Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Innovative and sensitive detection of a cancer cell line using a GMR sensor-based biochip prototype for diagnosis purposes†
A.
Trillat‡
 *ab,
M.
Deroo‡
a,
M.
Giraud
a,
E. Fabre
Paul
a,
A.
Solignac
a,
P.
Bonville
a,
F.
Coneggo
a,
A.
Afroun
a,
M.
Thévenin
*ab,
M.
Deroo‡
a,
M.
Giraud
a,
E. Fabre
Paul
a,
A.
Solignac
a,
P.
Bonville
a,
F.
Coneggo
a,
A.
Afroun
a,
M.
Thévenin
 a,
A.
Wijkhuisen
b,
C.
Fermon
a,
S.
Simon
b,
A.
Duret
a,
G.
Cannies
a,
V.
Padilla
a,
F.
Doucet-Populaire
c,
G.
Jasmin-Lebras‡
a and
C.
Féraudet-Tarisse‡
b
a,
A.
Wijkhuisen
b,
C.
Fermon
a,
S.
Simon
b,
A.
Duret
a,
G.
Cannies
a,
V.
Padilla
a,
F.
Doucet-Populaire
c,
G.
Jasmin-Lebras‡
a and
C.
Féraudet-Tarisse‡
b
aSPEC UMR 3680 CEA-CNRS, Université Paris Saclay, CEA Saclay Orme des merisiers, 91191 Gif-sur-Yvette, France. E-mail: agathe.trillat@cea.fr
bCEA, INRAE, Medicines and Healthcare Technologies Department (DMTS), Paris-Saclay University, 91191 Gif-sur-Yvette, France
cI2BC UMR 9198, Team: “Host–pathogen interactions in sepsis”, Université Paris Saclay, France
First published on 22nd May 2025
Abstract
For several years now, the development of rapid, sensitive, portable and inexpensive early diagnosis techniques has been the focus of increasing attention in the healthcare field, for both primary care and emergency medicine. We have previously demonstrated the proof-of-concept of a patented microfluidic biochip integrating a giant magnetoresistance (GMR)-based sensor, placed on either side of the channel, allowing for the one-by-one dynamic detection of single magnetically labeled biological targets, in a continuous flow mode. In this article, we implemented this two-stage GMR sensor to improve the readiness level of this technology and move towards point-of-care (POC) analysis. We used semi-complex culture medium samples spiked with a murine cancer cell line, pre-labeled with functionalized magnetic particles, to evaluate the biochip performances in detail. The quantitative detection of target cells in low concentrated samples was achieved, with a sensitivity of 5 × 102 cells per mL at a 2 mL per hour flow rate and good specificity, even after addition of irrelevant cells to the sample. Finally, we demonstrated that these performances are competitive with existing techniques such as ELISA tests and flow cytometry analysis, paving the way for new GMR-based POC tests.
Introduction
Micro- and nano-biotechnologies have allowed the development of efficient and increasingly competitive lab-on-chips for rapid, sensitive and easy-to-use diagnostic tests.1–3 In particular, biosensors, which are analytical devices that convert a biological response into a measurable signal, have been improving our healthcare system.4,5 For the accurate detection of target analytes in a complex matrix (e.g., blood, urine), a biosensor usually combines a bioreceptor that specifically recognizes the analyte (e.g., antibody, aptamer, molecular imprinted polymer (MIP), enzyme, complementary nucleic acid) with a transducer which converts the bio-recognition event into a measurable signal (optical, electrochemical, magnetic, etc.). The amplitude of this signal can then be directly linked to the presence, and also usually the concentration, of the target analyte in the analyzed matrix. Biosensors can be divided into several categories according to their detection method,1,2 with the main ones being optical3 or electrochemical biosensors.4 Magnetic sensors have been expanding rapidly in the last decades, with applications in precision medicine, and particularly, in the development of point-of-care (POC) systems.5 Among them, giant magnetoresistance (GMR) sensors, which have been developed in a wide variety of spintronic applications (automotive industry, computer industry, etc.), have demonstrated real potential in the field of health, in particular for the development of early diagnosis devices at the point of care.6–9 They have the advantages of very high detectivity (around 50 to 200 pT Hz−1/2), low cost and easy integration into lab-on-a-chip systems. GMR sensors can be used for both static and dynamic measurements,10–13 with the latter one being our focus of interest. As biological objects are usually not naturally magnetic, they must be previously labeled using superparamagnetic particles (MPs) functionalized with bioreceptors such as monoclonal antibodies specifically directed against the target of interest.14 The one-by-one dynamic detection of the magnetically labeled biological objects flowing in a microfluidic channel is achieved using GMR sensors.We previously integrated this detection approach into a first simple biochip prototype, which was composed of a series of sensors disposed on the bottom wall of a microfluidic channel.15 The main limitation of this first device was its limited specificity due to false positive detection. Indeed, GMR sensors detect the dipole field Hdip (proportional to μ/z3) produced by the magnetic object of moment μ flowing in the channel at a height of z from the sensor in the channel. Thus, our first prototype biochip did not allow discrimination of a strongly labeled targeted biological object (such as a eukaryotic cell labeled by numerous MPs) flowing high in the channel (remote from the sensor) from a small aggregate of magnetic beads flowing closer to the sensors, generating false positives. This problem was overcome by heightening the floor of the channel above the magnetic sensors to increase the z height so that single beads or small aggregates could not be detected. But such an issue was thus limiting the sensitivity of our first device for the detection of magnetically labeled eukaryotic cells. In order to remove this lock and therefore increase sensitivity, a new prototype of a patented biochip16 called the “two-stage GMR sensor lab on a chip”, has been developed in which the GMR sensors are arranged face to face on both sides (top and bottom) of the microfluidic channel, allowing each magnetic object to be detected simultaneously by both sensors. For the first time, thanks to this technique, it is possible to determine the following physical constants of the detected objects: their height in the flow of the microfluidic channel, their magnetic moment and therefore the number of magnetic beads contained in each detected object. This technique has therefore real potential for various applications. First, in our study case, it is then possible to differentiate biological objects magnetically labeled by numerous beads from bead aggregates (made of a few magnetic particles). The principle of this device has been described in detail in previous work17 and will be briefly reviewed in the Materials and methods section for better understanding. A second example of application lies in performing local magnetometry. Indeed, as this two-stage GMR sensor allows determination of the number of magnetic objects detected as a function of their moment or height of passage, it is possible to characterize the moment distribution of magnetic particles diluted in solution and thus know the state of aggregation.
In this article, we further characterize the biological detection performances of this two-stage GMR biochip. We have performed a detailed study using a murine myeloma cell line (NS1) labeled by commercial magnetic beads functionalized with specific antibodies aiming at evaluating the strengths and weaknesses of the actual device in terms of sensitivity, specificity and potential for field use. The cell model that we used has been chosen because it is easy to handle and non-pathogenic, enabling the study to be performed outside a biosafety laboratory (BSL2/3), both under optimal noise conditions (a magnetic chamber) and in field use (standard biology laboratory). The sizes of the cells (7 to 10 μm diameter) and magnetic beads (1 μm) also make it possible to visually control magnetic cell labeling and bead aggregation at different steps of the experiments, using a standard optical microscope. It allows the consistency of the results obtained from the signals emitted by the biochip to be validated. The performances of this two-stage GMR biochip have been compared to the results obtained on the same model with routine laboratory tests such as ELISA or flow cytometry and show that it could be added to the range of tools ideally tailored for point-of-care early diagnosis.
Materials and methods
Principle of the GMR detection
The principle of the GMR effect comes from the fact that electrons have a spin which can acquire magnetic polarization able to modify the electrical resistance. A GMR sensor consists of a nanometric stack of thin layers, called a spin valve.18 The main structure consists of a so-called “hard” (or pinned) layer that is insensitive to the external magnetic field, and a so-called “free” layer that follows the external magnetic field. A copper metal spacer magnetically separates these two layers. The resistance then varies according to the angle of the magnetization between the hard and the free layers. These values range between the parallel and antiparallel configurations of the hard and free magnetization layers, corresponding to the minimum (RP) and maximum (RAP) resistance values, respectively. The sensors are yoke-shaped in order to have single-domain magnetization and allow very low magnetic noise19 (Fig. 1A). GMR sensors are characterized by their giant magnetoresistance (MR) and their sensitivity (S) (see below eqn (1a) and (1b)). In our experiments, MR varied between 5 and 10%, and the sensitivity between 0.7 and 2% mT−1. Fig. 1B shows the variation of the sensor resistance as a function of the magnetic field (sensitivity of the GMR sensor) applied along its sensitivity axis (the axis of the pinned layer). Between the two end configurations, in which the free and the pinned layers are respectively parallel and antiparallel, there is a linear zone which allows the conversion of resistances (or voltages) measured at the sensor terminals into magnetic fields. | (1a) |
 | (1b) |
Principle of the two-stage GMR sensor lab-on-chip
Our GMR biochip is composed of a microfluidic channel whose height is adapted to the hydrodynamic diameter of the studied biological target. Above and below this channel, GMR sensors are perfectly aligned in pairs all along the channel as described in previous work.17 In this latter study, only one pair of sensors is needed to detect the signals of the different samples and to evaluate the sensitivity of the lab-on-chip. The detection principle of the two-stage biochip is briefly reviewed below. A magnetic object of moment μ, located at point B in the channel creates a dipole field at point C of the GMR sensor which is written as in eqn (2). | (2) |
In the case of the 2-stage GMR sensor lab-on-chip, each detected magnetic object (labeled target cells, bead aggregates, etc.), polarized along Oz by a permanent field of 0.09 T, generates simultaneously a dipole field on each of the two sensors aligned face to face on either side of the channel, giving rise to two signals (Fig. 2A). This pair of signals for the same magnetic object is called a coincidence. The geometry used for the sensor, in the shape of a yoke, makes it insensitive to field variations along the x- or z-axis, so that only the component Hy of the dipole field is relevant. Each signal has a characteristic antisymmetric shape that allows it to be distinguished from background noise. This sinusoidal shape results from the sensor dynamic detection of magnetic objects flowing in the channel. The Hy tangential component of the dipole field acts symmetrically on the sensor's free layer as it flows from one side of the sensor to the other (Fig. 2A). As the GMR sensors are aligned face to face on either side of the channel (top and bottom), the signals emitted have the same shape but are of opposite sign. In order to analyze them, they must be in the same sense. By analysis convention, a valid peak is constituted by a negative peak followed by a positive peak. The Python analysis program therefore inverts the signal that is not in the right sense.17
As the GMR sensor is not a point-like object, the dipole field generated by a magnetic object flowing in the channel with a velocity vector along Oy must be integrated over the entire surface of the sensor located in the (xy) plane.15 As explained in detail in the ESI,† for each coincidence, the ratio of the amplitudes of the signals, related to dipole fields by sensitivity S, makes it possible to determine the height of passage zB of the magnetic object detected simultaneously by the two sensors (Fig. 2B). Once the height of the object has been obtained, the dipole field corresponding to the signal of one magnetic bead (whose moment μ has been determined by magnetization measurements m(H) at room temperature using a vibrating sample magnetometer) can be calculated. On the other hand, we have previously shown through simulations that a biological object with a non-negligible diameter (around 7–10 μm in the case of NS1 cells) labeled with N superparamagnetic beads randomly distributed on the surface gives a signal identical to that of a point core consisting of N beads.17 Therefore, once zB is determined, it is possible to know the number of beads that the detected object contains by calculating the ratio between the calculated signal of one bead and the signal of the detected object.
GMR sensors and biochip fabrication
The GMR layer manufacture, sensor design and biochip assembly were developed and described in a previous study.17 Briefly, the orientation of the hard layer (IrMn or PtMn) is established through annealing under a strong magnetic field at high temperatures (1 T, 200 °C for IrMn, 300 °C for PtMn). The free layer, designed for high sensitivity to external magnetic fields, requires a low coercive field and high spin polarization to enhance the giant magnetoresistance (GMR) effect. It is composed of a NiFe layer (low coercivity) and a CoFe layer (high spin polarization) coupled to follow the magnetization of the lower coercivity material. In order to obtain a linear response to the magnetic field of GMR sensors, the magnetization of the hard layer is fixed perpendicular to the axis of easy magnetization of the free layer (y axis is the sensitive axis as shown before). A thin copper layer separates and decouples the hard and free layers preventing the hard layer from being affected by the inversion of the free layer. The thickness of this layer must be sufficiently thin (2 nm) so that electrons do not lose their polarization and the GMR effect does not disappear. In this study, the GMR layers were deposited on Si wafers to fabricate the sensors located under the channel and on sapphire wafers for those at the top in order to align them perfectly with those at the bottom.To summarize the biochip fabrication, the channel skeleton is made of SU-8 2025 resin on a SU-8 2002 primer layer deposited on the Si wafer and its height can be adjusted using the spin coating parameters. The biochip is closed by gluing the sapphire wafer, containing the top sensors, to the channel walls using ‘Norland’ optical glue, perfectly aligning the sensors (top and bottom) on either side of the channel. PDMS dots are glued to the top of the channel reservoirs, on the sapphire wafer previously drilled using an excimer laser. The biochip is then positioned on a PCB support, allowing the electrical connections of the sensors, as well as connection to an electronic box that filters and amplifies the signals delivered by the GMR sensors.
Cells and antibodies
A non-pathogenic and easily manipulated biological study model has been chosen, namely a well-characterized cell line consisting of 7 to 10 μm diameter non-adherent eukaryotic murine myeloma cells, known as NS1 cells (ATCC no. TIB-18). These are murine B lymphocyte cells derived from the P3X63Ag8 line (ATCC TIB-19), used in laboratories as a fusion partner of B lymphocytes isolated from immunized mice for the generation of murine hybridomas, during the first stage of in vitro monoclonal antibody production.20 Another cell line has been used in this study as a negative control to mimic the reality of an unknown irrelevant biological sample with non-targeted cells. This control consists of the Chinese Hamster Ovary cell line (CHO, purchased from ECACC, Wiltshire, UK) which does not express the targeted membrane biomarker. These adherent epithelial cells are easy to culture and well-characterized, since they are routinely used in many laboratories to synthesize therapeutic compounds for example.NS1 cells are cultivated in RPMI-1640 medium with 15% of decomplemented fetal bovine serum, 1% of non-essential amino acids, 1% sodium pyruvate, 1% of antibiotics (penicillin and streptomycin) and 1% of L-glutamine at 37 °C under a controlled atmosphere containing 7% of CO2. CHO cells are cultivated in Ham's F-12 nutrient mixture with 10% of decomplemented fetal bovine serum, 1% of non-essential amino acids, 1% of antibiotics (penicillin and streptomycin) and 1% of L-glutamine at 37 °C under a controlled atmosphere containing 5% of CO2. All culture media and supplements for both cell lines are from Gibco, Life Technologies or Sigma-Aldrich. The antibody used to functionalize the magnetic beads is a purified rat IgG2a monoclonal antibody directed against the mouse CD138 membrane receptor, highly expressed at the surface of NS1 cells21 (but not of CHO cells). It is a commercial antibody purchased from BD Pharmingen (clone 281-2).
Biofunctionalization of antibodies on magnetic beads
Grafting antibodies onto magnetic beads involves affinity coupling between the streptavidin present on the bead surface (superparamagnetic Dynabead MyOne Streptavidin T1, Dynal) and the biotin conjugated antibody. The process begins thus with antibody biotinylation and ends with bead functionalization. These steps are described in detail in previous work.22Bioassay protocol
Previously to each experiment, cultured NS1 cells and trypsin–EDTA detached CHO cells are centrifuged at 1000 RPM (centrifuge diameter 344 mm) for 10 min at 10 °C and transferred to a fresh NS1 culture medium. Cell suspensions are counted using a TC20 automated cell counter (BioRad). In order to optimize the magnetic labeling of the cells (quantity and labeling time), the beads are added in excess. It has been previously determined that 1.5 × 107 beads are ideally adapted to magnetically label a maximum quantity of 105 NS1 cells in 2 hours.22 For all experiments, cells are diluted to the desired concentration in 1 mL of NS1 culture medium (semi-complex biological matrix containing 15% decomplemented fetal bovine serum). The culture medium is more complex than the PBS used in our very first experiments22 and has been chosen as a matrix of interesting composition to progress step-by-step towards the complexity of samples used in clinical diagnosis (as serum, blood, urine, etc.). Then, 1.5 × 107 functionalized magnetic beads are added for 2 h incubation at 20 °C under gentle rotation. The totality of the sample (1 mL) is injected into the biochip at a flow rate of 2 mL per hour. For each experiment, a visual verification of samples (correct specific labeling, control of magnetic particle aggregates) is also performed with an EVOS optical microscope at a 40× magnification. NS1 cells are labeled on average with at least 50–60 magnetic beads.22For each experiment, two negative controls are systematically prepared and analyzed to evaluate and control the detection specificity of the biochip. The first negative control consists only of 1.5 × 107 magnetic beads functionalized with the anti-CD138 antibody in 1 mL of culture medium. The second negative control is made of irrelevant CHO cells in the presence of 1.5 × 107 anti-CD138 antibody functionalized magnetic beads in 1 mL of culture medium. In this last control, the CHO cell concentration (varying from 103 to 105 CHO per mL) is adapted to the design of each experiment and is equal to the highest NS1 cell concentration used for the ongoing experiment. It enables us to assess the specificity of the measurements and to quantify any non-specific binding of the functionalized beads to the irrelevant cells.
The sensitivity of detection is assessed by performing serial dilutions of NS1 cell solution in culture medium (from 102 to 105 cells per mL, depending on the experiments) and by incubating these solutions for 2 h at 20 °C under rotation with 1.5 × 107 anti-CD138 functionalized magnetic beads before direct injection into the biochip at a 2 mL per hour flow rate. These positive samples containing different concentrations of NS1 cells are introduced into the biochip from the lowest concentration to the highest. Between each sample, 1 mL of cell culture medium is injected into the biochip to wash the microfluidic channel and recover any cells or any bead aggregates that may have been trapped.
The dynamic operating range is characterized by calculating a “detectability percentage” defined as the ratio of the number of events detected by the biochip to the number of NS1 cells counted (using a TC20 automated counter) introduced into the device.
Comparative ELISA and flow cytometry test
Comparative ELISA and flow cytometry tests are performed as described previously.22Experimental set-up and magnetic detection
As described in ref. 17 and 22, the biochip is inserted in a home-made support positioned in the center of a homogeneous 0.09 T permanent magnet23 that polarizes the magnetic moment of the superparamagnetic beads along Oz. The support contains three micrometric screws which allow the position of the GMR sensors to be adjusted in the directions (x, y, z) to optimize their sensitivity S. The samples are injected into the microfluidic channel of the biochip, using a Fluigent© pressure controller with a fixed pressure of 345 mbar or a syringe pump (BBraun) at a flow rate of 2 mL per hour, and flow along the y-axis at a speed between 20 and 30 cm s−1. This velocity can be determined by fitting the width of the experimental signals with a Python program developed previously.17The sensors are powered by a 1 V voltage generated by a compact electronic box, which also filters and amplifies by a factor of 8600 the signals (of the order of a few μV) emitted by the magnetic objects (labeled biological targets or magnetic bead aggregates) flowing in the channel. Data are acquired using a data acquisition card (data translation) at a frequency of 200 kHz interfaced with a program written in Pascal (under Lazarus). At the beginning of each experiment, the background noise of the setup is recorded during 20 seconds without flowing a sample. The measured voltage signal zero-peak (V0p) is then converted to a field using the sensitivity (S) of the GMR sensors. The limit of detection signal (LOD) is defined as the H-field inducing a signal-to-noise ratio (SNR) of 1 and is therefore very dependent on the noise environment. The experiments were performed in two different locations. The first one is a magnetic environment to prove the concept of this detection technique, with an average noise signal voltage V0p of about 40 mV (gain 8600). The corresponding LOD is around 400 nT. The setup has been then moved to a standard biology laboratory environment, where the experimental noise varies much more over the course of an experiment, but also from one experiment to another, as it is intrinsically linked to activity in the laboratory. For this reason, to optimize measurements in this new environment, a homemade aluminum shield has been built to isolate the biochip and the electronic box from external noise (copper was excluded as it is too heavy for the miniaturized assay). A detailed study of noise as a function of the environment is presented in the Results and discussion section (Fig. 9).
Python programs have been developed previously to analyze the data. The signals emitted simultaneously by the top and bottom sensors give rise to the coincidences (described previously) which are selected according to a number of criteria such as the shape, width and amplitude, described in detail previously.17 The introduction of a test of χ2 verifying the fit of the signals with the criteria enables the selection of the coincidences to be refined. In the magnetic environment, where the noise is low, the selection requirement was an amplitude greater than or equal to 2 × LOD for both signals forming the coincidence. In the conventional laboratory, where the noise and therefore the LOD are higher (Fig. 9), the selection requirement was an amplitude greater than or equal to 2 × LOD for one of the signals forming the coincidence and an amplitude greater than or equal to 1 × LOD for the other one. As the signal has a highly characteristic symmetrical shape that must satisfy the above criteria (with a fit validated by the test of χ2), it cannot be confused with random noise signals. Analysis of the signals forming the coincidences makes it possible to determine the height of passage in the channel of the magnetic objects detected, as well as the number of beads they contain.17
Results and discussion
Magnetic labeling of cells
To evaluate the performances of our GMR biochip and its potential relevance as a tool for in vitro diagnostic tests, a thorough study has been performed by using a cell model, namely the NS1 murine myeloma cell line, in a semi-complex matrix made of a culture medium. The magnetic detection of these cells is possible thanks to the use of magnetic Dynabeads MyOne functionalized with monoclonal antibodies directed against the CD138 membrane receptor, highly expressed on the target cells.24 We have previously observed that NS1 cells are labeled on average with at least 50–60 magnetic beads17,22 by optical microscopy. Here, we used environmental scanning electron microscopy (ESEM) images to confirm the level of magnetic labeling of NS1 cells with the same beads (Fig. 3). These measurements have also been confirmed by GMR-sensing using our biochip, which allows us to determine the magnetic moment of each detected object, and therefore the number of magnetic particles it contains.17,22Magnetic signal detection
The biochip enables dynamic detection of magnetic objects at a 2 mL h−1 flow rate, such as not only NS1 target cells specifically labeled with anti-CD138 functionalized magnetic beads (green box in Fig. 4B), but also aggregates of beads flowing through the microfluidic channel (black box in Fig. 4B). The face-to-face arrangement of GMR sensors on both sides (top and bottom) of the microfluidic channel allows each magnetic object to be detected simultaneously giving rise to top and bottom signals (called coincidences). Then, a distribution of events according to its magnetic labeling can be represented graphically similar to flow cytometry monoparametric histograms. Fig. 4A shows such examples of histograms obtained for two samples of 1 mL from one experiment after data processing. They represent the number of coincidences obtained as a function of the number of beads contained in each detected magnetic object. The first sample (represented in black) is a negative control containing 1.5 × 107 anti-CD138 functionalized magnetic beads, the second (in red) is a specific sample with 7 × 103 NS1 cells and the same quantity of beads in a 1 mL volume of culture medium. With the LOD criteria as defined in the Materials and methods section (more details in Fig. 10), the selected coincidences correspond to magnetic objects containing more than 10 beads (Fig. 4A–C). The negative control sample containing only the functionalized beads is used as a reference for the rest of the experiment to define the background noise resulting from aggregates. It is always the first sample tested in the biochip. Signals obtained with this bead negative sample are processed to eliminate 98% of coincidences and to keep only the 2% of them corresponding to the largest aggregates. This process gives us a limit on the maximal number of beads involved in aggregates, called Nlimit, which constitutes our positivity threshold. For most experiments, Nlimit is close to 50 beads (Nlimit = 48.2 ± 19.5 (n = 12)). Using this signal classification technique, all detected magnetic objects containing less than Nlimit beads are considered as bead aggregates or poorly labeled cells (Fig. 4C). A visual study of pictures of NS1 cells recorded systematically under an optical microscope shows that they are labeled by an average of at least 50–60 beads (Fig. 4B), which validates our data processing. When the Nlimit obtained is superior to 90 beads (i.e. much higher than 50 beads), this means that there are too many false positives (due to excessive bead aggregates) and the experiment is then considered as invalid (such a case has arisen occurring twice out of 15). Finally, the threshold chosen in this study, 98% confidence, is consistent with the confidence range (95 to 99%) classically used in biostatistical analysis. In conclusion, only coincidences corresponding to magnetic objects containing a number of beads greater than or equal to the 98% confidence bead aggregate limit (Nlimit) are kept. In the experiment shown in Fig. 4A and C, the threshold obtained with this method is 60 beads. It can be noticed that this method underestimates the number of specific events but allows the majority of the false positives to be eliminated.Evaluation of GMR biochip performances
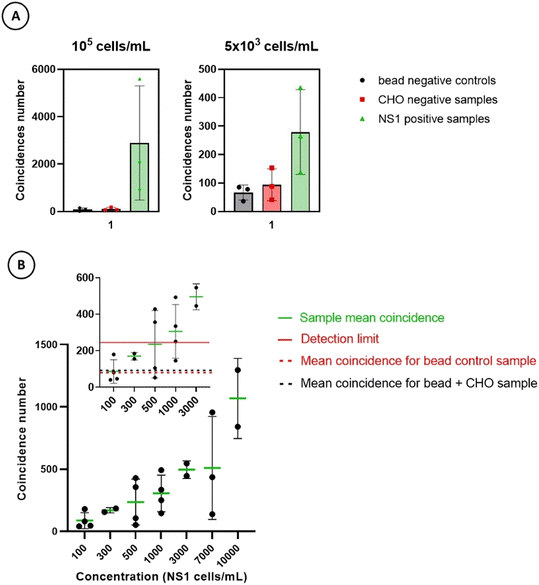
The first criterion to be assessed when developing a new detection assay is the specificity of the measurements. The specificity can be described as the assay's ability to detect the intended target, without false positives due to cross-reactivity.25 Therefore, if samples containing NS1 target cells are considered as “positive” and those without NS1 are considered as “negative”, specificity measures the test's ability to detect as negative the “negative” samples without any false positive. As described in the Materials and methods section, the experiments systematically contain a negative control sample with irrelevant CHO cells in the presence of 1.5 × 107 anti-CD138 antibody functionalized magnetic beads in 1 mL of culture medium. The CHO cell concentration corresponds to the maximum concentration of the specific sample range studied in the experiment. It therefore varies between 103 and 105 CHO cells per mL. It allows estimation of the non-specific binding of magnetic beads that could lead to false positive. Three experiments for two different cell concentrations were carried out to prove the specificity of our detection technique. It can be observed that the average number of coincidences measured for the positive samples containing NS1 cells is higher than that in the two negative controls by a factor 27 at the concentration of 105 cells per mL and, respectively, by a factor 3 at the concentration of 5 × 103 cells per mL (Fig. 5A). The p-values of the Kruskal–Wallis test at each concentration (6.6 × 10−2 and 7.9 × 10−2, respectively) are close to the significance threshold of 5%, further supporting the specificity of the assay (i.e. absence of cross-reactivity with other biological targets).Conversely, in the presence of NS1 cells, the results obtained are truly positive, showing specificity of detection in the culture medium semi-complex matrix provided that a minimum quantity of target cells is present in the sample. This last point is directly linked with the sensitivity of the method and its detection limit defined as the lowest NS1 concentration giving a number of coincidences that is significantly different (at the 99% confidence level) from the number of coincidences of the bead control sample alone. To assess this limit, several measurements yielding the number of coincidences for a bead control sample are performed and the theoretical detection limit is then equal to the mean of the coincidence number recorded plus three times the standard deviation.25,26 The sensitivity reached in culture medium is illustrated in Fig. 5B for five independent experiments performed with different ranges of NS1 concentrations. Our GMR biochip achieves an average detection limit of around 5 × 102 cells per mL making it a promising detection technique. It can be noticed that this detection limit is approximately 100 times better than that reached with the first GMR prototype described in ref. 22 which does not have double GMR detection. The ability to determine the number of beads contained in the magnetic objects detected is therefore a major advance in terms of distinguishing negative controls from positive controls, consequently, drastically increasing specificity and sensitivity. The main factors likely to influence these two last criteria require further optimization to ensure an important contrast between the colloidal stability of the beads on the one hand (to reduce false positives and increase specificity) and the strong magnetic labeling of the targets on the other hand (to reduce false negatives and increase sensitivity). Concerning this second point, in order to increase the magnetic labeling of target cells, the combined use of magnetic beads functionalized with different NS1 cell-specific antibodies (anti-CD38, anti-CD184, anti-CMHI (H-2Kd/H-2Dd) in addition to anti-CD138 antibodies) has been tested. However, no improvement has been observed (data not shown), probably due to both the low membrane expression of these new targeted receptors compared to the CD138 high one21,24 and an observed more important aggregation state of functionalized beads (data not shown).
Finally, Fig. 6 illustrates the biochip dynamic operating range as it shows the mean detection efficiency of NS1 cells by our GMR device through six independent experiments. The percentage of detection efficiency is higher when the cell concentration tested is low. There are different explanations for this result.
First, as shown in Fig. 7B, at high cell concentrations (105 NS1 cells per mL labeled with 1.6 × 107 DynaMyOne magnetic beads in culture medium), NS1 cells tend to group together in clusters because of possible bridging of cells by functionalized beads. These cell clusters are detected by the GMR sensors as a single magnetic object as they flow in the microfluidic channel (Fig. S4†). Secondly, when the cell concentration is high, it is more likely that two cells will flow simultaneously over each other or next to each other in the microfluidic channel. In this case, they are detected as a single cell by the GMR sensors. Finally and according to our simulations, two magnetic objects must be distant of at least 45 μm in the microfluidic channel to be correctly detected as separate objects by a pair of GMR sensors.17 At high concentrations, the distance between two cells may be smaller, and they are therefore detected as a single magnetic object by GMR sensors. Because of all these phenomena, the number of cells detected by the biochip decreases with increasing concentrations, indicating that the dynamic operating range of the biochip is optimal at low concentration (as illustrated in Fig. 7A) where cell-by-cell detection is facilitated. This characteristic makes the biochip more adapted for low target concentrations, which is one among the conditions required for early diagnosis.
Matrix complexification
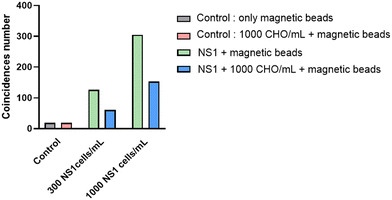
With the aim of using this biochip for diagnostic purposes, another assessment criterion of major importance is the selectivity of the detection assay. It can be defined as the ability of the method to discriminate the target analyte unequivocally from components that may be present in the sample matrix and that may alter assay results.26,27 In contrast to specificity that is an absolute characteristic, selectivity can be graded step by step. After the use of a very simple matrix (phosphate buffered saline) in the very first GMR prototype,22 we gradually make it more complex with the use of a culture medium semi-complex matrix containing 15% fetal bovine serum. In line with this matrix complexification, Fig. 8 illustrates the GMR biochip ability to detect target NS1 cells, in culture medium, in the presence (or absence) of an excess of CHO cells mimicking the presence of irrelevant biological analytes (as it will be the case on a larger scale in clinical samples for diagnostic purposes). These first results show that CHO cells do not contribute to increasing the number of coincidences and therefore to creating false positives. A positive sample containing irrelevant biological objects therefore appears to be correctly detected by our biochip although sensitivity seems to decrease as the matrix complexity increases. These preliminary results, needing further in-depth study, are nevertheless encouraging for future measurements in more complex environments. In the ESI† have been added pictures of labelling assays of NS1 cells in different complex matrix (fetal calf serum, mouse plasma, rabbit serum, rabbit plasma). The next step will be to use them in our biochip (Fig. S5†).Comparison with standard methods
In order to compare the results obtained using the GMR biochip with other well-known and widely used techniques, two other standard methods were tested using the same reagents (antibodies, samples, matrix): sandwich ELISA (enzyme-linked immunosorbent assay) and flow cytometry. The ELISA tests repeated in four independent experiments show a limit of detection of 3.5 × 103 ± 1.6 × 103 NS1 cells per mL in cell culture medium (Fig. 9A). Our GMR device improves detection by a 3-fold factor making it a promising technique. As can be seen in Fig. 9B, the agreement between results from flow cytometry and GMR detection is remarkably good for all concentrations over 3 × 103 NS1 cells per mL. At a lower concentration, the GMR biochip tends to be more sensitive as it counts more events than flow cytometry, although this last technique is not optimized to give absolute cell counts. When compared to the ELISA test or flow cytometry requiring several washing steps (that may trap cells) and qualified personnel, our GMR biochip offers the advantage of greater simplicity (no washing, possible real-time acquisition and parallelization of tests, device portability, etc.). The only preparation step is magnetic labeling which strongly depends on the target, the beads and the used antibody(ies): other research groups reported magnetic immunocapture times between 30 and 180 min.28,29GMR biochip optimization for field use
As mentioned in the Materials and methods section, in order to approach field or laboratory test conditions, the lab-on-a-chip has been transferred to a standard lab environment. A complete noise study has been performed in the shielded chamber and in the normal environment with or without shielding. At the start of an experiment, before the sample flows, 8 blank seconds (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600
600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 channels with acquisition at a 200 kHz frequency) are recorded to assess the Vop noise corresponding to both top and bottom GMR sensors. These values are used to evaluate the LOD (SNR = 1).
000 channels with acquisition at a 200 kHz frequency) are recorded to assess the Vop noise corresponding to both top and bottom GMR sensors. These values are used to evaluate the LOD (SNR = 1).
Fig. 10 shows the simulation of the dipole field emitted on the top and bottom sensors by different numbers of magnetic beads with a mean individual moment of 1.56 10−11 emu (moment of the DynaMyone beads polarized by the 0.09 T permanent magnet of our device), flowing in a channel of 23 micrometers of height. The 2 × LOD thresholds calculated under the three experimental conditions mentioned above are also represented. According to these simulations, selected coincidences correspond to magnetic objects containing more than 10 beads for experiments performed in a shielded chamber (as in Fig. 4), more than 15 beads for those carried out in a normal laboratory with aluminum shielding around the device and more than 25 beads in this last environment without shielding. In this last environment in the absence of shielding, the LOD is too high (green line) for optimum detection of coincidences arising from magnetically labeled cells.
Fig. 11 presents the comparison of two experiments performed in different magnetic environments. The first experiment represented by the blue curve has been performed in a shielded chamber that greatly reduces electrical and magnetic noise. The second experiment has been carried out in a biology standard laboratory environment with a home-made aluminum shield chamber around the device. The similarity of the results in both environments proves that our detection technique can work in a standard laboratory or in any medical analysis laboratory, as well as in hospitals. This is essential for the transfer and use of this biochip in the medical field as a future diagnostic test at the patient's bedside.
The device is currently miniaturized to be easily transportable, including electronic and magnetic developments. The electronic part will be integrated in a single shielded box with a graphical user interface on a touch screen. This box will contain a low noise amplifier and an analog to digital board for the GMR signal acquisition, a current source for the calibration coils which is already designed, and a control module with an embedded computer (RPI4). For the magnetic part, a very light and space-saving Halbach magnet (ferrite magnet) generating a homogeneous field of 550 G has been designed.30 It has an internal diameter of 10 cm and is composed of 16 ferrite magnets with a cross-section of 2 × 2 cm2 and a length of 8 cm (Fig. 12).
Conclusion
In this study, the biological performances of a two-stage GMR biochip previously developed17 have been evaluated with a murine myeloma cell line. The use of such a GMR sensor-based biochip seems to be promising for early diagnosis. Indeed, a sensitivity near 5 × 102 cells per mL has been reached in raw medium culture (without washing nor centrifugation steps) with satisfactory specificity, using a GMR prototype device which is more user-friendly than conventional detection techniques such as flow cytometry or ELISA. The advantages of the technique are, on the one hand, the simplicity of the various handling steps and, on the other hand, that all measurements can be performed in more complex matrices than the one used in this study, without washing steps. A number of challenges have been overcome, providing promising sensitivity for diagnostic purposes using this GMR biochip. The miniaturization, currently in progress, will allow the device to be transportable and to become a point-of-care in vitro diagnostic test. By adjusting the microfluidic channel size and antibodies coupled to the magnetic beads, it can be adapted to a variety of biological targets, such as bacteria or yeasts, showing the ease of implementation due to its versatility. It could open a wide range of applications in the health and environmental fields, as well as in magnetic characterization (local magnetometry).Live subject statement
The study was conducted in compliance with French and European regulations on care and protection of laboratory animals (European Community (EC) Directive 2010/63/UE, French law 2001-486, 6 March 2018) and with the agreement of the Ethics Committee of the Commissariat à l'Energie Atomique (CEtEA “Comité d'Etique en Expérimentation Animale” no 44) no. 12-026 and 15-046 delivered to S. Simon by the French Veterinary Services and CEA agreement D-91-272-106 from the Veterinary Inspection Department of Essonne (France).Data availability
Simulation data supporting this article have been included in the ESI.† The data concerning the experimental results supporting the results of this work are available in the article. Further data are available upon request from the authors.Author contributions
Conceptualization: AT, MD, MG, EFP, AS, PB, SS, GJL, CFT. Formal analysis: AT, MD, GJL, CFT. Funding acquisition: GJL, CFT, SS, FDP. Investigation: AT, MD, MG, AD. Methodology: PB, FC, AA, MT. Software: PB. Resources: FC, AA, MT, AW, CF, GC, VP. Supervision: GJL, CFT. Writing – review & editing: AT, PB, SS, FDP, GJL, CFT. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was possible thanks to the CEA transverse programs PTC “Instrumentation et Detection”. We would also like to thank Dominique Duet, Gérald Le Goff and Jean-Claude Tack for their regular help with technical issues. For cell culture, we would like to thank Marc Plaisance and Stéphanie Gelhaye. We give special thanks to Mathieu Jeckelmann for his precious help in signal processing and the design of the electronic box. We are grateful to Florent Malloggi and Hervé Volland for fruitful scientific exchanges. We are also greatly indebted to Etienne Thevenot for his expert assistance in statistical analysis. We would also like to thank Christophe Depagne and Franck Leleu from Zeiss for the photos with ESEM. Finally, we thank Karla Perez-Toralla for her precious scientific advices.References
- O. Pashchenko, T. Shelby, T. Banerjee and S. Santra, ACS Infect. Dis., 2018, 4, 1162–1178 CrossRef CAS PubMed.
- S. A. Walper, G. L. Aragonés, K. E. Sapsford, C. W. Brown, C. E. Rowland, J. C. Breger and I. L. Medintz, ACS Sens., 2018, 3, 1894–2024 CrossRef CAS PubMed.
- P. Damborský, J. Švitel and J. Katrlík, Essays Biochem., 2016, 60, 91–100 CrossRef PubMed.
- I.-H. Cho, D. H. Kim and S. Park, Biomater. Res., 2020, 24, 6 CrossRef CAS PubMed.
- D. J. Denmark, X. Bustos-Perez, A. Swain, M.-H. Phan, S. Mohapatra and S. S. Mohapatra, J. Electron. Mater., 2019, 48, 4749–4761 CrossRef CAS.
- K. Wu, D. Tonini, S. Liang, R. Saha, V. K. Chugh and J.-P. Wang, ACS Appl. Mater. Interfaces, 2022, 14, 9945–9969 Search PubMed.
- J. Loureiro, R. Ferreira, S. Cardoso, P. P. Freitas, J. Germano, C. Fermon, G. Arrias, M. Pannetier-Lecoeur, F. Rivadulla and J. Rivas, Appl. Phys. Lett., 2009, 95, 034104 CrossRef.
- K. Wu, T. Klein, V. D. Krishna, D. Su, A. M. Perez and J.-P. Wang, ACS Sens., 2017, 2, 1594–1601 CrossRef CAS PubMed.
- J. Choi, A. W. Gani, D. J. B. Bechstein, J.-R. Lee, P. J. Utz and S. X. Wang, Biosens. Bioelectron., 2016, 85, 1–7 CrossRef CAS PubMed.
- C. Fermon and M. H. van de Voorde, Nanomagnetism: Applications and Perspectives, Wiley-VCH, Weinheim, 2017 Search PubMed.
- D. Su, K. Wu, R. Saha, C. Peng and J.-P. Wang, Micromachines, 2019, 11, 34 CrossRef PubMed.
- J. Loureiro, P. Z. Andrade, S. Cardoso, C. L. Da Silva, J. M. Cabral and P. P. Freitas, Lab Chip, 2011, 11, 2255 RSC.
- T. Klein, W. Wang, L. Yu, K. Wu, K. L. M. Boylan, R. I. Vogel, A. P. N. Skubitz and J.-P. Wang, Biosens. Bioelectron., 2019, 126, 301–307 Search PubMed.
- E. Gheorghiu, J. Biomed. Res., 2021, 35, 277 CrossRef PubMed.
- G. Li, S. X. Wang and S. Sun, IEEE Trans. Magn., 2004, 40, 3000–3002 CrossRef CAS.
- C. Fermon, M. Giraud, F. D. Delapierre and G. Jasmin, WO Pat., WO2019/238857A1, 2019 Search PubMed.
- M. Deroo, M. Giraud, F.-D. Delapierre, P. Bonville, M. Jeckelmann, A. Solignac, E. Fabre-Paul, M. Thévenin, F. Coneggo, C. Fermon, F. Malloggi, S. Simon, C. Féraudet-Tarisse and G. Jasmin-Lebras, Lab Chip, 2022, 22, 2753–2765 RSC.
- B. Dieny, J. Magn. Magn. Mater., 1994, 136, 335–359 CrossRef CAS.
- M. Pannetier, C. Fermon, G. Le Goff, J. Simola and E. Kerr, Science, 2004, 304, 1648–1650 CrossRef CAS PubMed.
- G. Köhler and C. Milstein, Nature, 1975, 256, 495–497 CrossRef PubMed.
- E. A. Rosa, S. R. Lanza, C. R. Zanetti and A. R. Pinto, Hybridoma, 2012, 31, 1–6 CrossRef CAS PubMed.
- M. Giraud, F.-D. Delapierre, A. Wijkhuisen, P. Bonville, M. Thévenin, G. Cannies, M. Plaisance, E. Paul, E. Ezan, S. Simon, C. Fermon, C. Féraudet-Tarisse and G. Jasmin-Lebras, Biosensors, 2019, 9, 105 CrossRef CAS PubMed.
- N. Pamme, Lab Chip, 2006, 6, 24–38 RSC.
- M. Setoguchi, S. Yoshida, Y. Higuchi, S. Akizuki and S. Yamamoto, Somatic Cell Mol. Genet., 1988, 14, 427–438 CrossRef CAS PubMed.
- J. W. Lee and M. Hall, J. Chromatogr., B, 2009, 877, 1259–1271 CrossRef CAS PubMed.
- EMA/CVMP/VICH, 2009.
- U. Andreasson, et al. , Front. Neurol., 2015, 6 DOI:10.3389/fneur.2015.00179.
- R. Soares, V. C. Martins, R. Macedo, F. A. Cardoso, S. A. M. Martins, D. M. Caetano, P. H. Fonseca, V. Silvério, S. Cardoso and P. P. Freitas, Anal. Bioanal. Chem., 2019, 411, 1839–1862 Search PubMed.
- A. R. Soares, R. Afonso, V. C. Martins, C. Palos, P. Pereira, D. M. Caetano, D. Carta and S. Cardoso, Biosens. Bioelectron.:X, 2022, 11, 100149 CAS.
- H. Raich and P. Blümler, Concepts Magn. Reson., Part B, 2004, 23, 16–25 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of the calculation of the dipolar field and figure with 3 simulations of the signals for different configurations of targeted cells in the channel. See DOI: https://doi.org/10.1039/d5sd00029g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |