat Jaume I, Castellon de la Plana, Spain. E-mail: gavilan@uji.es
bDepartment of Stomatology, Valencia University Medical and Dental School, Valencia, Spain
cDepartment of Polymers and Advanced Materials: Physics, Chemistry and Technology, Universidad del País Vasco, San Sebastián, Spain
dProteomics Platform, CIC bioGUNE, Basque Research and Technology Alliance (BRTA), CIBERehd, ProteoRed-ISCIII, Bizkaia Science and Technology Park, Derio, Spain
First published on 7th April 2025
Abstract
Quercetin (QUE), a natural flavonoid found in various fruits and vegetables, has diverse biological functions, including anti-inflammatory effects, regulation of cell adhesion and oxidative stress mitigation. In this study, sol–gel materials with increasing concentrations of quercetin (0.5, 1 and 2 wt%) were synthesised and applied onto titanium (Ti) surfaces as coatings. The materials were characterised physiochemically, and in vitro responses were examined using HOb osteoblastic cells and THP-1 macrophages. Human serum protein adsorption was evaluated using nLC-MS/MS. The incorporation of quercetin did not affect the sol–gel network cross-linking, and a controlled release of quercetin was achieved. The materials exhibited no cytotoxicity at any concentration. The HOb cells cultured on quercetin-doped materials were more elongated than those grown on QUE-free coatings, with protruding lamellipodia and increased cell surface. QUE-doped surfaces enhanced the expression of BMP-2, RANKL, and cell adhesion-related genes CTNNB1 and β-actin. In the THP-1 cells, pro-inflammatory gene expression (IL-1β, MCP-1 and iNOS) was down-regulated on 0.5QUE material, while it increased on 2QUE, as did the cytokine liberation. These changes correlated with altered protein adsorption patterns. The 2QUE coatings enhanced the adsorption of acute-phase proteins (SAA1, SAA2 and SAA4), indicating an inflammatory response; this behaviour was not seen on 0.5QUE. Moreover, cell adhesion (COF1, PROF1) and oxidative stress proteins (GPX3, SEPP1, AMBP) were preferentially adsorbed onto QUE-doped coatings. These results highlight the significance of optimising quercetin concentration in sol–gel coatings to modulate the immune response and enhance cell adhesion effectively.
1. Introduction
Enhanced bone healing and post-surgical recovery remain the pivotal objectives in the field of oral health. The quest for obtaining better clinical outcomes continues, aiming to reduce the process of bone resorption and improve bone growth. To achieve this goal, new biomaterials with bioactive properties1 are being developed. Ti is commonly used in the field of medicine and orthodontics due to its mechanical and anticorrosive properties and its capacity to osseointegrate.2 However, despite the high rate of success, it is typically classified as a bioinert material; it has significantly lower bioactivity than bioactive ceramics.To overcome this problem, the researchers are focusing on the development of new coatings that enhance the bioactivity of Ti.3 The sol–gel method is a promising approach among the many surface treatments being explored; it produces materials with high stability, adhesion strength and a homogeneous composition.4 Moreover, the sol–gel coatings can be used as a delivery system for the release of both organic5 and inorganic6 molecules. Martínez-Ibáñez et al.7 have developed a sol–gel coating based on alkoxysilane precursors, made using the mixture of methyltrimethoxysilane (MTMOS) and tetraethyl orthosilicate (TEOS) in a proportion of 70% MTMOS to 30% TEOS. The authors have reported that the coating has high biocompatibility.8 Its osteoinductive properties have been analysed both in vivo and in vitro.9
Flavonoids, a group of compounds well known for their wide range of potential health benefits, are now being investigated with a view to using them in biomedical applications.10 Quercetin, (3,3′,4′,5,7-pentahydroxyflavone) belongs to the flavonoid subfamily of flavones. This polyphenol is not produced in the human body but is found in many fruits (mainly citrus) and vegetables.8 It has some interesting anticancer, angiogenic, antioxidant and anti-inflammatory properties;11 it causes downregulation of several inflammatory factors such as TNF-α, IL-1β and IL-6.12 Zhou et al.13 have reported that quercetin can promote cell proliferation and osteogenic differentiation.14 It can also improve cell adhesion in hFOB osteoblastic cells15 and mitigate oxidative stress by reducing reactive oxygen species (ROS) levels within human osteoblasts.16
After an implant comes into contact with living tissue, one of the first events is protein adsorption onto the material surface. The composition of this initial protein layer can determine the response to the implant.14 Variations in the biomaterial surface can lead to changes in protein adsorption and, consequently, affect cellular responses such as cell adhesion, proliferation, and differentiation processes.17 Analysing the adsorption of proteins on QUE-doped biomaterials could help us understand the body response to this type of bioactive molecules.
The aim of this study was to develop new sol–gel coatings doped with increasing percentages of quercetin to bioactivate Ti implants. The synthesised biomaterials were physiochemically characterised. They were also evaluated biologically in vitro using human osteoblasts (HOb) to examine their osteogenic potential. A human monocytic cell line (THP-1) was used to study their immune responses. Moreover, the human serum protein adsorption was analysed utilising nLC-MS/MS. The results presented here should help to form a comprehensive overview of the potential applications of QUE in the field of oral health.
2. Materials and methods
2.1. Sol–gel synthesis and sample preparation
For the sol–gel synthesis, organically-modified alkoxysilanes (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), methyltrimethoxysilane (MTMOS) and tetraethyl orthosilicate (TEOS) were used as precursors in a proportion of 70% MTMOS to 30% TEOS. 2-Propanol was employed as a solvent to carry out the sol–gel reactions, with a volume ratio of alcohol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siloxane 1
siloxane 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Different amounts of QUE hydrate (TCI Europe; Zwijndrecht, Belgium) dissolved in 2-propanol were incorporated into the sol–gel mixtures. The sol–gel compositions with 0, 0.5, 1 and 2 wt% QUE were obtained. The maximum percentage of quercetin was established based on the solubility limit of this molecule in the sol–gel mixture. Hydrolysis was carried out by adding the corresponding stoichiometric quantity of H2O at a rate of 1 drop s−1. H2O was acidified with nitric acid (0.1 M HNO3) as a catalyst for sol–gel reactions. The sol–gel mixtures were stirred for 1 hour and kept at rest for another hour. Once the sol–gel materials were synthesised, grade-4 Ti discs (10- and 12-mm diameter, 1-mm thick) were used as substrates for the coatings. The Ti discs were sandblasted and acid-etched (SAE; Ilerimplant-GMI SL., Lleida, Spain) following the procedure described in Romero-Gavilán et al.18 The coatings were manufactured using a dip-coater (KSV DC; KSV NIMA, Espoo, Finland). The Ti discs were immersed for 1 minute in the sol–gel mixture at a velocity of 60 cm min−1 and removed at a speed of 100 cm min−1. Hydrolytic degradation and QUE release were evaluated using glass slides as a substrate for sol–gel coatings. The glass slides were first treated with HNO3 solution (25% v/v) in an ultrasonic bath (Sonopuls HD 3200) for 20 min at 30 W and then washed again with distilled H2O under the same conditions. The slides were coated with a controlled amount of sol–gel by casting. For their chemical characterisation, sol–gel-free films were obtained by pouring 5 mL of each composition into non-stick Teflon moulds. Finally, all samples were heat-treated for 2 h at 80 °C.
1. Different amounts of QUE hydrate (TCI Europe; Zwijndrecht, Belgium) dissolved in 2-propanol were incorporated into the sol–gel mixtures. The sol–gel compositions with 0, 0.5, 1 and 2 wt% QUE were obtained. The maximum percentage of quercetin was established based on the solubility limit of this molecule in the sol–gel mixture. Hydrolysis was carried out by adding the corresponding stoichiometric quantity of H2O at a rate of 1 drop s−1. H2O was acidified with nitric acid (0.1 M HNO3) as a catalyst for sol–gel reactions. The sol–gel mixtures were stirred for 1 hour and kept at rest for another hour. Once the sol–gel materials were synthesised, grade-4 Ti discs (10- and 12-mm diameter, 1-mm thick) were used as substrates for the coatings. The Ti discs were sandblasted and acid-etched (SAE; Ilerimplant-GMI SL., Lleida, Spain) following the procedure described in Romero-Gavilán et al.18 The coatings were manufactured using a dip-coater (KSV DC; KSV NIMA, Espoo, Finland). The Ti discs were immersed for 1 minute in the sol–gel mixture at a velocity of 60 cm min−1 and removed at a speed of 100 cm min−1. Hydrolytic degradation and QUE release were evaluated using glass slides as a substrate for sol–gel coatings. The glass slides were first treated with HNO3 solution (25% v/v) in an ultrasonic bath (Sonopuls HD 3200) for 20 min at 30 W and then washed again with distilled H2O under the same conditions. The slides were coated with a controlled amount of sol–gel by casting. For their chemical characterisation, sol–gel-free films were obtained by pouring 5 mL of each composition into non-stick Teflon moulds. Finally, all samples were heat-treated for 2 h at 80 °C.
2.2. Physicochemical characterisation
A Thermo Nicolet 6700 Fourier-transform infrared spectrometer (FT-IR; Thermo Fisher Scientific, NY, US) with an attenuated total reflection system (ATR) was used for the FTIR characterisation of the QUE-doped sol–gel materials. The spectra were obtained in the wavenumber range of 4000–400 cm−1. The silicon nuclear magnetic resonance spectroscopy (29Si-NMR) technique was employed to examine the reticular level of the sol–gel networks. For this purpose, a Bruker 400 AVANCE III WB Plus spectrometer (Bruker, Billerica, MA, US) with a cross-polarisation magic-angle spinning (CP-MAS) was used with a probe for solid samples. The standard pulse sequence was fixed at 79.5 MHz frequency, the spectral width was 55 kHz, with 2-ms contact time, 5-s delay time and a spinning speed of 7.0 kHz. Mass loss was evaluated before and after soaking the coated glass slides in 50 mL of distilled H2O for 1, 2, 4 and 8 weeks at 37 °C. The hydrolytic degradation kinetics were obtained by measuring the percentages of weight loss. The assay was made in triplicate. Additionally, to assess the stability of the coatings under more realistic conditions, coated Ti discs were incubated with 2 mL of artificial saliva (Sigma-Aldrich) in a 24-well plate (Thermo Fisher Scientific, Waltham, MA, USA) for 7, 14, and 21 days (37 °C, 95% H2O, 5% CO2). After incubation, the samples were washed with distilled water and dried at 50 °C. The effects of degradation on the material's morphology were then analyzed using SEM (Leica-Zeiss LEO equipment, Leica). QUE release was examined by submerging coated glass slides in 50 mL of Milli-Q water at 37 °C. After 0, 1, 3, 5, 8, 24, 48, 72, 96, 168 and 336 h of incubation, the absorbance of the medium was measured with a Helios Omega UV-VIS (Thomas Scientific, New Jersey, USA). Three samples were evaluated for each condition. Surface topography modifications caused by QUE-doping were examined using a scanning electron microscope (SEM) with Leica-Zeiss LEO equipment under vacuum (Leica, Wetzlar, Germany). To enhance conductivity, the materials were pretreated with a ∼2 nm layer of platinum via sputter deposition. Surface roughness was evaluated employing an optical profilometer (interferometric and confocal) PLm2300 (Sensofar, Barcelona, Spain); the assays were carried out in triplicate using 12-mm Ti discs coated with the sol–gel materials. The results were expressed as the arithmetic average roughness parameter (Ra). Contact angles were obtained using an automatic contact angle meter OCA 20 (DataPhysics Instruments, Filderstadt, Germany). For this purpose, 10-μL aliquots of Milli-Q H2O were deposited on the disc surfaces and contact angles were obtained employing the SCA 20 software (DataPhysics Instruments, Filderstadt, Germany). Six discs of each material were studied after depositing two drops on each disc.2.3. In vitro assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−1 and cultured for 24 hours. The samples were then washed with PBS and fixed with 4% paraformaldehyde (PFA) at room temperature for 20 minutes. Following fixation, the samples were permeabilised with 0.1% Triton X-100 for 5 minutes. Subsequently, they were incubated with phalloidin (diluted 1
000 cells cm−1 and cultured for 24 hours. The samples were then washed with PBS and fixed with 4% paraformaldehyde (PFA) at room temperature for 20 minutes. Following fixation, the samples were permeabilised with 0.1% Triton X-100 for 5 minutes. Subsequently, they were incubated with phalloidin (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 0.1% w/v bovine serum albumin-PBS; Abcam, Cambridge, UK) for 1 hour at room temperature. To stain the nuclei, the samples were washed twice with PBS and then incubated for 5 minutes in a mounting medium containing DAPI (Abcam). Fluorescence detection was performed using a Leica TCS SP8 Confocal Laser Scanning Microscope equipped with 20× dry lenses. The images were captured with LAS X software (Leica) and analysed using the ImageJ tool (National Institutes of Health, Maryland, USA).
100 in 0.1% w/v bovine serum albumin-PBS; Abcam, Cambridge, UK) for 1 hour at room temperature. To stain the nuclei, the samples were washed twice with PBS and then incubated for 5 minutes in a mounting medium containing DAPI (Abcam). Fluorescence detection was performed using a Leica TCS SP8 Confocal Laser Scanning Microscope equipped with 20× dry lenses. The images were captured with LAS X software (Leica) and analysed using the ImageJ tool (National Institutes of Health, Maryland, USA).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C). The top layer (aqueous phase) was mixed with 550 μL of isopropanol and incubated at room temperature for 10 min. Samples were centrifuged (20 min, 14
000 rpm, 4 °C). The top layer (aqueous phase) was mixed with 550 μL of isopropanol and incubated at room temperature for 10 min. Samples were centrifuged (20 min, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C), washed twice with 0.5 mL of 70% EtOH and air-dried. Finally, the resulting pellet was dissolved in 20 μL of RNAse-free water and frozen at 80 °C. RNA concentration and quality were measured with a nanodrop NanoVue® Plus Spectrophotometer (GE Healthcare Life Sciences, Little Chalfont, UK).
000 rpm, 4 °C), washed twice with 0.5 mL of 70% EtOH and air-dried. Finally, the resulting pellet was dissolved in 20 μL of RNAse-free water and frozen at 80 °C. RNA concentration and quality were measured with a nanodrop NanoVue® Plus Spectrophotometer (GE Healthcare Life Sciences, Little Chalfont, UK).
For cDNA synthesis, approximately 1 μg of total RNA was converted into cDNA using PrimeScript RT Reagent Kit (Perfect Real Time; Takara Bio Inc., Shiga, Japan) in a reaction volume of 20 μL. The reaction was conducted under the following conditions: 37 °C for 15 min, 85 °C for 5 s, and a final hold at 4 °C. The resulting cDNA quality and concentration were measured using a NanoVue® Plus Spectrophotometer; then, the DNA was diluted in DNAse-free water to a concentration suitable for qRT-PCR analysis and stored at −80 °C.
Quantitative real-time PCRs (qRT-PCR) were carried out in 96-well plates (Applied Biosystems®, Thermo Fisher Scientific). The genes of interest were quantified in each sample using a housekeeping gene (glyceraldehyde phosphate dehydrogenase (GAPDH)) as a reference. Primers for each gene were designed using the Primer3plus software tool (https://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Specific DNA sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov/nucleotide/) and purchased from Thermo Fisher Scientific. Specific primers for each gene are shown in Table S1 (ESI†). Individual reactions contained 1 μL of cDNA, 0.2 μL of specific primers (forward and reverse, 10 μM), 5 μL of SYBR Premix Ex Taq (Tli RNase H Plus; Takara) and autoclaved distilled water in a final volume of 10 μL. Reactions were carried out in a StepOnePlus Real-Time PCR System (Applied Biosystems®, Thermo Fisher Scientific) at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 34 s, 95 °C for 15 s and 60 °C for 60 s. The data were obtained using the StepOnePlus Software 2.3 (Applied Biosystems®, Thermo Fisher Scientific). Fold changes were calculated using the 2−ΔΔCt method, and the data were normalised against the blank wells (without any material). Six technical replicates of each sample were examined.
The total protein concentration in the serum was quantified using the Pierce BCA assay kit (Thermo Fisher). For protein analysis, an Evosep ONE chromatograph coupled with a hybrid trapped ion mobility-quadrupole time-of-flight mass spectrometer (timsTOF Pro with PASEF; Bruker, Billerica, MA, USA) was employed. Each sample was analysed in quadruplicate. Protein identification was performed using MaxQuant software (https://maxquant.org/). Differential analysis of protein layers deposited on the QUE-doped coatings and the 0QUE coating was conducted using the Perseus platform (https://www.maxquant.org/perseus/).
Functional characterisation of proteins was conducted using PANTHER (https://www.pantherdb.org/), DAVID v6.8 (Database for Annotation, Visualisation and Integrated Discovery; https://david.ncifcrf.gov/) and the UniProt database (https://www.uniprot.org/), utilising UniProt ID codes as protein identifiers.
3. Results
3.1. Psysicochemical characterisation
Sol–gel materials doped with QUE were successfully synthesised and applied as coatings on the Ti substrates. The samples were morphologically evaluated using SEM. As we can see on the micrographs (Fig. 1a–e), sol–gel networks doped with QUE covered the whole area of the discs, showing a good adhesion to the Ti substrate. However, some sol–gel accumulated in the cavities of Ti introduced by sandblasting, smoothing the initial SAE irregularities. Nevertheless, accumulations of QUE on the doped coatings were not detected, and no pores or cracks were observed. The Ra value was used in the evaluation of surface roughness. Fig. 1f shows that the application of the sol–gel network as coating decreased the roughness in comparison with the uncoated Ti. Moreover, the addition of QUE to the sol–gel structure caused a reduction in Ra for 0.5QUE and 2QUE compared to 0QUE.29Si-solid NMR spectra are shown in Fig. 2a. Tn signals detected between −50 and −75 ppm are related to MTMOS, while the Qn, between −90 and −115 ppm, corresponds to TEOS.19 For MTMOS, the T2 and T3 signals at −58 and −66 ppm, respectively, indicate a high grade of MTMOS cross-linking (no signals for T0 and T1 were found). The Q2, Q3 and Q4 signals appeared at −95, −103 and −112 ppm, respectively. These shifts are associated with the condensation of TEOS, with the Q3 being the most intense signal. Moreover, the incorporation of QUE into the network did not affect the cross-linking degree, as we can see by comparing the QUE-doped materials with the 0QUE coatings.
Fig. 2b shows FT-IR spectra for the QUE-doped sol–gel networks. The hydrolysis was successful; the siloxane chain signals were detected. These bands are due to Si–O–Si bond, appearing close to 1090 cm−1 (asymmetric tension) and 770 cm−1 (symmetrical tension and vibration of deformation).20,21 Nevertheless, the condensation was not complete, as shown by the related signal at 970 cm−1, indicating the presence of the Si–OH bond from silanol.20 Moreover, there is a wide band around 3400 cm−1, related to OH groups.20,21 Signals related to Si–CH3 bonds were found at 1275 cm−1.22 The peaks close to 3000 cm−1 are attributed to C–H bonds typically associated with MTMOS, corresponding to non-hydrolysable methyl groups.23 The spectra for the different evaluated materials showed the same patterns regardless of the presence of QUE in the network.
Contact angle results for the coatings are displayed in Fig. 3a. The addition of QUE to the sol–gel network produced an increase in the contact angles, indicating a reduction in the surface hydrophilicity. 2QUE coating showed a larger contact angle than 0.5QUE. Fig. 3b shows QUE liberation kinetics for the doped coatings. The release of QUE demonstrated a dose-dependent trend, rising with increasing QUE concentration in the sol–gel structure. Significantly more QUE was released from 2QUE-coated materials than from 0.5QUE and 1QUE. The maximum concentrations of released QUE were 0.0052 mg mL−1, 0.0094 mg mL−1 and 0.0498 mg mL−1 for 0.5QUE, 1QUE and 2QUE, respectively. Fig. 3c displays the hydrolytic degradation kinetics of the developed sol–gel coatings. For all formulations, the greatest mass loss occurred in the first week of the degradation assay. All coatings showed similar degradation over time. However, the materials that incorporated QUE reached almost 40% mass loss after 7 days, while it took 14 days for 0QUE to reach the same level. To evaluate the stability of the coatings developed on titanium discs under physiologically relevant conditions, the samples were incubated in artificial saliva in a controlled environment (95% humidity, 5% CO2, and 37 °C). Fig. S1 (ESI†) presents SEM micrographs of the QUE coatings after 7, 14, and 21 days of incubation. At day 0, the sol–gel networks displayed a uniform and smooth morphology. However, following incubation, the coatings developed cracks—likely due to thermal stresses during sample drying—and surface irregularities indicative of degradation. Notably, the 1QUE and 2QUE coatings showed signs of mass loss as early as day 7, whereas the 0QUE and 0.5QUE coatings remained intact until day 14.
3.2. Osteogenic results in vitro
3.3. Immune response in vitro
3.4. Proteomic evaluation
The nLC-MS/MS analysis revealed 68 proteins (Table S1, ESI†) differentially adsorbed on the coatings containing QUE compared to 0QUE. These proteins were classified according to their biological functions. This classification was made using the DAVID proteomic tool (Table 1). In the immune response group, 19 proteins showed increased adsorption to QUE coatings compared to 0QUE, while 2 proteins were less adsorbed. A rise in immunoglobulin abundance (HV315, HV70D, HV434, KV108, HV226, HV428 and HV315) was noticed on all the compositions with QUE except for 1QUE, which showed no change in immunoglobulin adsorption compared to 0QUE. Moreover, reduced adsorption of another immunoglobulin, HV70, was observed for 2QUE relative to 0QUE. Proteins related to the activation of the complement system (CO8G, CO5 and CO2) were found preferentially adsorbed on 0.5QUE and 2QUE, while proteins linked to its regulation (FHR1 and FHR5) were more abundant on 0.5QUE and 1QUE surfaces than on 0QUE. Moreover, serum amyloid A proteins, involved in the acute immune response (SAA2, SAA1 and SAA4), were preferentially adsorbed on 1QUE and 2QUE. Finally, other proteins related to the immune system (RARR2, CFAD, PRG4 and DCD) were more abundant on 1QUE and 2QUE, whereas the adsorption of TUT4 was reduced for 1QUE and 2QUE. A significantly increased adsorption of cell adhesion-related proteins was observed on the QUE-doped coatings (ALS, COMP, IBP5, COF1, IBP2, PLAK, CDSN, PROF1, COLA1, COFA1 and TSK). However, a reduction in the adsorption of FILA2 was seen on 1QUE. Proteins with function in the blood coagulation process, such as pro-coagulants KLKB1, HRG, FIBA and FA5, as well as PLMN protein related to fibrinolysis, were more abundant on the QUE coatings. In contrast, there was a decrease in the adsorption of 3 proteins related to the coagulation pathway (SPB12, THRB, F13B) to 1QUE compared with 0QUE. Some proteins playing a key role in oxidative stress also modified their adsorption onto the QUE-doped surfaces. GPX3, SEPP1 and AMBP proteins, involved in the protection of cells and enzymes from oxidative damage, increased their affinity to 1QUE and 2QUE in comparison with 0QUE.| 0.5QUE vs. 0QUE | 1QUE vs. 0QUE | 2QUE vs. 0QUE | ||
|---|---|---|---|---|
| Immune response | UP | HV315, CO8G, HV70D, FHR1, HV434, KV108, HV226,CO5 | RARR2, SAA2, FHR5 | RARR2, SAA2, HV428, HV315, CO8G, SAA4, CFAD. G3P, SAA1, PRG4, CO2, DCD |
| DOWN | — | TUT4 | TUT4, HV70D | |
| Cell adhesion | UP | ALS, COMP | IBP5, COF1 | IBP5, COF1, IBP2, PLAK, CDSN, ENOB, COIA1, COFA1, MMRN2, TSK |
| DOWN | — | FILA2 | — | |
| Coagulation | UP | KLKB1 | FIBA, CXCL7, HRG | FIBA, CXCL7, HRG, FA5, PLMN |
| DOWN | — | SPB12, THRB, F13B | ||
| Oxidative stress | UP | — | GPX3 | GPX3, SEPP1, AMBP |
| DOWN | — | — | — |
PANTHER analysis was employed to classify differentially adsorbed proteins based on their respective protein class and functions in pathways. In Fig. 7, the proteins differentially adsorbed to the QUE surfaces are classified by protein class. Thus, the common differentially adsorbed protein types were defence/immunity proteins, gene-specific transcriptional regulators, protein-binding activity modulators, cell adhesion molecules, protein modifying enzymes, transmembrane signal receptors, cytoskeletal proteins, metabolite interconversion enzymes, intercellular signal molecules, transfer/carrier, extracellular matrix proteins, RNA metabolism and scaffold/adaptor organisation proteins. The proteins with reduced adsorption to QUE-doped surfaces were classified as chaperones, defence/immunity proteins, metabolite interconversion enzymes, protein modifying enzymes, cytoskeletal proteins and protein-binding activity modulators. PANTHER analysis also supplied valuable insights into the pathways affected by the proteins differentially adsorbed on QUE-doped coatings. Fig. 8 shows pathways associated with the functions of these proteins. Proteins with augmented adsorption on the QUE-doped coatings enhance blood coagulation, plasminogen activation, cytoskeletal regulation by Rho GTPase, glycolysis, integrin signalling and toll receptor signalling pathways. The proteins with reduced affinity to 0.5QUE are related to vitamin D metabolism and blood coagulation pathways.
 | ||
| Fig. 7 PANTHER pie-chart classification by protein class for the proteins differentially adsorbed to the QUE-doped sol–gel coatings (relative to 0QUE). | ||
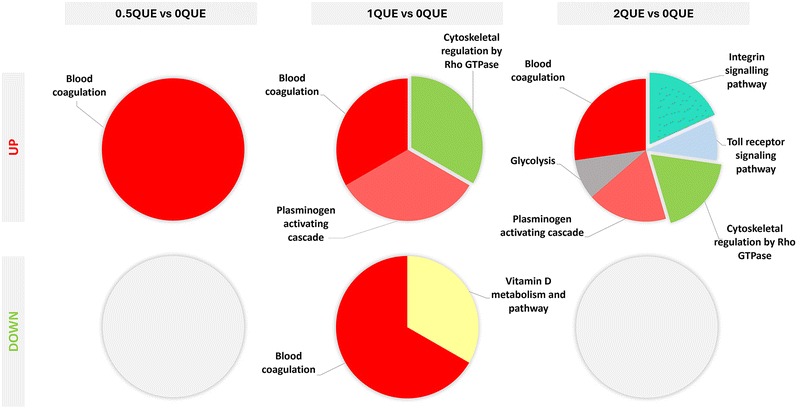 | ||
| Fig. 8 PANTHER pie-chart classification by pathways associated with the proteins differentially adsorbed on the QUE-doped sol–gel coatings relative to 0QUE. | ||
4. Discussion
Effective cell adhesion ensures that cells can attach to the implant surface, promoting stability and reaching good osseointegration levels,24 which is essential for the long-term success of the implant.25 Furthermore, a balanced immune response is crucial to prevent inflammation and ensure proper healing.26 QUE is known for its anti-inflammatory and osteogenic abilities,12 and its addition to the sol–gel network might result in improved osseointegration of implants. The aim of this study was to develop QUE-doped sol–gel coatings to bioactivate Ti implants. Their effect on in vitro cell responses and their proteomic adsorption patterns were examined to investigate the potential of QUE in bone regeneration. Sol–gel networks containing 0.5, 1 and 2 wt% QUE were successfully synthesised and used to coat the SAE-grade 4 Ti discs. Chemical analysis confirmed that the desired sol–gel reactions occurred, achieving a significant level of cross-linking. Moreover, the 29Si solid-state NMR analysis indicated that the incorporation of QUE did not affect the formation of the sol–gel network. However, the QUE-doping produced an increase in the hydrophobicity of the material. Padial-Molina et al.27 have examined the effects of changes in contact angles on the morphology and proliferation of MG-63 osteoblasts. Their results have indicated that increased contact angles improve both the morphology and proliferation of these osteoblasts, which is in agreement with our in vitro findings.In this study, the incorporation of QUE did not affect the hydrolytic degradation rate of the coatings. The maximum degradation (over 40%) was reached after 7 days for all compositions except for 0QUE, which reached this maximum value after 14 days.
A controlled release of QUE was observed; the amounts of released QUE rose with increasing content of this component in the coatings. The 2QUE material showed a significant increase in the amount of QUE released after 168 h compared to 0.5QUE and 1QUE. The QUE-doped materials maintained a steady release over the entire duration of the assay. The flavonoid incorporated in this sol–gel network has OH groups that could participate in sol–gel condensation reactions,28 which would explain the weak release of this compound from the 0.5QUE and 1QUE coatings compared with 2QUE. As the 2QUE contained the highest amount of QUE, an increased proportion of free QUE would be expected in that network; this should cause an additional rise in the release of this compound.
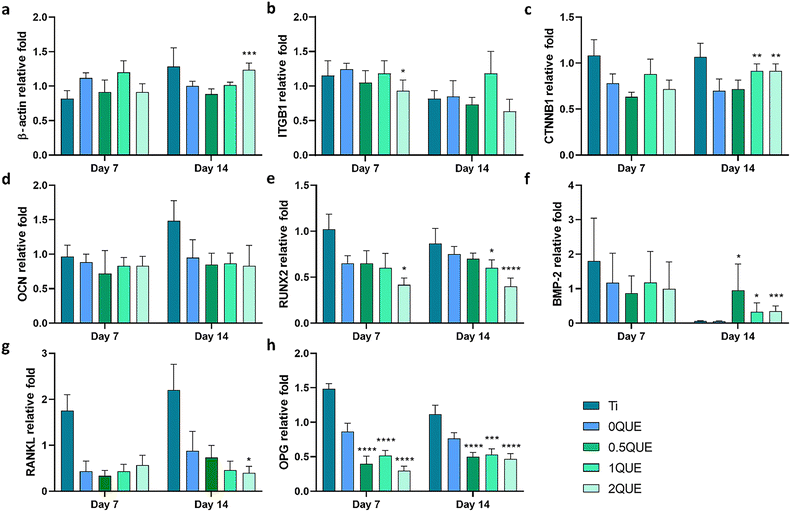
In their study of cell adhesion, Lezcano et al.29 have shown that incubation with 1 μM QUE improves cellular adhesion of the human foetal osteoblastic cell line (hFOB). Similarly, culturing HOb cells with our QUE-doped sol–gel coatings (0.5QUE, 1QUE and 2QUE) increased cell length and significantly enlarged the cell surface area. Moreover, a rise in the numbers of filopodia and lamellipodia structures was observed in these cultures, further enhancing cell adhesion. The cells incubated with QUE-doped materials showed an increased expression of β-catenin (CTNNB1) and β-actin. Katz et al.30 have reported that β-actin can directly interact with the interface between the actin filament and adhesion sites, thereby reinforcing the stability of adhesion. Bian et al.31 have noted that quercetin-treated BMSCs had elevated CTNNB1 levels and increased expression of mRNAs for BMP-2 and RUNX2 (both associated with osteogenic differentiation). Here, we also observed augmented expression of BMP-2 for QUE compositions after 14 days, although the material with the highest amount of QUE provoked a decrease in RUNX2 expression. These changes, observed on the studied genes, may contribute to enhanced cell adhesion and improved osteointegration.
Most factors that stimulate osteoclast formation and activity work by inducing RANKL expression in osteoblastic cells.32 Thus, a reduction in RANKL levels may lead to a diminished osteoclast formation. The OPG protein is a cytokine receptor in charge of inhibiting osteoclastogenesis.33 In this study, a reduction in OPG expression levels was detected in cultures with QUE-doped materials. However, a decrease in the expression of RANKL was also observed for 2QUE after 14 days of culture, which might lead to reduced osteoclast activity.
Inflammation triggered during immune response heavily depends on the activity of macrophages. These cells, playing a vital role in immunity, can differentiate, giving two phenotypes: M1 and M2.34 M1 macrophages are linked to a pro-inflammatory response and the secretion of cytokines such as TNF-α, IL-1β or MCP-1. The M2 macrophages are associated with anti-inflammatory and pro-regenerative functions and the secretion of TGF-β or IL-10, among other regulatory cytokines.6 Numerous studies have highlighted QUE for its anti-inflammatory effects,12 including its ability to inhibit pro-inflammatory cytokines. This could reduce the expression of M1-related genes, e.g. TNF-α,35 decreasing the activity of pathways such as NF-κB signalling.36 Our study showed that the QUE-doping affected the immune response in a dose-dependent manner. It significantly reduced the expression of TNF-α, IL-1β and MCP-1 in 0.5QUE cultures. However, at the highest concentration (2QUE), there was a notable increase in the expression of TNF-α and IL-1β. The same tendency was observed in the ELISA assays, where an increase in TNF-α secretion was noted in 2QUE samples. Culturing the cells with QUE-doped materials also resulted in an increase in IL-10 levels after day 1, correlating with the overall rise in TNF-α levels observed on the same day. Conversely, a decrease in IL-10 levels, noted on day 3 for 0.5QUE and 1QUE, which coincided with a reduction in the expression of pro-inflammatory genes, might reveal the immunomodulatory effect of QUE.
The inducible nitric oxide synthase (iNOS) is a characteristic marker of M1 macrophages, associated with the pro-inflammatory behaviour of these cells.37 In our study, there was a significant reduction in iNOS expression for 0.5QUE and 1QUE. Our results indicate that the incorporation of QUE in the sol–gel coatings affects the inflammation in a dose-dependent manner. Small amounts of QUE increased the expression of pro-inflammatory genes (TNF-α, MCP-1) on day 1. This was followed by a rise in the expression of the anti-inflammatory gene IL-10. After 3 days, there was a clear reduction in the expression of pro-inflammatory genes (IL-1B, TNFA, MCP-1, iNOS), accompanied by a decrease in IL-10 expression, indicating an immunomodulatory response for 0.5QUE and 1QUE. However, a pro-inflammatory behaviour was observed for the highest concentration of QUE (2QUE).
One of the main events after contact of an implant with biological fluids is the adsorption of proteins onto the biomaterial surfaces. The formation of this protein layer plays a crucial role in determining the implant–tissue interactions.6 Proteomic methods make it possible to acquire the profiles of protein adsorption on materials exposed to human serum, overcoming the limitations of conventional in vitro characterisation methods.38 In this study, the nLC-MS/MS analysis revealed 68 proteins preferentially adsorbed onto the sol–gel coatings incorporating QUE. A preferential immunoglobulin adsorption was observed for 0.5QUE (HV315, HV70D, HV434, KV108 and HV226) and 2QUE (HV428 and HV315). Immunoglobulins are glycoproteins produced by plasma cells in charge of the immune response.39 The complement system is a key component of innate immunity and host defence.40 Proteomic analysis revealed (for samples incorporating QUE) increased adsorption of CO5, CO8G, CO2 and CFAD, the proteins associated with the activation of the complement cascade. FHR1 and FHR5, involved in the regulation of the complement system41 increased their adsorption on 0.5QUE and 1QUE.40 Serum amyloids A (SAA) belong to a highly conserved, acute-phase protein family.42 The increased adsorption of SAA1, SAA2 and SAA4 on 2QUE may lead to the expression of pro-inflammatory genes by activation of the transcription NFKB,43 as it was found in the in vitro tests of this QUE-doped coating. In contrast, the 0.5QUE material, which did not exhibit an increased affinity for SAA proteins, caused a significant reduction in the expression of pro-inflammatory genes. Hirai et al.44 have reported that SAA serves as an indicator of systemic inflammation in individuals with chronic periodontitis; the SAA proteins show a crucial damage-associated molecular pattern in periodontal lesions. Furthermore, Oh et al. have found that SAA family acts as a strong inhibitor of RANKL activity, which in turn reduces osteoclast formation.45 This behaviour was consistent with the decrease (for 2QUE) in the expression of RANKL in HOb cells.
Cell adhesion is a crucial factor in cell proliferation and migration; it is affected by proteins such as integrins and cadherins, transmembrane glycoproteins responsible for cell–cell and cell–extracellular matrix interactions.46 Proteomics revealed preferential adsorption of cell adhesion-related proteins on the QUE-doped sol–gel coatings. Among these, we observed insulin-like growth factor binding proteins (ALS, IBP5 and IBP2) associated with extending the half-life of IGFs and cell adhesion functions.47 Collagen type I (COLA1) protein also preferentially adsorbed onto the QUE-coatings. This protein is an essential component of the bone extracellular matrix and has been used to bioactivate Ti bone implants to improve their osseointegration.48 In addition, COLA1 facilitates cell adhesion, proliferation, and osteoblast differentiation, which in turn accelerates mineralisation.49 Cytoskeletal assembly proteins COF1 and PROF1, also preferentially adhering to QUE-doped surfaces, are recognised as regulators of actin filament dynamics through the Rho GTPase system.50 The Panther analysis results demonstrated an up-regulation in the arrangement of the cytoskeleton via Rho GTPase and integrin signalling pathways, leading to the improvement in cell adhesion functions.51 The proteomic results correlated with the increased cell adhesion and augmented formation of filopodia and lamellipodia structures observed in in vitro QUE cultures.
Blood clotting is essential for tissue healing, not only preventing excessive blood loss but also creating a natural scaffold for tissue repair and regeneration.52 The coagulation process is also closely linked to inflammation, with both processes playing a role in maintaining homeostasis during wound healing.53 Our proteomic analysis revealed that a group of proteins involved in coagulation had an increased affinity for QUE surfaces, particularly notable for 2QUE; however, the significance of these changes is still unclear.
In oxidative stress studies, QUE has been recognised for its antioxidant properties, removing free radicals and strengthening antioxidant defence systems in the body.54 In contrast, iNOS is known for its inflammatory and oxidative stress properties. The addition of QUE to the coatings led to a considerable reduction in iNOS expression in the cultured cells in a dose-dependent manner. A similar tendency was observed in the proteomics results. An increase in QUE concentration in the coatings led to augmented adsorption of proteins associated with the inhibition of oxidative stress (GPX3, SEPP1 and AMBP), particularly evident for 2QUE. Proteins such as GPX3 and SEPP1 are crucial for combating oxidative stress and maintaining redox balance.55,56
The findings of this study highlight the potential of QUE in the development of biomaterials, demonstrating its impact on in vitro cell responses and protein adsorption patterns. The results suggest that QUE incorporation can improve the biomaterials by affecting cell adhesion, osteogenesis, and inflammation response in a dose-dependent manner. Among the three compositions tested, the 0.5QUE appears to be the optimal formulation. This composition was associated with significant improvements in cell adhesion, oxidative stress and anti-inflammatory behaviour. A marked reduction in the expression of pro-inflammatory genes and a decrease in the adsorption of proteins associated with acute inflammation was observed. Incorporating larger amounts of QUE into the sol–gel coating (2QUE) also enhanced cell adhesion. However, it considerably increased the expression of pro-inflammatory genes and the adsorption of proteins linked to an acute immune response.
5. Conclusion
The aim of this study was to examine the behaviour of sol–gel coatings capable of releasing quercetin (QUE) at varying concentrations, to analyse protein adsorption on coated Ti surfaces and the effects of QUE-doping on cell responses in vitro. The sol–gel coatings doped with QUE were successfully synthesised and applied onto Ti surfaces. The QUE-doped coatings exhibited preferential adsorption of proteins related to the immune system, cell adhesion, oxidative stress and coagulation. In terms of immune response, QUE affected the expression of immune-associated genes, displaying immunomodulating properties at low concentrations (0.5QUE) but pro-inflammatory effects at higher concentrations (2QUE). Proteomic analysis revealed that 2QUE caused preferential adsorption of proteins linked to acute immune responses (SAA1, SAA2 and SAA4). The QUE-doped compositions promoted the elongation of human osteoblasts (HOb) and the formation of structures such as lamellipodia and filopodia. These findings were consistent with the results of proteomic analysis. The data obtained in the analysis indicated preferential adsorption of proteins associated with enhanced cell adhesion (e.g. COF1 and PROF1), up-regulation of integrin and cytoskeleton reorganisation by Rho GTPase pathways. The QUE compositions showed increased adsorption of proteins related to oxidative stress inhibition; they also decreased the iNOS expression in macrophages in vitro. Our results suggest that the 0.5QUE composition may be optimal for further study due to its combination of anti-inflammatory effects, enhancement of cell adhesion capacity and protective potential against oxidative stress.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. Carlos Arias-Mainer: writing – original draft, data curation, investigation, formal analysis. Francisco Romero-Gavilán: data curation, investigation, formal analysis, writing – review & editing. Andreia Cerqueira: data curation, investigation, formal analysis, writing – review & editing. David Peñarocha-Oltra: funding acquisition, resources. Iñaki García-Arnáez: data curation, investigation. Oihana Amorrotu: data curation, investigation. Mikel Azkargorta: data curation, investigation Felix Elortza: re-sources, investigation Marilo Gurruchaga: writing – review & editing, funding acquisition, project administration. Isabel Goñi: writing – review & editing, funding acquisition, project administration.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that there are no conflicts to declare.Acknowledgements
This work was supported by Ministerio Ciencia e Innovación [PID2020-113092RB-C21/AEI/10.13039/501100011033; CPP2021-008492/AEI/10.13039/501100011033/Unión Europea NextGenerationEU/PRTR], Generalitat Valenciana [CIGE/2021/082; PROMETEO/2020/069], Universitat Jaume I [UJI-B2021-25]. The authors would like to thank Raquel Oliver, José Ortega, and Iraide Escobés for their valuable technical assistance and GMI-Ilerimplant for making the titanium discs.Notes and references
- Z. Zhang, M. J. Gupte and P. X. Ma, Biomaterials and stem cells for tissue engineering, Expert Opin. Biol. Ther., 2013, 13(4), 527–540, DOI:10.1517/14712598.2013.756468.
- X. Liu, S. Chen, J. K. H. Tsoi and J. P. Matinlinna, Regener. Biomater., 2017, 4(5), 315–323, DOI:10.1093/rb/rbx027.
- A. Jaafar, C. Hecker, P. Árki and Y. Joseph, Bioengineering, 2020, 7(4), 127, DOI:10.3390/bioengineering7040127.
- J. A. Oshiro, M. P. Abuçafy, E. B. Manaia, B. L. Da Silva, B. G. Chiari-Andréo and L. A. Chiavacci, Polymers, 2016, 8(4), 91, DOI:10.3390/polym8040091.
- A. Cerqueira, F. Romero-Gavilán, N. Araújo-Gomes, I. García-Arnáez, C. Martinez-Ramos, S. Ozturan, M. Azkargorta, F. Elortza, M. Gurruchaga, J. Suay and I. Goñi, Mater. Sci. Eng., C, 2020, 116, 111262 Search PubMed.
- A. Cerqueira, F. Romero-Gavilán, I. García-Arnáez, C. Martinez-Ramos, S. Ozturan, R. Izquierdo, M. Azkargorta, F. Elortza, M. Gurruchaga, J. Suay and I. Goñi, Mater. Sci. Eng., C, 2021, 125, 112114, DOI:10.1016/j.msec.2021.112114.
- M. Martínez-Ibáñez, M. J. Juan-Díaz, I. Lara-Saez, A. Coso, J. Franco, M. Gurruchaga, J. Suay Antón and I. Goñi, J. Mater. Sci.:Mater. Med., 2016, 27, 80 Search PubMed.
- N. Araújo-Gomes, F. Romero-Gavilán, I. García-Arnáez, C. Martínez-Ramos, A. M. Sánchez-Pérez, M. Azkargorta, F. Elortza, J. J. M. de Llano, M. Gurruchaga, I. Goñi and J. Suay, JBIC, J. Biol. Inorg. Chem., 2018, 23, 459–470 Search PubMed.
- A. Ullah, S. Munir, S. L. Badshah, N. Khan, L. Ghani, B. G. Poulson, A. H. Emwas and M. Jaremko, Molecules, 2020, 25(22), 5243, DOI:10.3390/molecules25225243.
- A. V. Anand David, R. Arulmoli and S. Parasuraman, Pharmacogn. Rev., 2016, 10(20), 84–89, DOI:10.4103/0973-7847.194044.
- A. V. Anand David, R. Arulmoli and S. Parasuraman, Pharmacogn. Rev., 2016, 10(20), 84–89, DOI:10.4103/0973-7847.194044.
- S. K. Wong, K. Y. Chin and S. Ima-Nirwana, Int. J. Mol. Sci., 2020, 21(17), 6448, DOI:10.3390/ijms21176448.
- Y. Zhou, Y. Wu, W. Ma, X. Jiang, A. Takemra, M. Uemura, L. Xia, K. Lin and Y. Xu, J. Mater. Chem. B, 2017, 5, 612–625 RSC.
- V. Lezcano, S. Morelli and V. González-Pardo, Biochimie, 2022, 199, 46–59 CrossRef CAS PubMed.
- K. F. Braun, S. Ehnert, T. Freude, J. T. Egaña, T. L. Schenck, A. Buchholz, A. Schmitt, S. Siebenlist, L. Schyschka, M. Neumaier, U. Stöckle and A. K. Nussler, Sci. World J., 2011, 11, 2348–2357 CrossRef CAS PubMed.
- Z. Othman, B. Cillero Pastor, S. van Rijt and P. Habibovic, Biomaterials, 2018, 167, 191–204, DOI:10.1016/j.biomaterials.2018.03.020.
- H. P. Felgueiras, J. C. Antunes, M. C. L. Martins and M. A. Barbosa, Peptides and Proteins as Biomaterials for Tissue Regeneration and Repair, Elsevier, 2018, pp. 1–27 Search PubMed.
- F. Romero-Gavilán, N. C. Gomes, J. Ródenas, A. Sánchez, M. Azkargorta, I. Iloro, F. Elortza, I. García Arnáez, M. Gurruchaga, I. Goñi and J. Suay, Biofouling, 2017, 33, 98–111 CrossRef PubMed.
- F. Romero-Gavilán, I. García-Arnáez, A. Cerqueira, L. Scalschi, B. Vicedo, A. Villagrasa, R. Izquierdo, M. Azkargorta, F. Elortza, M. Gurruchaga, I. Goñi and J. Suay, Biomater. Sci., 2022, 11, 1042–1055 RSC.
- F. Romero-Gavilán, S. Barros-Silva, J. García-Cañadas, B. Palla, R. Izquierdo, M. Gurruchaga, I. Goñi and J. Suay, J. Non-Cryst. Solids, 2016, 453, 66–73 CrossRef.
- H. Aguiar, J. Serra, P. González and B. León, J. Non-Cryst. Solids, 2009, 355, 475–480 CrossRef CAS.
- A. Jitianu, G. Amatucci and L. C. Klein, J. Mater. Res., 2008, 23, 2084–2090 CrossRef CAS.
- L. B. Capeletti, I. M. Baibich, I. S. Butler and J. H. Z. Dos Santos, Spectrochim. Acta, Part A, 2014, 133, 619–625 CrossRef CAS PubMed.
- S. Chen, Y. Guo, R. Liu, S. Wu, J. Fang, B. Huang, Z. Li, Z. Chen and Z. Chen, Colloids Surf., B, 2018, 164, 58–69, DOI:10.1016/j.colsurfb.2018.01.02225.
- V. Campos-Bijit, N. C. Inostroza, R. Orellana, A. Rivera, A. Von Marttens, C. Cortez and C. Covarrubias, Int. J. Mol. Sci., 2023, 24(23), 16686, DOI:10.3390/ijms242316686.
- M. Rahnama-Hezavah, P. Mertowska, S. Mertowski, J. Skiba, K. Krawiec, M. Łobacz and E. Grywalska, Int. J. Mol. Sci., 2023, 24(24), 17620, DOI:10.3390/ijms242417620.
- M. Padial-Molina, P. Galindo-Moreno, J. E. Fernández-Barbero, F. O’Valle, A. B. Jódar-Reyes, J. L. Ortega-Vinuesa and P. J. Ramón-Torregrosa, Acta Biomater., 2011, 7, 771–778 CrossRef CAS PubMed.
- L. P. Singh, S. K. Bhattacharyya, R. Kumar, G. Mishra, U. Sharma, G. Singh and S. Ahalawat, Adv. Colloid Interface Sci., 2014, 214, 17–37, DOI:10.1016/j.cis.2014.10.00729.
- V. Lezcano, S. Morelli and V. González-Pardo, Biochimie, 2022, 199, 46–59 CrossRef CAS PubMed.
- Z. B. Katz, A. L. Wells, H. Y. Park, B. Wu, S. M. Shenoy and R. H. Singer, Genes Dev., 2012, 26, 1885–1890 CrossRef CAS PubMed.
- W. Bian, S. Xiao, L. Yang, J. Chen and S. Deng, BMC Complementary Med. Ther., 2021, 21(1), 243, DOI:10.1186/s12906-021-03418-8.
- B. F. Boyce and L. Xing, Arch. Biochem. Biophys., 2008, 473(2), 139–146, DOI:10.1016/j.abb.2008.03.018.
- B. F. Boyce and L. Xing, Arch. Biochem. Biophys., 2008, 473(2), 139–146, DOI:10.1016/j.abb.2008.03.018.
- J. M. Anderson, Principles of Regenerative Medicine, Elsevier, 2nd edn, 2010, pp. 693–716 Search PubMed.
- S. M. Huang, C. H. Wu and G. C. Yen, Mol. Nutr. Food Res., 2006, 50, 1129–1139 Search PubMed.
- P. A. Ruiz, A. Braune, G. Hö, L. Quintanilla-Fend and D. Haller, The Journal of Nutrition Nutritional Immunology Quercetin Inhibits TNF-Induced NF-kB Transcription Factor Recruitment to Proinflammatory Gene Promoters in Murine Intestinal Epithelial Cells 1,2, 2007, vol. 137 Search PubMed.
- Q. Xue, Y. Yan, R. Zhang and H. Xiong, Int. J. Mol. Sci., 2018, 19(12), 3805, DOI:10.3390/ijms19123805.
- A. Cerqueira, F. Romero-Gavilán, H. Helmholz, M. Azkargorta, F. Elortza, M. Gurruchaga, I. Goñi, R. Willumeit-Römer and J. Suay, ACS Biomater. Sci. Eng., 2023, 9, 3306–3319 CrossRef CAS PubMed.
- L. T. Roumenina, J. Rayes, S. Lacroix-Desmazes and J. D. Dimitrov, Trends Mol. Med., 2016, 22(3), 200–213 CrossRef CAS PubMed.
- J. R. Dunkelberger and W. C. Song, Cell Res., 2010, 20, 34–50 CrossRef CAS PubMed.
- C. Skerka, Q. Chen, V. Fremeaux-Bacchi and L. T. Roumenina, Mol. Immunol., 2013, 56(3), 170–180, DOI:10.1016/j.molimm.2013.06.001.
- K. K. Eklund, K. Niemi and P. T. Kovanen, Immune Functions of Serum Amyloid A, 2012, 32, 335–348 CAS.
- A. P. Lane, Q.-A. Truong-Tran, A. Myers, C. Bickel and R. P. Schleimer, Serum amyloid A, properdin, complement 3, and toll-like receptors are expressed locally in human sinonasal tissue, 2006 Search PubMed.
- K. Hirai, H. Furusho, N. Kawashima, S. Xu, M. C. de Beer, R. Battaglino, T. Van Dyke, P. Stashenko and H. Sasaki, J. Dent. Res., 2019, 98, 117–125 CrossRef CAS PubMed.
- E. Oh, H. Y. Lee, H. J. Kim, Y. J. Park, J. K. Seo, J. S. Park and Y. S. Bae, Exp. Mol. Med., 2015, 47, e194 CrossRef CAS PubMed.
- T. Matsunaga, T. Iyoda and F. Fukai, Colloid and Interface Science in Pharmaceutical Research and Development, Elsevier Inc., 2014, pp. 243–260 Search PubMed.
- J. B. Allard and C. Duan, Frontiers Media S.A., 2018Front Endocrinol, Lausanne, 2018, vol. 9, p. 117 DOI:10.3389/fendo.2018.00117.
- T. C. Silva de Almeida, T. M. Valverde, T. M. da Mata Martins, F. de Paula Oliveira, P. da Silva Cunha, M. A. B. Tavares, E. M. Rodrigues, J. D. S. Albergaria, G. M. Vieira, D. A. Gomes, P. L. Gastelois, R. L. de Souza, A. M. de Góes, G. T. Kitten and M. D. Martins, Mater. Today Commun., 2024, 39, 108535 CrossRef CAS.
- F. Nudelman, K. Pieterse, A. George, P. H. H. Bomans, H. Friedrich, L. J. Brylka, P. A. J. Hilbers, G. De With and N. A. J. M. Sommerdijk, Nat. Mater., 2010, 9, 1004–1009 CrossRef CAS PubMed.
- M. Raftopoulou and A. Hall, Dev. Biol., 2004, 265(1), 23–32, DOI:10.1016/j.ydbio.2003.06.003.
- X. Pang, X. He, Z. Qiu, H. Zhang, R. Xie, Z. Liu, Y. Gu, N. Zhao, Q. Xiang and Y. Cui, Signal Transduction Targeted Ther., 2023, 8, 1 CrossRef CAS PubMed.
- Y. Yang and Y. Xiao, Adv. Healthcare Mater., 2020, 9(23), 2000726 Search PubMed.
- K. Oikonomopoulou, D. Ricklin, P. A. Ward and J. D. Lambris, Semin. Immunopathol., 2012, 34(1), 151–165, DOI:10.1007/s00281-011-0280-x.
- D. Xu, M. J. Hu, Y. Q. Wang and Y. L. Cui, Molecules, 2019, 24(6), 1123, DOI:10.3390/molecules2406112355.
- J. Pei, X. Pan, G. Wei and Y. Hua, Front. Pharmacol., 2023, 14, 1147414, DOI:10.3389/fphar.2023.1147414 , https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2023.1147414.
- V. Mostert, Arch. Biochem. Biophys., 2000, 376, 433–438 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb02821j |
| This journal is © The Royal Society of Chemistry 2025 |