Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bioinspired hybrid DNA/dendrimer-based films with supramolecular chirality†
Rita
Castro
 a,
Pedro L.
Granja
a,
Pedro L.
Granja
 bc,
João
Rodrigues
bc,
João
Rodrigues
 a,
Ana Paula
Pêgo
a,
Ana Paula
Pêgo
 bcd and
Helena
Tomás
bcd and
Helena
Tomás
 *a
*a
aCQM-Centro de Química da Madeira, MMRG, Universidade da Madeira, Campus da Penteada, 9020-105 Funchal, Portugal. E-mail: lenat@staff.uma.pt
bInstituto de Investigação e Inovação em Saúde (i3S), Universidade do Porto, 4200-135 Porto, Portugal
cInstituto de Engenharia Biomédica (INEB), Universidade do Porto, 4200-135 Porto, Portugal
dInstituto de Ciências Biomédicas Abel Salazar (ICBAS), Universidade do Porto, 4050-343 Porto, Portugal
First published on 11th March 2025
Abstract
Bioinspired hybrid DNA/dendrimer films were obtained by heating long double-stranded DNA (dsDNA) above its melting temperature and, while in the denatured state, mixing it with poly(amidoamine) (PAMAM) dendrimers, followed by controlled cooling. The formation of these new types of films was found to be dependent on several parameters, including the initial heating temperature, pH, buffer composition, dendrimer generation, amine/phosphate (N/P) ratio, and cooling speed. In addition to the PAMAM dendrimers (generations 3, 4, and 5), films could also be produced with branched poly(ethylenimine) with a molecular weight of 25 kDa. The results indicated that these films were formed not only through electrostatic interactions established between the negatively charged DNA molecules and the positive dendrimers, as expected, but also through random rehybridization of the single-stranded DNA (ssDNA) during the cooling process. The resulting films are water-insoluble, transparent when thin, highly elastic when air-dried, exceptionally stable over extended periods, cytocompatible, and easily scalable. Notably, the slow cooling process allowed for the establishment of at least a partially ordered structure in the films, as revealed by circular dichroism, providing evidence of supramolecular chirality. It is envisioned that these films may have significant potential in biomedical applications, such as drug/gene delivery systems, platforms for cell-free DNA transcription and components in biosensors.
Introduction
Dendrimers may be regarded as a special type of branched polymer that grows radially from a central core, layer by layer, increasing their generation each time a layer is built.1,2 They differ from classical polymers in their characteristics and well-defined architecture, size, shape, and controlled number of terminal groups. Poly(amidoamine) (PAMAM) dendrimers, which have an ethylenediamine (EDA) core with tertiary amines at the branching points and terminal primary amines, were initially synthesized by Tomalia et al. in the 1980s.3,4 These multivalent molecules display a number of terminal groups that increase exponentially with each generation (2n + 2, where “n” is the generation number).5 Depending on the solution's pH value, PAMAM dendrimers may present different protonation degrees, resulting in conformational changes.6,7 The pKa is in the range of 3 to 6 for the internal tertiary amines, and from 7 to 9 for the terminal primary amines.5 Consequently, at a very high pH (pH > 10), PAMAM dendrimers are non-charged. They become slightly cationic at a near-neutral pH (pH ≃ 7) due to the protonation of the primary amines and finally form very strongly cationic polymers at a low pH (pH < 6) due to the full protonation of the tertiary amines. This characteristic of PAMAM dendrimers opened the possibilities for their use as vehicles to deliver genetic material into cells. Indeed, a vast body of work shows that cationic dendrimers can interact with anionic RNA or DNA molecules, neutralizing their charge and serving as condensing and transfection agents.8–12 The use of dendrimers in nucleic acid delivery was based on their resemblance to histones, the proteins that condense DNA in the nucleus of eukaryotic cells to form the chromatin.13 Histones, highly basic proteins, establish several distinct interactions with DNA (e.g., electrostatic forces, hydrogen bonds, and non-polar interactions) that together overcome the energy barrier for compaction.14,15 Approximately 147 base pairs (bp) of DNA are wrapped around a group of 8 positively charged histones (histone octamer) in a left-handed superhelix to form a nucleosome core particle.16 Each nucleosome is then connected to the next by linker DNA, forming a linear array of nucleosomes, a structure known as the “beads-on-a-string” that constitutes chromatin.15 In fact, chromatin has been shown to be not only a highly complex biological structure but also a dynamic one that responds to environmental changes, and dendrimers have been used as models to study DNA–histone interactions.17Inspired by nature and exploring the distinctive interactions between dendrimers and nucleic acids, this work introduces a novel approach to the preparation of DNA-based films. The idea was to subject long DNA to temperatures exceeding its melting temperature (Tm), inducing its denaturation and formation of single-stranded DNA (ssDNA). Then, while keeping the denatured DNA molecules at high temperature and before allowing a slow cool down, introduce dendrimers into the system. Under strict experimental conditions, the application of this methodology resulted in the formation of soft 2D films composed of DNA and dendrimers, which exhibited at least partial internal ordering, as indicated by circular dichroism analysis. Herein, we present the optimized conditions for the preparation of these films, as well as the characterization of the end materials. By exploring different experimental conditions, insights into the type of interactions responsible for film formation were obtained, which may also be valuable for understanding other DNA-based constructs, including natural or biomimetic systems such as the previously mentioned nucleosome arrays in chromatin.
Results and discussion
Optimal experimental conditions for film formation
Hybrid DNA/dendrimer-based films were successfully prepared following the strategy depicted in Fig. 1 and using PAMAM dendrimers of generation 3 (G3), 4 (G4) and 5 (G5). An optimal experimental protocol was first established, which involved long dsDNA (≃ 12 kbp, as determined by agarose gel electrophoresis, ESI,† Fig. S1). In this context, dsDNA was initially dissolved in phosphate-buffered saline solution (PBS, without Ca2+ and Mg2+) and the pH was adjusted to 6. After heating the dsDNA to 99 °C to promote denaturation into ssDNA (above its Tm of 87.5 °C), a determined volume of the specific dendrimer solution was added, and the two oppositely charged polymers were left to interact for a few minutes. Following this, the solution underwent a slow cooling process to room temperature using a controlled temperature ramp (CTR: 99 °C (5 min) → 87 °C (5 min) → 68 °C (5 min) → 50 °C (5 min) → 24 °C (5 min) → room temperature), allowing sufficient time for the system components to organize. Thin and homogeneous films were always formed at the bottom of the tube, which could be removed using a micropipette tip and easily characterized (ESI,† Movie). These films could be prepared at N/P ratios ranging from 3 to 5, where the N/P ratio represents the molar ratio between the number of primary amine groups in the dendrimers and the number of phosphate groups in the DNA. An illustrative example of the optimized protocol used for film preparation is shown in detail in the ESI,† Fig. S2, beginning with 500 μg of dsDNA and using G3 PAMAM dendrimers at an N/P ratio of 4.Confirmation of dendrimers and DNA presence in the films
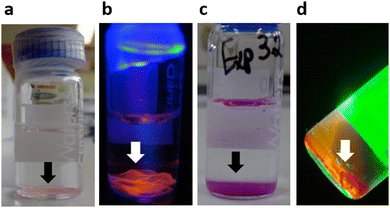
Experiments were conducted to visually confirm the actual presence of DNA and dendrimers in the film structure. To verify the presence of DNA, the films were immersed in a solution of ethidium bromide, a fluorescent molecule that intercalates between the base pairs of DNA. Upon contact with the solution, the DNA gradually became stained and showed a pale pink/orange colour (Fig. 2a), while the solution itself gradually lost its colour. When exposed to UV light, these films exhibited fluorescence in the orange range (Fig. 2b). The presence of dendrimers in the films was confirmed using rhodamine-functionalized dendrimers (Fig. 2c) as part of the film preparation process (see details of the functionalization process in the ESI,† Fig. S3). Under green light illumination, these films emitted a bright orange/red fluorescence characteristic of the rhodamine dye (Fig. 2d). Moreover, the DNA content in freshly prepared films was indirectly quantified by spectrophotometry (absorbance at 260 nm) using the supernatant, revealing a very high DNA incorporation efficiency, namely 97.7 ± 0.6%, 97.8 ± 0.3% and 98.5 ± 0.4%, for films prepared at an N/P = 4 with G3, G4 and G5 dendrimers, respectively.Impact of several experimental parameters on film formation
Studies on the effect of different experimental conditions on the successful preparation of the hybrid DNA/dendrimer-based films allowed for a comprehensive understanding of the underlying processes and the nature of the interaction between DNA and PAMAM dendrimers. These parameters included the maximum temperature applied (for dsDNA melting), the ionic composition of the dsDNA solution, the cooling program, the DNA concentration, the initial pH of the dsDNA solution and the N/P ratio, the dendrimer generation and the use of other polymers with varying molecular structures and weights.Moreover, the addition of MgCl2 at a very low concentration (0.5 mM) to the standard PBS solution prevented film formation, although the change in ionic strength was minimal. In fact, divalent Mg2+ ions are known to interact with the phosphate groups present in the DNA backbone, increasing double helix stability and the Tm value.21 Yet, agarose gel electrophoresis studies showed that the possible increase in Tm promoted by the presence of Mg2+ ions was not sufficient to impair the melting of DNA when 99 °C was applied, nor its partial hybridization upon controlled cooling (ESI,† Fig. S4). Hence, the shielding of DNA's negative charge and the decrease in interaction with the dendrimers should explain the impairment of film formation in the presence of the Mg2+ ions. Again, the importance of the electrostatic interactions between DNA and dendrimers is evidenced.
| a A quasi-film refers to a continuous layer of material that cannot be removed from the tube without breaking into fine particles. |
|---|

|
First, results showed that stable films were only formed when adjusting the initial pH to 6, regardless of the dendrimer generation or the N/P ratio. Second, at a pH of 6, film formation was restricted to three N/P ratios, namely 3, 4, and 5, regardless of the dendrimer generation. Therefore, it seems that film formation depends on the one hand on the initial global protonation level of the dendrimers in solution (determined by the initial pH of 6), and on the other hand, on the charge density associated with the individual dendrimers which is affected by the N/P ratio. Indeed, since the amount of DNA is kept constant, low N/P ratios correspond to fewer dendrimers in solution, whereas high N/P ratios correspond to a greater number. Thus, considering that the total number of protonated amines in solution remains constant at a given pH, the dendrimer's charge density is expected to be higher at lower N/P ratios and vice versa. An optimal positive charge density in the dendrimers is therefore required to achieve the right electrostatic conditions for film formation.
Also, during the course of the work, unsuccessful attempts were made to prepare the films using the 12 kbp dsDNA after subjecting it to sonication for different durations (ranging from 10 to 70 minutes). Since sonication causes DNA fragmentation, it was concluded that the DNA molecule size is crucial for film formation, most likely due to the extent of DNA rehybridization required in the process. That is, the DNA must be sufficiently long to, on the one hand, interact with the dendrimers and, on the other hand, contain chain segments capable of rehybridizing to ensure the spatial continuity of the films.
As previously mentioned, dendrimers have been used as models of histones and, in this context, protamine from salmon sperm was also assayed for film preparation. Protamine is an arginine-rich protein known for condensing DNA in sperm, as it strongly binds to and wraps around the DNA helix, forming supramolecular toroid structures.31 Due to its high content in arginine residues (pKa = 12.5), protamine is a very basic molecule. It contains many positively charged guanidinium groups that are protonated at pH values lower than its pKa value.32,33 Moreover, the binding of protonated arginine residues to DNA's phosphate groups is mainly of electrostatic nature and base-sequence independent, contrary to what happens with histones. When protamine was tested for preparing films with DNA, a very fragile film was formed at the bottom of the tube that could not be removed intact. Indeed, the MW of the used protamine (4.2 kDa) is between those of G2 and G3 PAMAMs, which may explain this result. Another possibility could be temperature-induced denaturation of protamine during the heating step, which may alter its conformation and, consequently, its interaction with DNA, affecting DNA condensation.
Characterization of DNA/dendrimer-based films
Some literature studies from the 1970s and 1980s, as well as other more recent studies, have reported this type of CD spectra for condensed forms of DNA, such as DNA particles of biological origin (e.g., sperm heads, nucleosomes),39,40 DNA aggregates produced by several condensing agents (e.g., salts, polymers),41–43 and cholesteric liquid crystalline dispersions of nucleic acids.44,45 Originally, these spectra were designated as ‘‘psi-spectra’’, meaning “polymer and salt-induced” CD spectra.41 Keller and Bustamante analysed these “abnormal” bands in the CD spectra of DNA aggregates and suggested that the anomalously large signals in CD spectra (which can be positive or negative) result from the presence of a long-range chiral structure in the aggregate, together with a “collective response” of the chromophores due to delocalization throughout the entire system of the light-induced excitations.46 This means that distant parts of the aggregate must be coupled to each other to generate abnormally high CD signals, although the distances in question are small, i.e., comparable to the wavelength of the incident light. Furthermore, based on experimental evidence, they concluded that the contribution of differential scattering to the overall CD signal in this type of system is relatively small and does not compromise the existence of differential left and right circularly polarized light absorption. Considering these findings, although Fig. 3b and c reveal the presence of long tails on the right side of the CD bands, which are indicative of differential scattering of the incident light, the observed large CD bands should be mainly due to light absorption processes. Nevertheless, both differential left- and right-circularly polarized light absorption and differential left- and right-circularly polarized light scattering have been associated with long-range chiral structures.46 Consequently, in the present case, the large bands in the CD spectra of DNA/dendrimer films should be a manifestation of the formation of a supramolecular arrangement of the condensed DNA molecules (at least partially), resulting in a chiral system. Drawing a parallel with what happens with cholesteric liquid crystalline dispersions of DNA, right-handed superhelices should form inside the films, as the observed large CD bands are positive.47 This long-range order was likely potentiated by the slow cooling program applied during film formation, which gave the system enough time for self-organization. Indeed, several studies report that dendrimers bind to DNA, bending the DNA helix and creating ordered mesophases containing DNA superhelices.13,17,28,48–50 Mansel et al. referred to this phenomenon as “dendrimer-induced DNA superhelicity”.48 Interestingly, for our DNA/dendrimer films, although the DNA underwent a denaturation process (ssDNA formation) before contact with the dendrimers, superhelices were also generated after cooling. Possibly, there are ordered domains within the films formed by dendrimers and dsDNA (domains containing DNA superhelices) that are linked by dsDNA segments resulting from the random cross-hybridization of different DNA molecules i.e., a crosslinked 2D material is formed that is partially crystalline.
While film formation seems to depend on the dendrimer's initial protonation state and the global proportion of positive and negative charges in solution, the extent of order achieved within the film appears to be generation-dependent (Fig. 3c). Possibly, G3 dendrimers are the most efficient at bending DNA, thus leading to films with CD signals of higher amplitude than those obtained with films made from G4 and G5 dendrimers. Indeed, research on the interaction between DNA and dendrimers has reported the formation of structures with different shapes (e.g., rods, toroids, and beads-on-string) depending on the dendrimer generation used.13,17,28,48–50 Therefore, it is not surprising that different generations of dendrimers also influence the characteristics of the formed films and impact CD signals. Moreover, the presence of a higher quantity of dendrimers in the film (higher N/P ratio) likely stabilizes the supramolecular structure formed. In this regard, films prepared with G3 dendrimers at an N/P ratio of 5 were those that achieved a higher degree of crystallinity. Interestingly, these films were also the ones that proved to be more physically resistant when manipulated.
Scale-up of the films
Although the optimized protocol for film formation was established using microcentrifuge tubes and a thermoblock to control the temperature, the scaling up of films was shown to be possible using glass flasks with a progressively increasing flat-bottom surface area and a simple incubator (in this case, the temperature was varied from 100 °C to RT at a rate of 15 °C h−1). Films readily formed at the bottom of the flasks (ESI,† Fig. S10). Their DNA content was controlled by adjusting the volume of the starting solutions, while their thickness was regulated by modifying the ratio between solution volumes and the deposition area. Thinner films exhibited a good level of transparency.Conclusions and perspectives
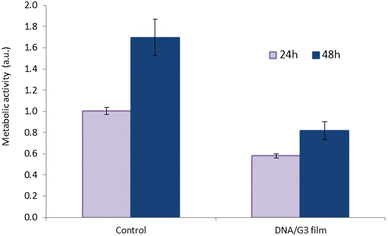
In summary, novel DNA-based films were created using a heating/cooling process driven by electrostatic interactions between DNA and PAMAM dendrimers (G3 to G5), alongside the concurrent random base pairing of ssDNA segments. The preparation of these films required carefully optimized experimental conditions, and this study provided valuable insights into their nature. Circular dichroism analysis indicates that the films exhibit supramolecular chirality, while electron microscopy also points to the presence of ordered structures. Films were also found to be stable over time in nuclease-containing media and exhibited elastic behaviour when dehydrated.We anticipate that films of this kind could find applications in biomedicine and biotechnology as drug or gene delivery systems, as platforms for in vitro cell-free DNA transcription, or in combination with DNAzymes for biosensing and diagnostic purposes.51,52 Additionally, their supramolecular chirality could be harnessed for sensor development or the creation of novel 2D materials with unique chemical and physical properties through integration with other chemical entities, such as DNA intercalators, metallic ions, or various nanomaterials. Clearly, the strong interaction between PAMAM dendrimers and DNA suggests the possibility of creating countless new materials with interesting technological and biomedical applications.
Author contributions
The manuscript was written with contributions from all authors. All authors have approved the final version of the manuscript.Data availability statement
The data supporting this article are included either in the main text or as part of the ESI.† The ESI† includes detailed experimental methods, additional figures/tables and a movie showing a film (i) being manipulated, (ii) prepared from fluorescently labelled dendrimers, (iii) stained with ethidium bromide and (iv) being degraded at high pH values.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported at CQM by the Portuguese Government through Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Inovação (Base Fund: UIDB/00674/2020, DOI: 10.54499/UIDB/00674/2020; Programmatic Fund: UIDP/00674/2020, DOI: 10.54499/UIDP/00674/2020). R. Castro acknowledges FCT for the PhD grant (Ref. SFRH/BD/87465/2012). This work was supported at i3S by POCI-01-0145-FEDER-007274, as well as by the European Regional Development Fund (ERDF) through COMPETE 2020 – Operational Programme for Competitiveness and Internationalisation (POCI), Portugal 2020. Circular dichroism measurements were conducted at the Biochemical and Biophysical Technologies i3S Platform with the assistance of Frederico Silva (PhD), and zeta potential measurements were performed at the Biointerfaces and Nanotechnology i3S Scientific Platform, a member of the Portuguese Platform of Bioimaging (PPBI; PPBI-POCI-01- 0145-FEDER-022122), with the assistance of Ricardo Vidal (MSc). A. P. P. also acknowledges the MOBILIsE Project, which has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 951723. This study was also financed by project IBEROS+ (0072_IBEROS_MAIS_1_E, Interreg-POCTEP 2021-2027).References
- D. Astruc, E. Boisselier and C. Ornelas, Chem. Rev., 2010, 110, 1857–1959 CAS.
- S. Mignani, J. Rodrigues, H. Tomas, M. Zablocka, X. Shi, A. M. Caminade and J. P. Majoral, Chem. Soc. Rev., 2018, 47, 514–532 CAS.
- D. A. Tomalia and J. M. J. Fréchet, J. Polym. Sci., Part A:Polym. Chem., 2002, 40, 2719–2728 CAS.
- D. A. Tomalia, A. M. Naylor and W. A. Goddard, Angew. Chem., Int. Ed. Engl., 1990, 29, 138–175 Search PubMed.
- D. A. Tomalia, H. Baker, J. Dewald, M. Hall, G. Kallos, S. Martin, J. Roeck, J. Ryder and P. Smith, Polym. J., 1985, 17, 117–132 CAS.
- W. Chen, D. A. Tomalia and J. L. Thomas, Macromolecules, 2000, 33, 9169–9172 CAS.
- P. K. Maiti, T. Çaǧin, G. Wang and W. A. Goddard, Macromolecules, 2004, 37, 6236–6254 CAS.
- D. Pandita, J. L. Santos, J. Rodrigues, A. P. Pêgo, P. L. Granja and H. Tomás, Biomacromolecules, 2011, 12, 472–481 CAS.
- J. L. Santos, H. Oliveira, D. Pandita, J. Rodrigues, A. P. Pêgo, P. L. Granja and H. Tomás, J. Controlled Release, 2010, 144, 55–64 CrossRef CAS PubMed.
- J. L. Santos, D. Pandita, J. Rodrigues, A. P. Pêgo, P. L. Granja, G. Balian and H. Tomás, Mol. Pharmaceutics, 2010, 7, 763–774 CrossRef CAS PubMed.
- Y. Dong, T. Yu, L. Ding, E. Laurini, Y. Huang, M. Zhang, Y. Weng, S. Lin, P. Chen, D. Marson, Y. Jiang, S. Giorgio, S. Pricl, X. Liu, P. Rocchi and L. Peng, J. Am. Chem. Soc., 2018, 140, 16264–16274 CrossRef CAS PubMed.
- V. Leiro, A. P. Spencer, N. Magalhães and A. P. Pêgo, Biomaterials, 2022, 281, 121356 CrossRef CAS PubMed.
- Y.-C. Huang, C.-J. Su, C.-Y. Chen, H.-L. Chen, U.-S. Jeng, N. V. Berezhnoy, L. Nordenskiöld and V. A. Ivanov, Macromolecules, 2016, 49, 4277–4285 CrossRef CAS.
- K. Zhou, G. Gaullier and K. Luger, Nat. Struct. Mol. Biol., 2019, 26, 3–13 CrossRef CAS PubMed.
- S. Baldi, P. Korber and P. B. Becker, Nat. Struct. Mol. Biol., 2020, 27, 109–118 CAS.
- C. A. Davey, D. F. Sargent, K. Luger, A. W. Maeder and T. J. Richmond, J. Mol. Biol., 2002, 319, 1097–1113 CAS.
- R. Dootz, A. C. Toma and T. Pfohl, Soft Matter, 2011, 7, 8343–8351 CAS.
- M. Kéri, Z. Nagy, L. Novák, E. Szarvas, L. P. Balogh and I. Bányai, Phys. Chem. Chem. Phys., 2017, 19, 11540–11548 RSC.
- K. Fant, E. K. Esbjörner, P. Lincoln and B. Nordén, Biochemistry, 2008, 47, 1732–1740 CAS.
- M. L. Örberg, K. Schillén and T. Nylander, Biomacromolecules, 2007, 8, 1557–1563 Search PubMed.
- R. Owczarzy, B. G. Moreira, Y. You, M. A. Behlke and J. A. Wälder, Biochemistry, 2008, 47, 5336–5353 CAS.
- A. M. Naylor, W. A. Goddard, G. E. Kiefer and D. A. Tomalia, J. Am. Chem. Soc., 1989, 111, 2339–2341 CAS.
- P. K. Maiti and B. Bagchi, Nano Lett., 2006, 6, 2478–2485 CrossRef CAS PubMed.
- B. Nandy and P. K. Maiti, J. Phys. Chem. B, 2011, 115, 217–230 CrossRef CAS PubMed.
- C. S. Braun, J. A. Vetro, D. A. Tomalia, G. S. Koe, J. G. Koe and C. R. Middaugh, J. Pharm. Sci., 2005, 94, 423–436 CrossRef CAS PubMed.
- M. L. Ainalem and T. Nylander, Soft Matter, 2011, 7, 4577–4594 RSC.
- Y. Su, X. Quan, L. Li, J. Zhou, Y. Su, X. Quan, L. Li and J. Zhou, Macromol. Theory Simul., 2018, 27, 1700070 CrossRef.
- K. Qamhieh, T. Nylander, C. F. Black, G. S. Attard, R. S. Dias and M. L. Ainalem, Phys. Chem. Chem. Phys., 2014, 16, 13112–13122 Search PubMed.
- L. B. Jensen, G. M. Pavan, M. R. Kasimova, S. Rutherford, A. Danani, H. M. Nielsen and C. Foged, Int. J. Pharm., 2011, 416, 410–418 Search PubMed.
- D. Fischer, Y. Li, B. Ahlemeyer, J. Krieglstein and T. Kissel, Biomaterials, 2003, 24, 1121 CrossRef CAS PubMed.
- O. A. Ukogu, A. D. Smith, L. M. Devenica, H. Bediako, R. B. McMillan, Y. Ma, A. Balaji, R. D. Schwab, S. Anwar, M. Dasgupta and A. R. Carter, Nucleic Acids Res., 2020, 48, 6108–6119 CrossRef CAS PubMed.
- B. Scheicher, A. L. Schachner-Nedherer and A. Zimmer, Eur. J. Pharm. Sci., 2015, 75, 54–59 CrossRef CAS PubMed.
- I. Ruseska, K. Fresacher, C. Petschacher and A. Zimmer, Nanomaterials, 2021, 11, 1508 CrossRef CAS PubMed.
- E. Bivehed, B. Hellman, Y. Fan, J. Haglöf and S. Buratovic, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2023, 891, 503680 CrossRef CAS PubMed.
- M. Liu, L. Zhang and T. Wang, Chem. Rev., 2015, 115, 7304–7397 CrossRef CAS PubMed.
- B. Ranjbar and P. Gill, Chem. Biol. Drug Des., 2009, 74, 101–120 Search PubMed.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- M. Vorlíčková, I. Kejnovská, K. Bednářová, D. Renčiuk and J. Kypr, Chirality, 2012, 24, 691–698 CrossRef PubMed.
- I. Tinoco, M. F. Maestre, C. Bustamante and D. Keller, Pure Appl. Chem., 1984, 56, 1423–1428 CrossRef CAS.
- M. K. Cowman and G. D. Fasman, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 4759 CrossRef CAS PubMed.
- A. V. Pietrini and P. L. Luisi, Biochim. Biophys. Acta, Biomembr., 2002, 1562, 57–62 Search PubMed.
- M. F. Maestre and C. Reich, Biochemistry, 1980, 19, 5214–5223 CrossRef CAS PubMed.
- Y. Yevdokimov, V. Salyanov, S. Skuridin, A. Dembo, Y. Platonov, A. Il’ina and V. Varlamov, Dokl. Biophys., 2000, 374, 47–49 CrossRef PubMed.
- C. F. Jmdan, L. S. Lerman and J. H. Venable, Nature, New Biol., 1972, 236, 67–70 Search PubMed.
- Y. M. Yevdokimov, S. G. Skuridin, S. V. Semenov, L. A. Dadinova, V. I. Salyanov and E. I. Kats, J. Biol. Phys., 2017, 43, 45–68 Search PubMed.
- D. Keller and C. Bustamante, J. Chem. Phys., 1986, 84, 2972–2980 CrossRef CAS.
- S. V. Semenov and Y. M. Yevdokimov, Biophysics, 2015, 60, 188–196 CrossRef CAS.
- B. W. Mansel, C. J. Su, C. Y. Chen, C. M. Young, Y. C. Huang, C. C. Yang and H. L. Chen, Soft Matter, 2021, 17, 7287–7293 RSC.
- C. Y. Chen, C. J. Su, S. F. Peng, H. L. Chen and H. W. Sung, Soft Matter, 2011, 7, 61–63 CAS.
- C. J. Su, H. L. Chen, M. C. Wei, S. F. Peng, H. W. Sung and V. A. Ivanov, Biomacromolecules, 2009, 10, 773–783 CAS.
- J. Yan, X. Ma, D. Liang, M. Ran, D. Zheng, X. Chen, S. Zhou, W. Sun, X. Shen and H. Zhang, Nat. Commun., 2023, 14, 1–21 Search PubMed.
- W. Ma, Y. Zhan, Y. Zhang, C. Mao, X. Xie and Y. Lin, Signal Transduction Targeted Ther., 2021, 6, 1–28 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb02761b |
| This journal is © The Royal Society of Chemistry 2025 |