Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Regenerable photonic aptasensor for detection of bacterial spores with stacks of GaAs–AlGaAs nanoheterostructures†
Ishika
Ishika
a,
Walid M.
Hassen
a,
René
St-Onge
a,
Houman
Moteshareie
ab,
Azam F.
Tayabali
ab and
Jan J.
Dubowski
 *a
*a
aInterdisciplinary Institute for Technological Innovation (3IT), CNRS IRL-3463, Quantum Semiconductors and Photon-based BioNanotechnology Group, Department of Electrical and Computer Engineering, Université de Sherbrooke, 3000, boul. de l'Université, Sherbrooke, Québec J1K 0A5, Canada. E-mail: Jan.J.Dubowski@usherbrooke.ca
bBiotechnology Laboratory, Environmental Health Science and Research Bureau, Healthy Environments and Consumer Safety Branch, Health Canada, 251 Sir Frederick Banting Driveway, Ottawa, Ontario K1A 0K9, Canada
First published on 20th February 2025
Abstract
The reusability of biosensors is a crucial advancement in environmental monitoring and laboratory efficiency. In this study, we introduce the concept of a regenerable aptasensor based on digital photocorrosion (DIP) of a GaAs–AlGaAs biochip, designed with alternating nanolayers of GaAs (12 nm) and AlGaAs (10 nm). Each GaAs–AlGaAs bilayer acts as an independent sensing unit. By employing a specific thiolated aptamer, we achieve efficient detection of Bacillus thuringiensis spp. kurstaki spores. The interaction between the thiolated aptamers with the targeted spores leads to the formation of aptamer-spore hybrids, which bind to the GaAs surface. The GaAs–AlGaAs nanoheterostructure biochip supports multiple biosensing cycles. After consumption of the first GaAs–AlGaAs bilayer, a simple regeneration step with a high ionic strength buffer releases the bound spores and prepares subsequent nanolayers of the same biochip for reuse. The capability to regenerate and reuse individual nanolayers presents a novel and practical solution for reducing biosensor waste while improving operational efficiency. We further explore the conditions necessary for sustainable DIP operation in biochips containing multiple GaAs–AlGaAs nanolayer pairs, ensuring reliable performance over numerous biosensing cycles. Our findings establish a cost-effective and durable biosensing platform. This work marks a significant step toward quasi-autonomous biosensing technologies, paving the way for cost-effective and robust reusable biosensors suitable for remote and field applications.
1. Introduction
Typical biosensors consist of three main components: a biorecognition element (bioreceptor), a transducer, and a signal-processing device.1 When a biomolecular target interacts with the biorecognition element, a quantifiable physical signal variation is measured and compared to the reference. Key attributes of an ideal biosensor include cost-effectiveness, user friendliness defined by operational readiness and portability, or even the ability to operate semi-autonomously in out-of-laboratory settings. The reusability of a biosensor is particularly important for devices designed for remote environmental sensing and monitoring. This requires either a regeneration procedure to overcome bioreceptor-analyte binding forces or washing of the biosensor followed by a re-functionalization procedure.Chemical regeneration, achieved by submerging a transducer in a regeneration buffer, is the most frequently applied method. This method often involves acids/bases, low concentration detergents, and high ionic strength solutions.2 For instance, a relatively inexpensive regeneration method demonstrated a five-fold regeneration of a ZnO/bulk GaAs acousto-optic biosensor for detection of Escherichia coli.3 The use of a pH 2 buffer can break antibody–antigen binding, while high ionic content can eliminate aptamer-target interaction.4 Heating 2% sodium dodecyl sulfate (SDS) at 50 °C enabled the regeneration of a fluorescence biosensor with a cyclodextrin–mannose entity designed to capture E. coli5 whereas heating at 80 °C was used for regenerating an aptamer-based impedimetric biosensor of E. coli.6 Up to seven regeneration steps were demonstrated with a surface plasmon resonance (SPR) biosensor designed for detecting small molecules, such as ochratoxin A.7 However, the sensitivity of SPR biosensors is typically limited by the penetration depth of evanescent waves, which does not exceed a few tens of nm.8 This results in relatively poor responses to large bio-objects, such as bacteria.9 Additionally, sensitive SPR devices are relatively bulky and prohibitively expensive. Fiber optic-based biosensors also suffer from the high cost of the reading equipment. Regenerable configuration of fluorescence,5 quartz crystal balance,10 acoustic wave3 and electrochemical11,12 biosensors have been reported for detecting bacteria. However, common challenges include bulky equipment,3 the risk of damaging recognition elements,11 and the loss of sensitivity.5,10,12 These issues could be related to the deterioration of the quality of sensing surface, such as gold in SPR biosensors,7 or the denaturation of recognition linkers.11 Examples of biosensors investigated for their regeneration characteristics are listed in Table S1 (ESI†).
Our interest concerns detection of Bacillus cereus (Bc), a foodborne pathogen13 with its infectious dose at 103 CFU mL−1,14,15 and Bacillus anthracis (Ba) known for its use as a biological weapon.16 In that context, we investigated detection of B. thuringiensis (Bt) to act as a surrogate for Ba and Bc, based on the high similarity of spore structures among members of the Bacillus cereus group.17 Although there are a few reports of Bt toxicity in humans, distinguishing between Bt and Bc is challenging, leading to an underestimation of Bt's real impact, particularly in relation to enterotoxins.18
Detection of B. anthracis toxin was demonstrated with an aptamer-based electrochemical biosensor.19 However, the detection of toxins requires sample processing that may not ensure rapid and cost-effective pathogen detection compared to identifying whole bacteria.17 Also, it may provide limited information of the infection as there could be many bacterial species producing structurally and functionally similar toxins.20
Biosensors based on digital photocorrosion (DIP) of GaAs–AlGaAs nanoheterostructures have recently been introduced for detecting bacteria and spores.21–27 The operation of the DIP biosensor is based on the controlled photocorrosion of GaAs–AlGaAs nanoheterostructures, which is monitored via the photoluminescence (PL) signal originating from GaAs.28,29 The DIP process is highly sensitive to the presence of charged molecules/macromolecules/bacteria in the vicinity of the chip surface. The distinct surface recombination rates of GaAs and AlGaAs29 result in the formation of a PL intensity maximum (PLmax) when the photocorrosion front reaches the interface of a ∼20 nm thick GaAs–AlGaAs nanolayer pair. The timing of the PLmax appearance is influenced by charge transfer interactions between the biochip and detected target, allowing for precise calibration of bacterial concentrations. This approach has been successfully demonstrated for the detection of Legionella pneumophila22,23,25 and E. coli,21 and was recently applied to aptamer-based spore detection.27 Up-to-date DIP biosensing results have been based on recording the GaAs PL signal originating from a single pair of GaAs (12 nm)–AlGaAs (10 nm) nanolayers. Given that a series of PLmax can be produced by DIP of a series of GaAs–AlGaAs nanolayers,28,30 we hypothesized that each pair of GaAs–AlGaAs nanolayers could be employed for repetitive biosensing with the same biochip. Here we discuss this concept and present the results of repetitive detection of Bacillus thuringiensis kurstaki (Btk) spores.
This work represents a significant advancement in biosensor technology by integrating nanoscale heterostructures with aptamer-based recognition for highly specific and repetitive detection of spores. The innovative concept of utilizing multiple GaAs–AlGaAs nanolayer pairs within a single biochip for sequential biosensing cycles not only extends the operational lifespan of the sensor but also reduces material consumption, improving cost-efficiency. By applying the DIP methodology for Btk spore detection, this study broadens the scope of autonomous biosensing to include reusable platforms, an essential step toward practical, field-deployable solutions.
2. Experimental methods
2.1. Materials and reagents
Semiconductor grade acetone was purchased from Allied and Chemical Products Inc. (ACP, Montréal, Canada). OptiClear was sourced from National Diagnostics (Mississauga, Canada), and isopropanol was obtained from Fisher Scientific (Ottawa, Canada). The 28% ammonium hydroxide (NH4OH) was provided by Anachemia (Richmond, Canada) and used without further purification. Anhydrous ethanol, from Commercial Alcohols, Inc. (Brampton, Canada), was degassed by flushing with high-purity nitrogen gas (99.9995%) that was obtained from Praxair, Canada. 11-Mercapto-1-undecanol (MUDO) thiol and phosphate buffered saline (PBS) solution (10×, pH 7.4) were purchased from Sigma-Aldrich (Oakville, Canada). Bacillus thuringiensis kurstaki (strain HD1) was drawn from the Foray 48B. Thiolated aptamers (Apt) against Btk spores were obtained from IDT (Ontario, Canada). Deionized (DI) water with a resistivity of 18.2 MΩ was produced using a customized Millipore purification system assembled by Culligan (Québec, Canada). A GaAs–AlGaAs wafer, obtained through CMC Microsystems (Ottawa, Ontario, Canada), comprised a stack of 7 bilayers of GaAs (12 nm) - Al0.35Ga0.65As (10 nm). The first two pairs of these bilayers were investigated for repetitive biosensing.2.2. Chip preparation
2 mm × 2 mm chips were cut out from the GaAs–AlGaAs wafer (D3422) using a high-precision diamond saw (DAD 320, Disco). The chips were sequentially cleaned with acetone, OptiClear, acetone again, and isopropanol (IPA), with each step performed for 5 minutes under sonication. Native oxides were removed from the chips by etching in 28% ammonium hydroxide (NH4OH) solution for 2 minutes, followed by a quick rinse with degassed ethanol. Immediately after, the chips were immersed in 1 mM 11-mercapto-1-undecanol (MUDO) thiol solution in degassed ethanol for 20 hours. The chips were then sonicated for 1 minute, rinsed with degassed ethanol to remove the unbound thiols, and dried using high-purity nitrogen gas. This results in the immobilization of thiols on the chip surface, which is tilted at approximately 19°, due to hydrophobic interactions.31 MUDO thiols form a self-assembled monolayer (SAM) on the GaAs surface, which partially passivates to the chip surface. The binding of thiols via sulphur with GaAs atoms decreased the photocorrosion rate decreases.21 The resulting decrease in DIP leads to a desirable resolution of PLmax plots and accurate detection of spores. Aptamers against Btk spores were premixed with spores at concentrations of 103, 104, and 105 CFU mL−1 and incubated for 1 hour at room temperature to form aptamer–spore hybrids. The MUDO thiolated chips were incubated in these suspensions for 1 hour and then washed three times with 1× PBS. Aptamers at 5 μM mL−1 were premixed with spores at concentrations of 103, 104, and 105 CFU mL−1 and incubated for 1 hour at room temperature to form aptamer–spore hybrids. Following that, MUDO thiolated chips were incubated in each of the prepared aptamer–spore suspensions for 1 hour and washed three times with PBS. Reference chips were prepared by incubating MUDO thiolated chips in pure 5 μM thiolated aptamers in 1× PBS, followed by washing three times with PBS. Fig. 1 schematically illustrates the formation of the aptamer–spore complexes (a), the structure of an aptamer used for specific capture of Btk spores (b), and functionalization of a biochip with aptamer–spore complexes (c).To perform a negative control test assessing the specificity of our regenerable aptasensor, we exposed the DIP biochip to a non-Bc group, Bacillus megaterium (Bm) spores, at a concentration of 105 CFU ml−1, using the same aptamer-spore hybrid architecture.
To evaluate the efficiency of our regenerable DIP biosensor under real world conditions, we performed two consecutive detections of Btk spores in a water sample from Magog River. A 10 ml sample of river water was filtered using a Millex GV' PVDF membrane filter to collect suspended matter. The retained matter was washed with 10 mL of deionized water (DI water), followed by a 10 mL of 1× PBS backwash to recover the suspended particles. The backwashed sample was used as the reference and spiked with Btk spores at 105 CFU ml−1 concentration to functionalise detection samples.
2.3. Validation of the regeneration approach
The regeneration approach involved detaching spores from aptamers by submerging the biochip in 3× PBS buffer solution for 20 min, followed by quick wash with deionized (DI) water to avoid salt crystal formation. After drying under nitrogen flow, the efficiency of this approach was studied using a Zeiss optical microscope (Oberkochen, Germany) under 500× magnification. All measurements were repeated at least three times for statistical analysis.The photocorrosion of the first two pairs of bilayers was investigated for the independent detection of spores. Fig. 2 illustrates the concept of a regenerable DIP biosensor. Following the consumption of the first GaAs–AlGaAs bilayer during the initial detection cycle, the biochip surface is treated with an aptamer regeneration buffer (3× PBS) for 20 minutes to facilitate the removal of captured spores. This step is followed by a rapid wash with 1× PBS to restore the aptamers to their original ternary structure. Once regenerated, the second bilayer of the same biochip is exposed to a fresh aptamer–spore hybrid suspension of unknown concentration. Calibration curves/testimonial plots are constructed based on PL intensity data obtained from biochips exposed to aptamer–spore complexes of known concentrations.
 | ||
| Fig. 2 Schematic illustration of a GaAs–AlGaAs biochip designed to detect Btk spores by sequential DIP of GaAs–AlGaAs nanolayers. | ||
2.4. PL monitored DIP process
Detection and reference runs were conducted using a custom-designed quantum semiconductor photonic biosensing (QSPB-3) reader.32 The chips were irradiated with a 375 nm LED, and PL emission from GaAs (maximum intensity at 870 nm) was collected with a CCD camera.22 Experiments were conducted with intermittent 1.5 second irradiation at power density of 15 mW cm−2, separated by a 5 s dark phase. Functionalized biochips underwent DIP under continuous flow of 1× PBS at 100 μL min−1. The schematic diagram of the physical prototype of our DIP biosensor is presented as Fig. S1 in the ESI† section. For repetitive detection, after revealing the first PL intensity peak, the DIP process was stopped at the intermediate PL intensity minimum, and the spores were released using the aptamer regeneration buffer. Reference chips were also exposed to the regeneration buffer for the same duration. Afterwards, the reference and detection chips were incubated in aptamer solution and aptamer–spore hybrids, respectively, for 1 hour, followed by washing with 1× PBS before conducting the second series of the DIP cycle.The effect of UV irradiation (375 nm) on aptamer reactivity was evaluated by exposing a 20 μL drop of 100 μM aptamer solution to UV light for 5 minutes. The UV-treated aptamer was then tested for spore capture at 105Btk spores per mL. Spore immobilization was quantified using optical microscopy and compared to results obtained from non-UV-treated aptamers. Blind tests were performed with biochips exposed sequentially to two suspensions of spores at unknown concentrations (UC1 and UC2) determined by the first and second GaAs–AlGaAs bilayer.
3. Results
3.1. Validation of the regeneration efficiency
The efficiency of releasing the spores by an aptasensor submerged in PBS buffer solution is illustrated with examples of optical microscopic images shown in Fig. 3. The MUDO functionalized GaAs chip exposed to Apt–Btk hybrids at 105 spores per mL shows an average 1920 ± 108 spores per mm2 (Fig. 3a), which is compared to 115 ± 30 spores per mm2 for the chip exposed for 20 min to 3× PBS (Fig. 3b). Thus, this regeneration procedure corresponds to 94% efficiency of removing Btk spores. | ||
| Fig. 3 Optical microscopy images of MUDO functionalized GaAs chips exposed to Apt–Btk hybrids at 105 CFU mL−1 (a), and after regeneration involving submersion in 3× PBS for 20 minutes (b). | ||
3.2. Repetitive detection of Btk spores using the GaAs–AlGaAs DIP aptasensor
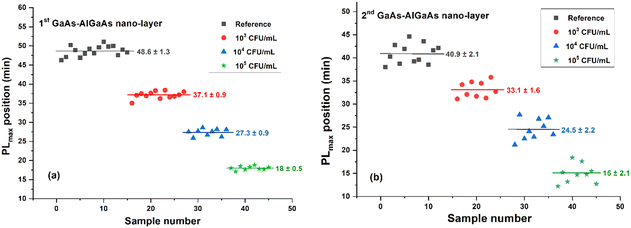
Temporal PL intensity plots collected during DIP of two consecutive pairs of GaAs–AlGaAs nanolayers are shown in Fig. 4. Increasing spore concentration accelerated the formation of PL intensity maxima observed with the first and second pairs of GaAs–AlGaAs nanolayers. The statistical error of 45 samples, summarized in Fig. 5, revealed PLmax values for the first pair of GaAs–AlGaAs nanolayers at 48.6 ± 1.3 min, 37.1 ± 0.9 min, 27.3 ± 0.9 min, and 18 ± 0.5 min for the reference run and spore suspensions at 103, 104, and 105 CFU mL−1, respectively. For the second pair of GaAs–AlGaAs nanolayers, PLmax values were 40.9 ± 2.1 min, 33.1 ± 1.6 min, 24.5 ± 2.2 min, and 15 ± 2.1 min, respectively, under nominally the same experimental conditions. These findings are further detailed in Table S2 (ESI†). | ||
| Fig. 4 Examples of PL intensity plots collected for the first (a) and second (b) pairs of GaAs–AlGaAs nanolayers of the regenerable DIP aptasensor. | ||
The binding ability of the aptamer was not affected by UV irradiation. The observed spore immobilization was at 1735 ± 138 Btk spores per mm2 for UV-treated aptamers, compared to 1920 ± 108 Btk spores per mm2 for non-UV-treated aptamers. The variation between these conditions is statistically insignificant as shown in Fig. S2.† Calibration plots derived from DIP using the first and second pairs of GaAs–AlGaAs nanolayers were employed for blind tests with two unknown concentrations (UCs) as illustrated in Fig. 6. Experimental points (triangle up and down) show detection of Btk spores at 1.6 × 104 CFU mL−1 (UC1) and 4 × 103 CFU mL−1 (UC2). These results compare to the intended spore concentrations of 2 × 104 CFU mL−1 and 5 × 103 CFU mL−1, respectively, indicating an error not exceeding 20%.
We note that detection errors of bacteria achieved with relatively advanced technologies, such as fluorescence-based biosensing, reported the related errors ranging between 5 and 23%, respectively, by Li et al.,33 and Chattopadhyay et al.,34 Thus, our approach offers attractive results in that context while taking advantage of the relatively early stage of development of the underlying technology.
Negative control experiments were conducted using non-Bc group spores (Bm). The PL maximum for the Bm-aptamer hybrid is observed at 45.9 ± 2.7 min, overlapping with the reference PL maximum at 48.6 ± 1.3 min. This result confirms the specificity of the aptamer used, and consequently, the reliability of our biosensing method (Fig. S3, ESI†). Optical microscopy imaging of GaAs chips exposed to the Bm aptamer–spore hybrid showed that captured spore densities did not exceed 277.6 ± 48 spores per mm2. This was 7 times lower than what was obtained for our target Btk spores (1920 ± 108 spores per mm2) under the same tested concentration of 105 spores per mL.
The successful detection of Btk spores in Magog River water is demonstrated in Fig. S4 (ESI†). The results indicate that the presence of Btk spores significantly accelerated the temporal position of the PL peak, shifting from 71.2 ± 2.3 min to 48.3 ± 3.3 min in the first bilayer and from 57.5 ± 3.1 min to 35.8 ± 3.8 min in the second bilayer. These findings align with the previously observed results under controlled laboratory conditions, further supporting the practical applicability of our biosensor.
4. Discussion
The immobilization of thiolated aptamer–spore hybrids on the GaAs–AlGaAs biochip induced charge exchange between the biochip and the semiconductor surface. The loss of electrons by the semiconductor biochip accelerates photocorrosion in proportion to the density of immobilized spores, which is facilitated by the relatively short length (∼3 nm) of the aptamer linker.35 The proximity of spores to the biochip surface is an important characteristic of a DIP GaAs–AlGaAs biosensor. Attempts to develop a similar device using antibody-functionalized GaAs–AlGaAs biochips were less successful, likely due to the significantly greater antigen-biochip surface distance (∼15 nm), resulting in excessively reduced photocorrosion rates and wider PLmax features.25The increased error in PLmax determination for the second pair of GaAs–AlGaAs nanolayers (Fig. 5) is attributed primarily to surface roughness changes resulting from DIP. While some residual spores might contribute to this effect, the primarily cause must be related to the quality of the DIP-produced surface. The principal sources of the increased roughness are Ga oxides that, normally, require processing with ammonia but accumulate on the surface of biochips photocorroding in PBS environment. This is in addition to Al-based products that accumulate even in an ammonia environment. A comparison of the atomic force microscopy surface roughness (σRMS) of GaAs–AlGaAs biochips functionalized with MUDO and aptamer showed the increase of σRMS from 0.81 nm to 1.32 nm and 2.12 nm after DIP of, respectively, the first and second pair of GaAs–AlGaAs nanolayers (Fig. S5, ESI†). Independent research of the similar GaAs–AlGaAs biochip functionalized with a 16-mercaptohexadecanoic acid self-assembled monolayer revealed σRMS increased from the initial 0.5 nm to 1.9 nm after DIP of the second pair GaAs–AlGaAs of nanolayers.32 Accurately determining the PLmax positions is important to the measurable differentiation of results originating from suspensions with different concentrations of spores, and to the development of regenerable DIP biosensors.
The specificity of our DIP aptasensor was further confirmed by the negligible variation in the PL maximum temporal position for Bm compared to the reference. This finding aligns with previous data reported by Moteshareie et al.,15 demonstrating that the aptamer used in this study exhibits significantly higher capture efficiency for Btk spores compared to Bm or B. subtilis spores.
The presence of impurities in the Magog River water influenced the temporal position of the PL intensity maxima, leading to a delay in the photocorrosion rate for both reference and detection samples compared to those in pristine conditions. This delay may be attributed to the presence of ionic species in the Magog River water sample. However, this effect did not compromise the ability of our biosensor to detect Btk spores, demonstrating its robustness and applicability for environmental monitoring.
Clearly, sustainable DIP of devices with numerous pairs of GaAs–AlGaAs nanolayers is crucial to operation of biochips capable of delivering a large number of biosensing runs.36 Given that neither water nor PBS environments support photoetching of Al-based products, this requires development of a dedicated chemical processing protocol.
5. Conclusions
This study demonstrated the functionality of a regenerable DIP biosensor incorporating two pairs of GaAs (12 nm)–AlGaAs (10 nm) nanolayers. The biochip surface was coated with low-density MUDO SAM, designed to reduce DIP rates and enable attractive detection of Btk spores at concentrations between 103–105 CFU mL−1. An enhanced limit of detection is still possible with this method, e.g., by pre-concentrating spores in tested water samples.The application of thiolated aptamers, specific to Btk spores, facilitated the immobilization of aptamer–spore hybrids on the biochip surface. After completing the biosensing run with the first pair of nanolayers, exposure to 3× PBS released the spores and prepared the same biochip for another run with the second pair of nanolayers.
The regenerable biosensor was blind tested for detection of Btk spores in samples spiked at 2 × 104 CFU mL−1 and 5 × 103 CFU mL−1. The results reveal the protocol of a repetitive detection involving the same GaAs–AlGaAs biochip with an error not exceeding 20%, i.e., comparable to such errors reported by other biosensing technologies. We argue that a chip with stacks of numerous pairs of GaAs–AlGaAs nanolayers, the production of which is readily available with the current semiconductor manufacturing technology, can deliver a large amount of biosensing data before getting swapped with a new unit. This should be of high interest to the development of biosensing workstations for quasi-continuous operation in out-of-laboratory settings attractive for environmental monitoring.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its ESI† file. Should any raw data files be needed in another format they are available from the corresponding author upon reasonable request.Author contributions
Conceptualization, methodology, and writing – original draft: Ishika Ishika and Walid M. Hassen. Formal analysis and investigation: Ishika Ishika, Walid M. Hassen, Houman Moteshareie (spore culture), and René St-Onge (AFM). Funding acquisition, supervising, and reviewing: Azam F. Tayabali and Jan J. Dubowski. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported through a grant from the Canadian Safety and Security Program of Defence Research and Development Canada, the Centre for Security Science (CSSP-2022-CP-2545) to Dubowski and Tayabali and subcontract No HT282-114353 to Dubowski; and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN-2020-05558 (JJD).References
- P. Bhalla and N. Singh, Eur. Phys. J. B, 2016, 89, 1–8 CrossRef CAS.
- J. Goode, J. Rushworth and P. Millner, Langmuir, 2015, 31, 6267–6276 CrossRef CAS PubMed.
- J. Chawich, W. M. Hassen, C. Elie-Caille, T. Leblois and J. J. Dubowski, Biosensors, 2021, 11, 145 CrossRef CAS PubMed.
- T. Hianik, V. Ostatná, M. Sonlajtnerova and I. Grman, Bioelectrochemistry, 2007, 70, 127–133 CrossRef CAS.
- Y. Qu, T. Wei, W. Zhan, C. Hu, L. Cao, Q. Yu and H. Chen, J. Mater. Chem. B, 2017, 5, 444–453 RSC.
- S. Brosel-Oliu, R. Ferreira, N. Uria, N. Abramova, R. Gargallo, F.-X. Muñoz-Pascual and A. Bratov, Sens. Actuators, B, 2018, 255, 2988–2995 CrossRef CAS.
- J.-H. Park, J.-Y. Byun, H. Mun, W.-B. Shim, Y.-B. Shin, T. Li and M.-G. Kim, Biosens. Bioelectron., 2014, 59, 321–327 Search PubMed.
- A. Shalabney and I. Abdulhalim, Laser Photonics Rev., 2011, 5, 571–606 CrossRef CAS.
- C. S. Zhu, J. Li and H. Wang, Bio-Protoc., 2023, 13(17), e4795 CAS.
- Y. Lian, F. He, H. Wang and F. Tong, Biosens. Bioelectron., 2015, 65, 314–319 CrossRef CAS PubMed.
- E. Yuhana Ariffin, L. Y. Heng, L. L. Tan, N. H. Abd Karim and S. A. Hasbullah, Sensors, 2020, 20, 1279 CrossRef.
- X. Xu, X. Lin, L. Wang, Y. Ma, T. Sun and X. Bian, Biosensors, 2023, 13, 868 CrossRef CAS.
- A. Okamoto and A. Okutani, Molecular Medical Microbiology, ed. M. Y. H. Yi-Wei Tang, D. Liu, A. Sails, P. Spearman and J.-R. Zhang, Elsevier, Academic Press, 3rd edn, 2024, ch. 47, pp. 957–986, DOI:10.1016/B978-0-12-818619-0.00152-0.
- P. E. Granum and T. Lund, FEMS Microbiol. Lett., 1997, 157, 223–228 CrossRef CAS.
- H. Moteshareie, W. M. Hassen, Y. Dirieh, E. Groulx, J. J. Dubowski and A. F. Tayabali, Microorganisms, 2022, 10, 1408 Search PubMed.
- S. Riedel, University Medical Center Proceedings, 2005, 18(3), 234–243 CrossRef PubMed.
- Y. Wu, C.-W. Wang, D. Wang and N. Wei, ACS Synth. Biol., 2021, 10, 333–344 CrossRef CAS PubMed.
- S. Johler, E. M. Kalbhenn, N. Heini, P. Brodmann, S. Gautsch, M. Bağcioğlu, M. Contzen, R. Stephan and M. Ehling-Schulz, Front. Microbiol., 2018, 9, 1915 CrossRef.
- L. N. Cella, P. Sanchez, W. Zhong, N. V. Myung, W. Chen and A. Mulchandani, Anal. Chem., 2010, 82, 2042–2047 CrossRef CAS.
- M. M. Lubran, Ann. Clin. Lab. Sci., 1988, 18, 58–71 CAS.
- E. Nazemi, S. Aithal, W. M. Hassen, E. H. Frost and J. J. Dubowski, Sens. Actuators, B, 2015, 207, 556–562 CrossRef CAS.
- M. R. Aziziyan, W. M. Hassen, D. Morris, E. H. Frost and J. J. Dubowski, Biointerphases, 2016, 11(1), 0119301 CrossRef PubMed.
- M. Aziziyan, W. Hassen, H. Sharma, E. S. Sani, N. Annabi, E. Frost and J. Dubowski, Sens. Actuators, B, 2020, 304, 127007 CrossRef CAS.
- M. A. Islam, W. M. Hassen, A. F. Tayabali and J. J. Dubowski, Biochem. Eng. J., 2020, 154, 107435 CrossRef CAS.
- M. A. Islam, W. M. Hassen, A. F. Tayabali and J. J. Dubowski, ACS Omega, 2021, 6, 1299–1308 CrossRef CAS.
- M. A. Islam, W. M. Hassen, I. Ishika, A. F. Tayabali and J. J. Dubowski, Biosensors, 2022, 12, 105 CrossRef CAS.
- I. Ishika, W. M. Hassen, H. Moteshareie, A. F. Tayabali and J. J. Dubowski, Proceedings Volume 12410, Nanoscale and Quantum Materials: From Synthesis and Laser Processing to Applications 2023, SPIE, 2023, 1241003, DOI:10.1117/12.2662421.
- S. Aithal, N. Liu and J. J. Dubowski, J. Phys. D: Appl. Phys., 2016, 50, 035106 Search PubMed.
- M. R. Aziziyan, H. Sharma and J. J. Dubowski, ACS Appl. Mater. Interfaces, 2019, 11, 17968–17978 CrossRef CAS PubMed.
- H. Sharma, K. Moumanis and J. J. Dubowski, J. Phys. Chem. C, 2016, 120, 26129–26137 Search PubMed.
- O. Voznyy and J. J. Dubowski, Langmuir, 2008, 24, 13299–13305 CrossRef CAS PubMed.
- R. St-Onge, J. Vermette, W. M. Hassen and J. J. Dubowski, Appl. Phys. Lett., 2021, 118, 222102 Search PubMed.
- X. Li, P. Xue, H. Wang, H. Li, R. Du, J. Gao, K.-Y. Wong and Y. Qin, J. Mater. Chem. C, 2024, 12, 16523–16532 Search PubMed.
- A. N. Chattopadhyay, M. Jiang, J. M. V. Makabenta, J. Park, Y. Geng and V. Rotello, Biosensors, 2024, 14, 360 CrossRef CAS PubMed.
- S. Yoon and J. J. Rossi, Adv. Drug Delivery Rev., 2018, 134, 22–35 Search PubMed.
- R. St-Onge, M. W. Hassen, H. Moteshareie, A. F. Tayabali and J. J. Dubowski, Phys. Status Solidi B, 2025, 2400567 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00367e |
| This journal is © The Royal Society of Chemistry 2025 |