Root-cause analyses for 3D intermolecular packing network formation in central unit extended small molecular acceptors†
Jiaxin
Guo‡
a,
Xiangjian
Cao‡
a,
Zheng
Xu
a,
Tengfei
He
a,
Xingqi
Bi
a,
Zhaoyang
Yao
 *a,
Yaxiao
Guo
b,
Guankui
Long
*a,
Yaxiao
Guo
b,
Guankui
Long
 c,
Chenxi
Li
a,
Xiangjian
Wan
c,
Chenxi
Li
a,
Xiangjian
Wan
 *a and
Yongsheng
Chen
*a and
Yongsheng
Chen
 *a
*a
aState Key Laboratory and Institute of Elemento-Organic Chemistry, The Centre of Nanoscale Science and Technology, Key Laboratory of Functional Polymer Materials, Renewable Energy Conversion and Storage Center (RECAST), College of Chemistry, Nankai University, Tianjin 300071, China. E-mail: zyao@nankai.edu.cn; yschen99@nankai.edu.cn; xjwan@nankai.edu.cn
bState Key Laboratory of Separation Membranes and Membrane Processes, School of Chemistry, Tiangong University, Tianjin 300387, China
cSchool of Materials Science and Engineering, National Institute for Advanced Materials, Renewable Energy Conversion and Storage Center (RECAST), Nankai University, Tianjin, 300350, China
First published on 15th November 2024
Abstract
The enhanced three-dimensional (3D) intermolecular packing network in central unit extended small molecular acceptors (SMAs) has boosted the performance of organic solar cells (OSCs) significantly by improving the inner exciton/charge photodynamics. However, the structural profiles that determine the formation of an efficient 3D packing network are still shrouded in mystery. Herein, a series of SMAs (CH1, CH2, CH3, CH20 and CH8F) with/without central conjugation extension and substitutions are systematically investigated at both single-molecule and aggregate levels. Notably, by examining the evolution of packing networks and modes from CH1 to CH8F, the determining role of central unit extension and halogenation in constructing an enhanced 3D intermolecular packing network is revealed for the first time. Additionally, binary OSCs of CH8F, which combine central unit extension with fluorination, achieve a first-class power conversion efficiency (PCE) of 19.02%, markedly outperforming their counterparts. These root-cause analyses unveil the essential structural elements for forming superior 3D packing networks and will further boost the rational design of SMAs.
1 Introduction
The power conversion efficiency (PCE) of organic solar cells (OSCs) has surpassed 20% owing to the extensive exploration of new conceptual light-harvesting small molecular acceptors (SMAs);1–8 nevertheless, it still lags far behind that of their perovskite or silicon counterparts.9–14 These PCE gaps primarily result from the unsatisfactory features of organic aggregates which are mainly bonded together by weak non-covalent bonds (such as π–π or hydrogen bonding interactions) rather than the much stronger covalent bonds.15–19 Moreover, the broad but highly flexible skeletons of organic molecules usually lead to a larger packing disorder and more amorphous domains in aggregates, in sharp contrast with inorganic materials that possess strict periodicity in spatial lattices.20–23 These intrinsic characteristics account for the poor molecular packing/crystalline ordering within the loose aggregation of organic materials, which theoretically hinders efficient exciton diffusion or charge carrier transfer/transport and may lead to severe charge recombination.24–27 Bearing these thoughts in mind, the rational optimization of molecular packing behaviors through unremitting structural innovations should be quite important if further breakthrough of OSCs is expected.Despite the dire scarcity of correlations between molecular structure and molecular packing features, we can still get a hint of inspiration from the leapfrog development of two generations of SMAs, typically from ITIC to Y6.28,29 As illustrated in Fig. 1a, “end-to-end” (E/E) is the most dominant packing mode for ITIC, thus forming a two-dimensional (2D) packing network.30 The central π-core of ITIC is relatively isolated from intermolecular interactions due to the bulky substituent on its quaternary carbons. Conversely, Y6 exhibits a unique three-dimensional (3D) stacking network structure, achieved by an emerging “end-to-end and central-to-central” (E/E + C/C) packing mode.31–33 Note that plenty of investigations have proven that such an exotic 3D stacking network significantly enhances charge transfer and transport dynamics compared to the 2D patterns in ITIC, thereby boosting the PCE of OSCs significantly.34,35 Following this logic, further carrying forward and optimizing the 3D stacking network of SMAs should be an effective pathway to yield more efficient OSCs. However, the structural profiles or essential structural elements that determine this packing network evolution from 2D to 3D remain shrouded in mystery. By comparing the structures of ITIC and Y6 in detail, the possible determining factors may be as follows: (1) the introduced thiadiazole central unit. The “E/E + C/C” packing occurs in Y6 due to the π–π stacking between two thiadiazole moieties. Therefore, the central units seem to be the most important factor that causes the above evolution from 2D to 3D. (2) A banana- or C-shaped molecular conformation. Two pyrrole rings of Y6 are fused to a benzothiadiazole unit with the same orientation, which results in a bent molecular geometry. This banana-shaped molecular conformation makes the crossed packing of two molecules possible and may determine the formation of a 3D intermolecular packing network. (3) The flatter molecular configuration. Y6 removes the quaternary sp3 carbon from the molecular backbone and is characterized by several flexible aliphatic side chains rather than the relatively rigid benzyl side groups found in ITIC. Given that the most conspicuous feature of the “E/E + C/C” packing mode is that the central unit participates in molecular packing greatly, lots of novel SMAs have been further explored by conducting central unit 2D expansion, such as CH6, CH17, CH22, etc.36–39 However, nearly no study has been carried out to unveil the underlying and likely close association between expanded central units and enhanced 3D molecular packing networks.
 | ||
| Fig. 1 (a) Single crystal structures of ITIC and Y6, including their dominant intermolecular packing modes and packing networks. (b) Chemical structures of CH1, CH2, CH3, CH20, and CH8F. | ||
With the aim of identifying the indispensable structural component in SMAs that boosts molecular packing evolving from 2D to 3D, and guiding further rational molecular design, a root-cause analysis was carried out by systematically comparing the SMAs of CH1, CH2, CH3, CH20 and CH8F with varying degrees of vertical conjugation extension and central substituents (Fig. 1b). For instance, CH1, which lacks the central thiadiazole unit, reduces the conjugation of the central unit. CH2 and CH3 were constructed by introducing electron-donating methoxy groups and electron-withdrawing fluorine atoms on the central unit of CH1. CH20 is characterized by a central 2D conjugation extension derived from CH1, and CH8F combines both the central conjugation extension and halogenation. The root-cause analyses of these SMAs at both single-molecule and aggregate levels firstly reveal that the central unit extension and halogenation synergistically play a determining role in constructing an enhanced 3D intermolecular packing network, rather than the banana-shaped or flat molecular conformation. As a result, CH8F, featuring both central unit extension and fluorination, achieves a first-class PCE of 19.02%, significantly outperforming its counterparts. By unveiling the essential structural elements for forming 3D packing networks, our work will further boost the rational design of SMAs.
2 Results and discussion
2.1 Synthesis and physicochemical properties
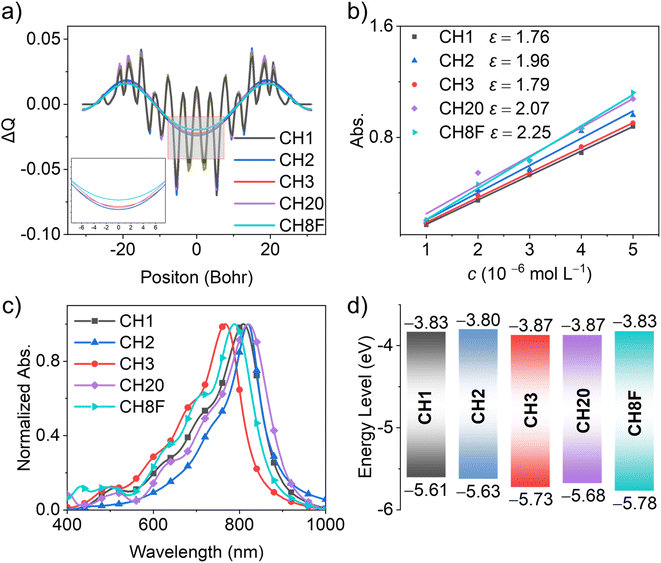
The synthesis of CH1, CH20 and CH8F was based on routes reported in the literature.7,40,41 For CH2 and CH3, the synthetic details are illustrated in Schemes S1 and S2,† with characterizations presented in Fig. S35–S53.† Despite the variation in electron-donating/electron-withdrawing substituents or central conjugation extension, a conspicuous A–D–A architecture can be observed for all SMAs, as indicated by the characteristic peak-valley-peak shape of ΔQ (Fig. 2a and S1†) and an electrostatic surface potential analysis (Fig. S2†). Such an A–D–A feature in SMAs could enhance molecular stacking, facilitate charge photodynamics and consequently improve the performance of OSCs.42,43 As displayed in Fig. S3,† it is interesting to note that the conjugation extension of the central unit significantly increases the molecular polarizability of SMAs, while the fluorination seems to reduce it marginally. Specifically, CH20 and CH8F possess the polarizabilities of 1328 and 1312 bohr3, respectively, much larger than those of the other three counterparts (1158, 1212 and 1142 bohr3 for CH1, CH2 and CH3, respectively). However, CH3 and CH8F exhibit a slightly decreased polarizability compared to CH1 and CH20 after fluorination on the central units. Theoretically, the enlarged polarizability is expected to increase the relative dielectric constant and electron mobility of SMAs in their solid films,38,44,45 as elaborated below.Thereafter, the optical absorption of SMAs was investigated using UV-vis spectroscopy. As anticipated, methoxy-substituted CH2 and conjugation-extended CH20 demonstrate subtle red-shifted absorption compared to that of CH1 in solution (Fig. S4†). In contrast, the fluorination on the central units causes notably blue-shifted absorption (e.g., CH1 vs. CH2, CH20 vs. CH8F), which should be caused by the electron-withdrawing feature of fluorine atoms. In addition, the central conjugation extension on CH20 and CH8F contributes to a larger molar extinction coefficient (for example, 2.07 × 105 and 2.25 × 105 M−1 cm−1 for CH20 and CH8F, respectively, compared to 1.76 × 105, 1. 96 × 105 and 1.79 × 105 M−1 cm−1 for CH1, CH2, and CH3), which will favor more efficient photon utilization (Fig. 2b). Regarding solid films, an obvious red shift of the maximum absorption peaks could be observed compared to that in solution, with the Δλ of 73, 70, 60, 67 and 60 nm for CH1, CH2, CH3, CH20, and CH8F, respectively (Fig. 2c and Table S1†). Note that the lower degree of red shifting for CH3, CH20 and CH8F may be ascribed to the packing preference for H-aggregates, implying more central units participating in molecular packing modes.46,47 Cyclic voltammetry (CV) measurements revealed the HOMO/LUMO energy levels for CH1, CH2, CH3, CH20, and CH8F in their solid films, being −5.61/−3.83 eV, −5.63/−3.80 eV, −5.73/−3.87 eV, −5.68/−3.87 eV, and −5.78/−3.83 eV, respectively (Fig. 2d and S5†). These are roughly consistent with the trends of HOMO/LUMO obtained from theoretical calculations (Fig. S6 and Table S1†). In light of the identical IC-2F end groups in SMAs, similar LUMO energy levels are afforded. Meanwhile, significant variations occur in the HOMOs due to the diverse structural modifications on the donor parts of S,N-heteroacene Especially for CH3 and CH8F, which possess electron-deficient fluorine atoms on the central unit, the HOMO energy levels are greatly downshifted by 0.12 and 0.10 eV compared to those of non-fluorinated CH1 and CH20, respectively. This further confirms that these types of SMAs resemble the A–D–A architecture, as also indicated in Fig. 2a. Note that the bandgap trend derived from CV aligns well with that obtained from the film absorption spectra (Table S1†).
2.2 Crystallographic analysis
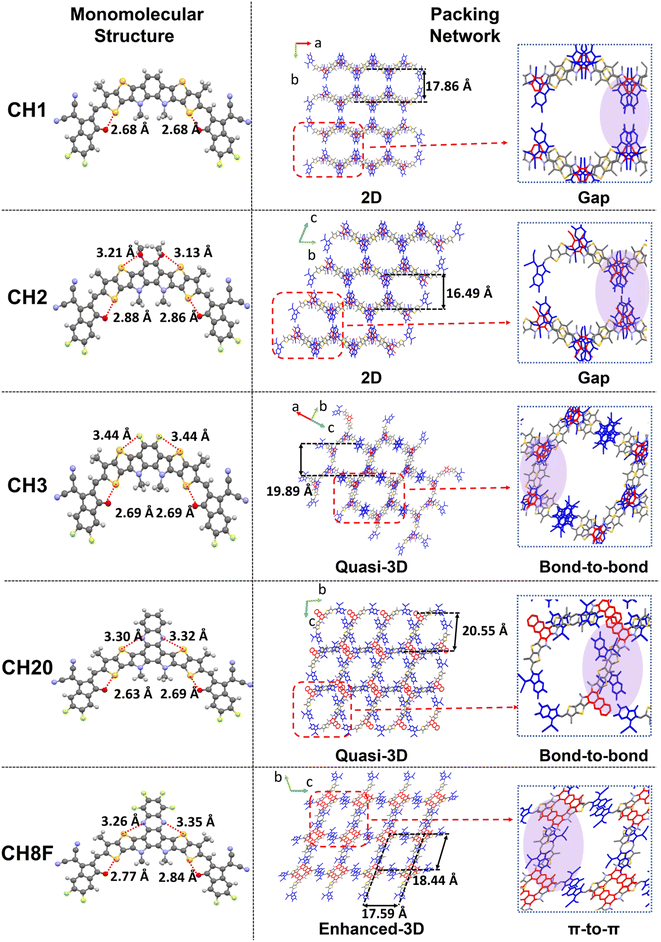
Molecules in single crystals are bonded together by balancing the attractive and repulsive strengths induced by the field of neighboring molecules. In general, a more thermodynamically stable molecular packing is inclined to form, obeying the principles of maximizing density while minimizing free volume and packing energy.48,49 Therefore, it is meaningful to simulate the real scenarios of molecular packing in spin-coating films through crystal analysis. Herein, diverse packing modes have been observed in selected SMAs which possess similar banana-shaped backbones but quite different central units (Fig. 3), thus providing a valuable opportunity to elucidate the structural factors determining 3D intermolecular packing network formation.The detailed parameters for new single crystals of CH2 and CH3 are enumerated in Tables S2 and S3.† Additionally, single crystal data of CH1, CH20 and CH8F were obtained from related literature.7,40,41 As illustrated in Fig. 3, all the studied SMAs exhibit similar banana-shaped and planar conformations, likely enforced by the rigid conjugated backbones of the central core and intramolecular S–O interactions with end groups. The torsion angles reflecting molecular planarity are quite small for CH1, CH2, and CH3, being 1.38°, 1.64° and 2.31°, respectively (Fig. S7†). In contrast, significantly increased torsion angles are observed in CH20 (3.86°) and CH8F (10.74°). This may be caused by the enlarged steric hindrance originating from the two elongated but conflicting alkyl chains on nitrogen sites. Since the excited state of organic molecules usually tends to be more planar than their ground state,50–53 such severe skeleton distortion in CH20 and CH8F will at least increase the reorganization energy at single molecular levels and most likely lead to more energy losses.54,55
As unveiled by the single crystal topological structures in Fig. 3, the variation in the central unit of SMAs results in dramatically different intermolecular packing networks. Overall, CH1 and CH3 can be classified as monoclinic systems, while the other three molecules are triclinic. It is quite interesting that CH1 and CH2 display a 2D connection, which is similar to ITIC. The inadequate interlayer interactions promote the molecular packing topological structure towards 2D brickwork. In sharp contrast, the fluorinated CH3 and conjugation-extended CH20 are capable of forming quasi-3D stacking networks, albeit mainly through relatively weak bond-to-bond interactions to link adjacent layers. Moreover, when combining conjugation extension of the central unit with adequate fluorination, a much-enhanced 3D intermolecular stacking network is afforded by CH8F. It seems that sufficient halogen bonding and π–π stacking of central units could work synergistically to play a dominant role in constructing optimized 3D networks.
Intrinsically, the distinct molecular packing frameworks above should arise from diverse intermolecular packing modes. This is why we have extracted the main packing modes with intermolecular potentials >|70| kJ mol−1 and presented them in Fig. 4 and S8–S12†. CH1 and CH2 exhibit two widely observed packing modes: “E/E” and “dual end-to-bridge” (dual E/b) modes, which contribute to the 2D brickwork packing similar to ITIC. Interestingly, central fluorination and pronounced conjugation expansion significantly enhance the interactions between the central unit and other moieties, leading to greater involvement of the central unit in molecular packing and a more diverse array of stacking modes. For example, in addition to the “E/E” and “dual E/b” modes, the typical packing mode “E/E + C/C” emerges in CH3, while CH20 possesses another two newly formed “central-to-central” (C/C) and “dual end-to-central” (dual E/C) modes with the disappearance of “E/E”. As regards CH8F, which combines central fluorination with conjugation expansion, two central units participating in packing modes, “dual central-to-bridge” (dual C/b) and “dual E/C”, can be observed besides the “E/E” mode. The increasing involvement of central units in packing from CH1 to CH8F is likely due to the gradually enhanced halogen bonding and π–π stacking of the central units, thus making them thermodynamically more stable during the slow crystallization process.38 As a result, the average π–π stacking distances for CH3 and CH20 decreased to 3.37 and 3.38 Å, respectively, compared to 3.64 Å for CH1 and 3.59 Å for CH2, indicating tighter molecular packing after central unit fluorination or conjugation expansion. Furthermore, CH8F affords the smallest average π–π packing distance of 3.33 Å among the five SMAs, with the largest intermolecular potential of −209.7 kJ mol−1 in the “dual E/C” packing mode and −193.0 kJ mol−1 in the “dual C/b” packing mode. This anticipated molecular packing of CH8F is expected to improve molecular crystallinity and facilitate charge transport in multiple directions within thin films.
In short, three new phenomena conducive to the rational design of SMAs have been highlighted. They are as follows: (1) compared to ITIC, the more planar and banana-shaped molecular conformation of SMAs, for example CH1 and CH2, can enable the π-bridge to participate in molecular stacking more effectively. Therefore, the new “dual E/b” packing mode appears in addition to maintaining the conventional “E/E” mode. (2) Central halogenation or conjugated extension is quite important for promoting the evolution of packing topological structures from 2D to 3D. This should be mainly attributed to the enhanced halogen bonding and π–π stacking interactions between the central units and other moieties. (3) Combining the sufficient halogenation with proper conjugated extension on the central unit of SMAs may be quite essential if much-optimized 3D intermolecular stacking networks are expected.
2.3 Molecular packing characteristics in aggregates
Fig. S13 and S14† show the X-ray diffraction patterns derived from single crystals of SMAs, which roughly correspond with the grazing-incidence wide-angle X-ray scattering (GIWAXS) patterns of neat films (Fig. S15†).56 This indicates that the main packing modes of SMAs in single crystals are largely retained in their neat films. Therefore, the diverse molecular packing modes and topological structures of SMAs could undoubtedly influence their aggregated properties in solid films. As shown in the line-cut profiles of GIWAXS in Fig. 5a and corresponding parameters in Table S4,† all the SMAs except CH2 demonstrate a desired face-on packing orientation, revealed by the obvious (010) peaks in the out-of-plane (OOP) direction. Note that CH8F possesses the smallest intermolecular π–π stacking distance of ∼3.63 Å compared to ∼3.67 Å for CH1, CH2, and CH20, while maintaining larger crystal coherence lengths (CCLs) in both OOP and in-plane (IP) directions. This suggests the improved molecular packing strength and crystalline ordering for CH8F, which will favor efficient electron transport. These positive results highlight the great advantages of increased fluorination and conjugated extension for the central unit of SMAs.As mentioned above, the increasingly twisted backbones are prone to increase the reorganization energies at the single molecular level from CH1 to CH8F, in light of the conventionally more planar excited states of SMAs compared to their ground states. This is in good accordance with the DFT-calculated electron reorganization energy values shown in Fig. 5b and Table S5.† In sharp contrast, CH8F exhibits the smallest Stokes shifts in its solid film (Fig. 5c and S16†), indicating lower reorganization energy compared to the other SMAs. This may mainly benefit from the formation of superior enhanced-3D intermolecular packing networks in CH8F. Similarly, theoretically predicted exciton binding energies (Eb) of all the SMAs in the gas phase fluctuate around 1.6 eV (Table S5†). However, the experimentally measured Ebs derived from temperature-dependent fluorescence show significant differences (Fig. 5d and S17†).50,54,57,58 It should be noted that while the absolute values of the exciton binding energies measured by this method may vary depending on the experimental conditions, the observed trend remains reliable. For instance, the conjugation-extended and sufficiently fluorinated CH8F exhibits a much smaller Eb of approximately 29 meV, compared to the values measured for CH1, CH2, CH3, and CH20 (80, 84, 92, and 62 meV, respectively). The effective exciton delocalization caused by more compact and ordered molecular packing in CH8F may account for the dramatically reduced Eb.52 Furthermore, such a small Eb is expected to ensure efficient exciton dissociation of the excited CH8F molecule even driven by a very small driving force. Finally, the electron mobility (μe) of neat films was evaluated by performing the space-charge limited current (SCLC) method (Fig. S18†).59 As displayed in Fig. 5e, CH8F exhibits a significantly higher μe compared to the other SMAs, underscoring its superiority as an electron-transporting material. The increased μe is primarily attributed to the superior packing behaviors of CH8F, including more compact and ordered molecular packing, enhanced-3D intermolecular packing networks, etc.
2.4 Photovoltaic performance
The central conjugation extension and fluorination have distinctly influenced the molecular stacking patterns and 3D assembly networks, which may also exert positive or negative effects on the photovoltaic performances of SMAs. In this study, a well-known donor material, PM6,60 was employed to blend with CH1, CH3, CH20, and CH8F to compose the active layers of OSCs. Given the elevated HOMO energy level of CH2, PBDB-T61 was chosen as the donor to fabricate devices. For a clear comparison, the corresponding photovoltaic performance of CH2 is displayed in Fig. S20 and S21† and will not be compared with the other four OSCs. Detailed device fabrication and characterization information, including optimization procedures, is provided in the ESI (Tables S6–S10†). The best-performing OSC based on CH1 achieved a PCE of 15.76%, with a VOC of 0.906 V, JSC of 24.06 mA cm−2, and a fill factor (FF) of 72.26% (Fig. 6a and Table 1). After central fluorination or conjugation extension, CH3 and CH20-based OSCs exhibited obviously improved PCEs of 16.59% and 16.45%, respectively. Encouragingly, despite the slightly blue-shifted absorption of CH8F, its OSC further reaches a first-class PCE of 19.02% with an impressive FF exceeding 80%. To validate the photovoltaic performance, the external quantum efficiencies (EQEs) of OSCs were also measured (Fig. 6b). CH1-, CH3-, CH20- and CH8F-based devices exhibited corresponding integrated JSC values of 23.42, 23.17, 24.94 and 25.74 mA cm−2, close to those afforded by J–V tests. In addition, we also evaluated the photovoltaic performance of D18:CH3-based OSCs, which afford an excellent PCE of 16.17%, accompanied by a VOC of 0.888 V, a JSC of 24.92 mA cm−2 and an FF of 73.06% (Fig. S19, Tables S11 and S12†).| Active layer | V OC (V) | J SC (mA cm−2) | J EQESC (mA cm−2) | FFa (%) | PCEa (%) |
|---|---|---|---|---|---|
| a Photovoltaic data in parentheses are reported as averages by averaging over 15 individual devices, where ± represents the standard deviation of the mean. The parameters outside parentheses are from the champion cells. | |||||
| PM6:CH1 | 0.906 (0.899 ± 0.004) | 24.06 (23.99 ± 0.26) | 23.42 | 72.26 (72.15 ± 0.58) | 15.76 (15.56 ± 0.12) |
| PBDB-T:CH2 | 0.802 (0.798 ± 0.004) | 18.19 (17.43 ± 0.40) | 17.29 | 67.12 (64.66 ± 1.74) | 9.79 (8.99 ± 0.30) |
| PM6:CH3 | 0.898 (0.901 ± 0.004) | 23.37 (23.31 ± 0.22) | 23.17 | 79.10 (78.73 ± 0.44) | 16.59 (16.53 ± 0.09) |
| PM6:CH20 | 0.888 (0.887 ± 0.002) | 25.80 (25.68 ± 0.17) | 24.94 | 71.80 (71.51 ± 0.51) | 16.45 (16.29 ± 0.10) |
| PM6:CH8F | 0.887 (0.885 ± 0.002) | 26.80 (26.71 ± 0.24) | 25.74 | 80.01 (79.88 ± 0.42) | 19.02 (18.88 ± 0.11) |
To understand the fundamental reasons for photovoltaic performance variations, the efficiencies of exciton dissociation (ηdiss) and charge collection (ηcoll) were evaluated (Fig. 6c). CH3 and CH8F have superior ηdiss/ηcoll values of 98.5%/89.3% and 99.0%/90.7% compared to 95.7%/84.1% for CH1 and 94.4%/81.5% for CH20. Furthermore, the dependence of JSC and VOC on light intensity (Fig. S22†) indicates similar and effectively suppressed bimolecular recombination, and gradually reduced trap-assisted recombination from CH1 to CH8F. Meanwhile, the superior charge transfer/transport dynamics in CH3- and CH8F-based OSCs should account for their larger EQE values and increased FFs, highlighting the crucial role of central fluorination in improving photovoltaic performance. In addition, the more efficient exciton dissociation in PM6:CH3 and PM6:CH8F blends can also be confirmed by their higher photoluminescence (PL) quenching efficiencies (Fig. S23 and S24†). As displayed in Fig. 6d and S25,† CH3 and CH8F-based devices yield higher relative dielectric constants (εr) compared to the others, benefiting from the strengthened fluorine-induced intermolecular secondary interactions. The larger εr is conducive to efficient charge transport. Thus, it is reasonable to observe the slightly improved μe for CH3 and CH8F-based devices (Fig. S26†). These results suggest that sufficient halogenation and proper conjugated extension on the central units of SMAs can improve charge separation and transport dynamics effectively in the resulting OSCs, leading to a marked improvement of JSCs, FFs and final PCEs.
To better understand the energy loss mechanisms in OSCs, we carried out a detailed analysis of the energy losses and their contributing factors. As depicted in Fig. S27,† the optical bandgaps (Eg) of blended films were determined by analysing the derivatives of EQE curves.62,63 Based on this, the total energy losses (Elosss) of OSCs were calculated to be 0.535, 0.617, 0.520 and 0.572 eV for CH1, CH3, CH20 and CH8F, respectively, all of which are at relatively low levels. To gain a more profound understanding of the underlying causes of Eloss, we evaluated all three parts of Eloss, as illustrated in Table S13.† The detailed calculation method for the three parts is provided in Note S3.† Among them, ΔE3 represents the non-radiative energy loss, which is a primary concern in current high-performance OSCs. Although some discrepancy can be observed for ΔE3 depending on the calculation method, the trend of ΔE3 aligns well with the values estimated from EQEEL (Fig. S28†),64 and they all remain at relatively low levels.
2.5 Morphology analysis
The morphology of active layers largely determines the charge transfer/transport/recombination dynamics and eventual photovoltaic performance of OSCs.65,66 Therefore, atomic force microscopy-infrared spectroscopy (AFM-IR) tests were performed to detect the 2216 cm−1 signal of acceptors (Fig. S29†) and unveil the morphological characteristics. As shown in Fig. 7a, S30, and S31,† all the blended films display smooth surfaces and obvious D/A interpenetrating morphologies. A statistical analysis of D/A domains shows a progressive enlargement from CH1- to CH8F-based blended films: 6.8 nm for PM6:CH1, 8.7 nm for PM6:CH3, 9.3 nm for PM6:CH20, and 10.6 nm for PM6:CH8F (Fig. 7b and S32†). This suggests that central fluorination and conjugation extension could potentially enhance molecular crystallinity and adjust the size of phase domains. These conclusions were further validated by GIWAXS measurements (Fig. 7c and S33, Table S14†). The sharp (010) diffraction peaks in the OOP direction imply that the preferential face-on packing orientation for SMAs was preserved in blended films. Among them, CH1, CH3 and CH20 show peaks located at 1.71 Å−1, assigned to a dπ–π of 3.67 Å. Moreover, a slightly smaller dπ–π for CH8F (q = 1.73 Å−1, dπ–π = 3.63 Å) can be further observed. The reduced dπ–π suggests the strong crystallinity of CH8F and is consistent with its larger and more suitable phase-separation in blended films. In order to compare the miscibility between PM6 and SMAs, we measured the contact angles and calculated the Flory–Huggins interaction parameters (χ). As shown in Fig. S34 and Table S15,† a gradually increased χD:A was observed from CH1 to CH8F (0.05 for CH1, 0.12 for CH3, 0.33 for CH20, and 0.52 for CH8F), indicating the reduced D/A miscibility with the introduction of central fluorine and conjugation extension. This is also consistent with the morphological evolution in their blended films discussed above. These improvements in D/A blending morphology fully validate the key role of central conjugation expansion and fluorination in regulating molecular packing and phase-separation. | ||
| Fig. 7 (a) AFM-IR phase images of blended films. (b) Statistical distribution of fibril diameters (listed in brackets). (c) 2D GIWAXS patterns of blended films. | ||
3 Conclusion
With the aim of identifying the essential structural components in SMAs for constructing 3D molecular packing networks and guiding further rational molecular design, a root-cause analysis was carried out by systematically comparing CH1, CH2, CH3, CH20, and CH8F with varying degrees of central conjugation extension and substituents. A comprehensive single crystal and GIWAXS analysis revealed several new findings to correlate molecular packing behaviors and chemical structures: (1) the planar and banana-shaped molecular conformation of SMAs, such as CH1 and CH2, can enable the π-bridge to participate in molecular stacking more effectively. (2) The halogenation or conjugated extension of central units is quite important for promoting the evolution of packing topological structures from 2D to 3D. (3) In view of designing more efficient SMAs, combining proper conjugated extension with sufficient halogenation on the central units may be quite essential for forming enhanced-3D intermolecular packing networks and yielding superior OSCs. Consequently, CH8F with both complete central fluorination and 2D conjugation extension afforded more compact and ordered molecular packing, increased dielectric constant, and significantly reduced exciton binding energy. Moreover, when blended with the PM6 donor, the PM6:CH8F blended film formed a pronounced fiber network with suitable phase-separation compared to other counterparts. These benefits collectively facilitate the more efficient charge generation and transport dynamics in CH8F-based OSCs. Eventually, OSCs of PM6:CH8F achieved a first-class efficiency of 19.02%, significantly exceeding the 15.76%, 16.59% and 16.45% for PM6:CH1, PM6:CH3 and PM6:CH20, respectively. Our work disclosed the critical role of central fluorination and conjugation extension in forming enhanced-3D intermolecular packing networks of SMAs for the first time and bridged the molecular structures and their packing behaviors to some extent. These findings could provide valuable insights into the structural profiles of high-efficiency SMAs and are likely to further boost the record efficiency of OSCs.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
J. G. and X. C. contributed equally to this work. The synthetic works were carried out by J. G.; the device optimizations and measurements were carried out by X. C.; T. H. and G. L. performed the DFT calculations. Y. C., X. W. and Z. Y. conceived and directed the study, and revised the manuscript. Z. X., X. B., Y. G. and C. L. helped to analyze the data and commented on the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors gratefully acknowledge the financial support from the MOST of China (2022YFB4200400, 2019YFA0705900, 2023YFE0210400), NSFC (21935007, 52025033, 22361132530, 22479081, 22309090), Tianjin city (20JCZDJC00740) and 111 Project (B12015). The authors gratefully acknowledge the cooperation of the beamline scientists at the BSRF-1W1A beamline.References
- Z. Chen, J. Ge, W. Song, X. Tong, H. Liu, X. Yu, J. Li, J. Shi, L. Xie, C. Han, Q. Liu and Z. Ge, Adv. Mater., 2024, 2406690 CrossRef CAS.
- X. Lu, C. Xie, Y. Liu, H. Zheng, K. Feng, Z. Xiong, W. Wei and Y. Zhou, Nat. Energy, 2024, 9, 793–802 CrossRef CAS.
- S. Guan, Y. Li, C. Xu, N. Yin, C. Xu, C. Wang, M. Wang, Y. Xu, Q. Chen, D. Wang, L. Zuo and H. Chen, Adv. Mater., 2024, 2400342 CrossRef CAS.
- Y. Sun, L. Wang, C. Guo, J. Xiao, C. Liu, C. Chen, W. Xia, Z. Gan, J. Cheng, J. Zhou, Z. Chen, J. Zhou, D. Liu, T. Wang and W. Li, J. Am. Chem. Soc., 2024, 146, 12011–12019 CrossRef CAS.
- J. Fu, Q. Yang, P. Huang, S. Chung, K. Cho, Z. Kan, H. Liu, X. Lu, Y. Lang, H. Lai, F. He, P. W. K. Fong, S. Lu, Y. Yang, Z. Xiao and G. Li, Nat. Commun., 2024, 15, 1830 CrossRef CAS PubMed.
- K. Liu, Y. Jiang, G. Ran, F. Liu, W. Zhang and X. Zhu, Joule, 2024, 8, 835–851 CrossRef CAS.
- Z. Yao, X. Cao, X. Bi, T. He, Y. Li, X. Jia, H. Liang, Y. Guo, G. Long, B. Kan, C. Li, X. Wan and Y. Chen, Angew. Chem., Int. Ed., 2023, 62, e202312630 CrossRef CAS.
- H. Lu, D. Li, W. Liu, G. Ran, H. Wu, N. Wei, Z. Tang, Y. Liu, W. Zhang and Z. Bo, Angew. Chem., Int. Ed., 2024, 63, e202407007 CrossRef CAS.
- S. Zhang, F. Ye, X. Wang, R. Chen, H. Zhang, L. Zhan, X. Jiang, Y. Li, X. Ji, S. Liu, M. Yu, F. Yu, Y. Zhang, R. Wu, Z. Liu, Z. Ning, D. Neher, L. Han, Y. Lin, H. Tian, W. Chen, M. Stolterfoht, L. Zhang, W. H. Zhu and Y. Wu, Science, 2023, 380, 404–409 CrossRef CAS.
- J. J. Yoo, G. Seo, M. R. Chua, T. G. Park, Y. Lu, F. Rotermund, Y.-K. Kim, C. S. Moon, N. J. Jeon, J.-P. Correa-Baena, V. Bulović, S. S. Shin, M. G. Bawendi and J. Seo, Nature, 2021, 590, 587–593 CrossRef CAS.
- H. Lu, W. Liu, G. Ran, Z. Liang, H. Li, N. Wei, H. Wu, Z. Ma, Y. Liu, W. Zhang, X. Xu and Z. Bo, Angew. Chem., Int. Ed., 2023, 62, e202314420 CrossRef CAS.
- B. Pang, C. Liao, X. Xu, L. Yu, R. Li and Q. Peng, Adv. Mater., 2023, 35, 2300631 CrossRef CAS PubMed.
- L. Rao, X. Meng, S. Xiao, Z. Xing, Q. Fu, H. Wang, C. Gong, T. Hu, X. Hu, R. Guo and Y. Chen, Angew. Chem., Int. Ed., 2021, 60, 14693–14700 CrossRef CAS PubMed.
- X. Y. Chin, D. Turkay, J. A. Steele, S. Tabean, S. Eswara, M. Mensi, P. Fiala, C. M. Wolff, A. Paracchino, K. Artuk, D. Jacobs, Q. Guesnay, F. Sahli, G. Andreatta, M. Boccard, Q. Jeangros and C. Ballif, Science, 2023, 381, 59–63 CrossRef CAS.
- X. Yan, J. Wu, J. Lv, L. Zhang, R. Zhang, X. Guo and M. Zhang, J. Mater. Chem. A, 2022, 10, 15605–15613 RSC.
- A. Gavezzotti and G. Filippini, J. Phys. Chem. A, 2002, 98, 4831–4837 CrossRef.
- Y. Wei, Z. Chen, G. Lu, N. Yu, C. Li, J. Gao, X. Gu, X. Hao, G. Lu, Z. Tang, J. Zhang, Z. Wei, X. Zhang and H. Huang, Adv. Mater., 2022, 34, 2204718 CrossRef CAS PubMed.
- Z. Wang, Y. Zhang, J. Zhang, Z. Wei and W. Ma, Adv. Energy Mater., 2016, 6, 1502456 CrossRef.
- H. Yang, C. Cui and Y. Li, Acc. Mater. Res., 2021, 2, 986–997 CrossRef CAS.
- L. Zhu, J. Zhang, Y. Guo, C. Yang, Y. Yi and Z. Wei, Angew. Chem., Int. Ed., 2021, 60, 15348–15353 CrossRef CAS PubMed.
- W. Zhu, A. P. Spencer, S. Mukherjee, J. M. Alzola, V. K. Sangwan, S. H. Amsterdam, S. M. Swick, L. O. Jones, M. C. Heiber, A. A. Herzing, G. Li, C. L. Stern, D. M. DeLongchamp, K. L. Kohlstedt, M. C. Hersam, G. C. Schatz, M. R. Wasielewski, L. X. Chen, A. Facchetti and T. J. Marks, J. Am. Chem. Soc., 2020, 142, 14532–14547 CrossRef CAS.
- J. Lin, M. Lai, L. Dou, C. S. Kley, H. Chen, F. Peng, J. Sun, D. Lu, S. A. Hawks, C. Xie, F. Cui, A. P. Alivisatos, D. T. Limmer and P. Yang, Nat. Mater., 2018, 17, 261–267 CrossRef CAS.
- L. D. Whalley, P. van Gerwen, J. M. Frost, S. Kim, S. N. Hood and A. Walsh, J. Am. Chem. Soc., 2021, 143, 9123–9128 CrossRef CAS PubMed.
- I. Ramirez, M. Causa, Y. Zhong, N. Banerji and M. Riede, Adv. Energy Mater., 2018, 8, 1703551 CrossRef.
- K. Nakano, Y. Chen, B. Xiao, W. Han, J. Huang, H. Yoshida, E. Zhou and K. Tajima, Nat. Commun., 2019, 10, 2520 CrossRef PubMed.
- D. Beljonne, G. Pourtois, C. Silva, E. Hennebicq, L. M. Herz, R. H. Friend, G. D. Scholes, S. Setayesh, K. Müllen and J. L. Brédas, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10982–10987 CrossRef CAS PubMed.
- K. Feron, X. Zhou, W. J. Belcher and P. C. Dastoor, J. Appl. Phys., 2012, 111, 044510 CrossRef.
- J. Yuan, Y. Zhang, L. Zhou, G. Zhang, H.-L. Yip, T.-K. Lau, X. Lu, C. Zhu, H. Peng, P. A. Johnson, M. Leclerc, Y. Cao, J. Ulanski, Y. Li and Y. Zou, Joule, 2019, 3, 1140–1151 CrossRef CAS.
- J. Wang, W. Wang, X. Wang, Y. Wu, Q. Zhang, C. Yan, W. Ma, W. You and X. Zhan, Adv. Mater., 2017, 29, 1702125 CrossRef.
- S. M. Swick, T. Gebraad, L. Jones, B. Fu, T. J. Aldrich, K. L. Kohlstedt, G. C. Schatz, A. Facchetti and T. J. Marks, ChemPhysChem, 2019, 20, 2608–2626 CrossRef CAS.
- L. Zhu, M. Zhang, G. Zhou, T. Hao, J. Xu, J. Wang, C. Qiu, N. Prine, J. Ali, W. Feng, X. Gu, Z. Ma, Z. Tang, H. Zhu, L. Ying, Y. Zhang and F. Liu, Adv. Energy Mater., 2020, 10, 1904234 CrossRef CAS.
- W. Zhu, A. P. Spencer, S. Mukherjee, J. M. Alzola, V. K. Sangwan, S. H. Amsterdam, S. M. Swick, L. O. Jones, M. C. Heiber, A. A. Herzing, G. Li, C. L. Stern, D. M. DeLongchamp, K. L. Kohlstedt, M. C. Hersam, G. C. Schatz, M. R. Wasielewski, L. X. Chen, A. Facchetti and T. J. Marks, J. Am. Chem. Soc., 2020, 142, 14532–14547 CrossRef CAS.
- G. Zhang, X.-K. Chen, J. Xiao, P. C. Y. Chow, M. Ren, G. Kupgan, X. Jiao, C. C. S. Chan, X. Du, R. Xia, Z. Chen, J. Yuan, Y. Zhang, S. Zhang, Y. Liu, Y. Zou, H. Yan, K. S. Wong, V. Coropceanu, N. Li, C. J. Brabec, J.-L. Bredas, H.-L. Yip and Y. Cao, Nat. Commun., 2020, 11, 3943 CrossRef CAS.
- T. J. Aldrich, M. Matta, W. Zhu, S. M. Swick, C. L. Stern, G. C. Schatz, A. Facchetti, F. S. Melkonyan and T. J. Marks, J. Am. Chem. Soc., 2019, 141, 3274–3287 CrossRef CAS.
- Z. Zhang, S. Guang, J. Yu, H. Wang, J. Cao, F. Du, X. Wang and W. Tang, Sci. Bull., 2020, 65, 1533–1536 CrossRef CAS.
- H. Chen, H. Liang, Z. Guo, Y. Zhu, Z. Zhang, Z. Li, X. Cao, H. Wang, W. Feng, Y. Zou, L. Meng, X. Xu, B. Kan, C. Li, Z. Yao, X. Wan, Z. Ma and Y. Chen, Angew. Chem., Int. Ed., 2022, 61, e202209580 CrossRef CAS PubMed.
- H. Chen, Y. Zou, H. Liang, T. He, X. Xu, Y. Zhang, Z. Ma, J. Wang, M. Zhang, Q. Li, C. Li, G. Long, X. Wan, Z. Yao and Y. Chen, Sci. China: Chem., 2022, 65, 1362–1373 CrossRef CAS.
- H. Z. Liang, X. Q. Bi, H. B. Chen, T. F. He, Y. Lin, Y. X. Zhang, K. Q. Ma, W. Y. Feng, Z. F. Ma, G. K. Long, C. X. Li, B. Kan, H. T. Zhang, O. A. Rakitin, X. J. Wan, Z. Y. Yao and Y. S. Chen, Nat. Commun., 2023, 14, 4707 CrossRef CAS.
- Z. Yao, X. Wan, C. Li and Y. Chen, Acc. Mater. Res., 2023, 4, 772–785 CrossRef CAS.
- Y. Zhu, Z. Zhang, W. Si, Q. Sun, G. Cai, Y. Li, Y. Jia, X. Lu, W. Xu, S. Zhang and Y. Lin, J. Am. Chem. Soc., 2022, 144, 12747–12755 CrossRef CAS PubMed.
- H. Chen, H. Liang, Z. Guo, Y. Zhu, Z. Zhang, Z. Li, X. Cao, H. Wang, W. Feng, Y. Zou, L. Meng, X. Xu, B. Kan, C. Li, Z. Yao, X. Wan, Z. Ma and Y. Chen, Angew. Chem., Int. Ed., 2022, 61, e202209580 CrossRef CAS.
- L. Meng, Y. Zhang, X. Wan, C. Li, X. Zhang, Y. Wang, X. Ke, Z. Xiao, L. Ding, R. Xia, H. Yip, Y. Cao and Y. Chen, Science, 2018, 361, 1094–1098 CrossRef CAS PubMed.
- X. J. Wan, C. X. Li, M. T. Zhang and Y. S. Chen, Chem. Soc. Rev., 2020, 49, 2828–2842 RSC.
- W. Jiang, H. Jin, M. Babazadeh, A. S. Loch, A. Raynor, N. Mallo, D. M. Huang, X. Jiao, W. L. Tan, C. R. McNeill, P. L. Burn and P. E. Shaw, Adv. Funct. Mater., 2022, 32, 2104259 CrossRef CAS.
- Z. Fu, X. Zhang, H. Zhang, Y. Li, H. Zhou and Y. Zhang, Chin. J. Chem., 2021, 39, 381–390 CrossRef CAS.
- S. Ma, S. Du, G. Pan, S. Dai, B. Xu and W. Tian, Aggregate, 2021, 2, e96 CrossRef CAS.
- X. Bi, X. Cao, T. He, H. Liang, Z. Yao, J. Yang, Y. Guo, G. Long, B. Kan, C. Li, X. Wan and Y. Chen, Small, 2024, 2401054 CrossRef CAS.
- J. Xu, S. B. Jo, X. Chen, G. Zhou, M. Zhang, X. Shi, F. Lin, L. Zhu, T. Hao, K. Gao, Y. Zou, X. Su, W. Feng, A. K. Y. Jen, Y. Zhang and F. Liu, Adv. Mater., 2022, 34, 2108317 CrossRef CAS.
- G. R. Desiraju, J. Am. Chem. Soc., 2013, 135, 9952–9967 CrossRef CAS PubMed.
- J. Benduhn, K. Tvingstedt, F. Piersimoni, S. Ullbrich, Y. Fan, M. Tropiano, K. A. McGarry, O. Zeika, M. K. Riede, C. J. Douglas, S. Barlow, S. R. Marder, D. Neher, D. Spoltore and K. Vandewal, Nat. Energy, 2017, 2, 17053 CrossRef CAS.
- X.-K. Chen, D. Qian, Y. Wang, T. Kirchartz, W. Tress, H. Yao, J. Yuan, M. Hülsbeck, M. Zhang, Y. Zou, Y. Sun, Y. Li, J. Hou, O. Inganäs, V. Coropceanu, J.-L. Bredas and F. Gao, Nat. Energy, 2021, 6, 799–806 CrossRef CAS.
- F. D. Eisner, M. Azzouzi, Z. Fei, X. Hou, T. D. Anthopoulos, T. J. S. Dennis, M. Heeney and J. Nelson, J. Am. Chem. Soc., 2019, 141, 6362–6374 CrossRef CAS PubMed.
- D. Qian, Z. Zheng, H. Yao, W. Tress, T. R. Hopper, S. Chen, S. Li, J. Liu, S. Chen, J. Zhang, X.-K. Liu, B. Gao, L. Ouyang, Y. Jin, G. Pozina, I. A. Buyanova, W. M. Chen, O. Inganäs, V. Coropceanu, J.-L. Bredas, H. Yan, J. Hou, F. Zhang, A. A. Bakulin and F. Gao, Nat. Mater., 2018, 17, 703–709 CrossRef CAS.
- A. Ashokan, T. Wang, M. K. Ravva and J.-L. Brédas, J. Mater. Chem. C, 2018, 6, 13162–13170 RSC.
- F. Huang, T. He, M. Li, L. Meng, W. Feng, H. Liang, Y. Zhou, X. Wan, C. Li, G. Long, Z. Yao and Y. Chen, Chem. Mater., 2022, 34, 6009–6025 CrossRef CAS.
- P. Müller-Buschbaum, Adv. Mater., 2014, 26, 7692–7709 CrossRef PubMed.
- J. Ji, L. Zhu, X. Xiong, F. Liu and Z. Liang, Adv. Sci, 2022, 9, 2200864 CrossRef CAS.
- M. Huang, Y. Zeng, G. Han and Y. Yi, J. Phys. Chem. C, 2023, 127, 5597–5603 CrossRef CAS.
- P. W. M. Blom, M. J. M. de Jong and J. J. M. Vleggaar, Appl. Phys. Lett., 1996, 68, 3308–3310 CrossRef CAS.
- M. Zhang, X. Guo, W. Ma, H. Ade and J. Hou, Adv. Mater., 2015, 27, 4655–4660 CrossRef CAS.
- D. Qian, L. Ye, M. Zhang, Y. Liang, L. Li, Y. Huang, X. Guo, S. Zhang, Z. a. Tan and J. Hou, Macromolecules, 2012, 45, 9611–9617 CrossRef CAS.
- Y. M. Wang, D. P. Qian, Y. Cui, H. T. Zhang, J. H. Hou, K. Vandewal, T. Kirchartz and F. Gao, Adv. Energy Mater., 2018, 8, 1801352 CrossRef.
- U. Rau, B. Blank, T. C. M. Muller and T. Kirchartz, Phys. Rev. Appl., 2017, 7, 044016 CrossRef.
- X. K. Chen, D. P. Qian, Y. M. Wang, T. Kirchartz, W. Tress, H. F. Yao, J. Yuan, M. Hulsbeck, M. J. Zhang, Y. P. Zou, Y. M. Sun, Y. F. Li, J. H. Hou, O. Inganas, V. Coropceanu, J. L. Bredas and F. Gao, Nat. Energy, 2021, 6, 799–806 CrossRef CAS.
- H. Yu, Y. Wang, X. Zou, H. Han, H. K. Kim, Z. Yao, Z. Wang, Y. Li, H. M. Ng, W. Zhou, J. Zhang, S. Chen, X. Lu, K. S. Wong, Z. Zhu, H. Yan and H. Hu, Adv. Funct. Mater., 2023, 33, 2300712 CrossRef CAS.
- L. Zhu, M. Zhang, J. Xu, C. Li, J. Yan, G. Zhou, W. Zhong, T. Hao, J. Song, X. Xue, Z. Zhou, R. Zeng, H. Zhu, C.-C. Chen, R. C. I. MacKenzie, Y. Zou, J. Nelson, Y. Zhang, Y. Sun and F. Liu, Nat. Mater., 2022, 21, 656–663 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: CCDC 2367742 and 2367743. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ta07485h |
| ‡ These authors contributed equally: Jiaxin Guo and Xiangjian Cao. |
| This journal is © The Royal Society of Chemistry 2025 |