Open Access Article
Open Access ArticleContrasting effects of mismatch locations on Z-DNA formation under bending force†
SoJung
Park‡
,
Jaehun
Yi‡
and
Nam Ki
Lee
 *
*
Department of Chemistry, Seoul National University, 08826, Seoul, Republic of Korea. E-mail: namkilee@snu.ac.kr
First published on 10th March 2025
Abstract
Z-DNA is a non-canonical, left-handed helical structure that plays crucial roles in various cellular processes. DNA mismatches, which involve the incorporation of incorrect Watson–Crick base pairs, are present in all living organisms and contribute to the mechanism of Z-DNA formation. However, the impact of mismatches on Z-DNA formation remains poorly understood. Moreover, the combined effect of DNA mismatches and bending, a common biological phenomenon observed in vivo, has not yet been explored due to technological limitations. Here, using single-molecule FRET, we show that a mismatch inside the Z-DNA region, i.e., the CG repeat region, hinders Z-DNA formation. In stark contrast, however, a mismatch in the B–Z junction facilitates Z-DNA formation. When the bending force is applied on double stranded DNA, a mismatch in the B–Z junction releases the bending stress more effectively than one in the CG repeat region. These findings provide mechanical insights into the role of DNA mismatches and bending forces in regulating Z-DNA formation, whether promoting or inhibiting it in biological environments.
Introduction
The dynamic nature of the right-handed B-DNA, which predominates under physiological conditions, can transition into various noncanonical forms during biological processes.1 Z-DNA, a well-known non-canonical form of DNA, exhibits a left-handed helical form with a zig-zag backbone caused by alternating anti- and syn-conformations of bases.2–5 This left-handed helical form of DNA forms transiently in nature but is known to play crucial roles in biological processes, such as transcription regulation,6,7 nucleosome positioning,8–10 and control of genetic instability.11–13 The formation of Z-DNA primarily occurs in regions rich in alternating purine and pyrimidine bases (e.g., CG repeats).2 It is promoted under specific conditions, including high salt conditions, which alleviate the electrostatic repulsion between phosphate backbone groups.14 Additionally, DNA-binding proteins can induce the transition to Z-DNA conformation.15–17 Mechanical stresses on DNA, such as supercoiling18,19 and torsional stress,20 also play a role in regulating Z-DNA formation. A recent study revealed that bending force, one of the most common types of mechanical stress experienced by DNA, facilitates Z-DNA formation under physiological salt conditions.21 It has been proposed that bending force induces the formation of a B–Z junction, accompanied by the extrusion of a single base pair at the interface between B-DNA and Z-DNA, which effectively releases the bending stress and stabilizes the Z-DNA structure in a lower bending energy state.21 This model, however, still requires further experimental proof. Conversely, a Z–Z junction forms when the alternating purine and pyrimidine bases in Z-DNA are disrupted by several defects, such as the insertion of a DNA mismatch, causing the two Z-DNA segments to become out of phase.22,23DNA mismatch, a ubiquitous phenomenon arising from various biological processes, is universal in nature and can be accommodated in any position within the Z-DNA.24 Previous studies have provided insights into the structure23,25 and energetic penalties22,26,27 of mismatches within Z-DNA regions. Ellison et al. obtained the energetic cost of the B–Z transition caused by base pair disruptions in CG repeats using two-dimensional gel electrophoresis and statistical mechanical analysis, reporting a value of 2.4–3.4 kcal per mol bp.26 Johnston et al. also reported that a Z–Z junction has an energetic penalty of 3.5 kcal mol−1 due to disruption of nearest-neighbor interactions, compared to a continuous Z-DNA sequence.22 Nichols et al. observed that mismatches in RNA also promote Z-formation in regions adjacent to Z-prone segments.28 However, the effect of DNA mismatch, introduced at the different locations within the duplex, on the B–Z transition remains largely unexplored. Moreover, the combined effect of mismatch and DNA bending, frequently observed in biological phenomena, has not yet been reported to date. To address these issues, we investigated the effect of mismatches, within the Z-DNA region and at the B–Z junction, on the B–Z transition using single-molecule fluorescence resonance energy transfer (smFRET).
Results and discussion
The FRET efficiency, which is inversely proportional to the sixth power of the distance between the donor and acceptor dyes, enables observation of conformational changes of dsDNA.29,30 The B–Z transition has been successfully observed in previous studies through the decrease in FRET efficiency,16,21,31,32 as Z-DNA has a longer pitch than B-DNA and the extrusion of a single base pair at the B–Z junction (Fig. 1a).3,21 In this work, the B–Z transition has been observed using alternating-laser excitation (ALEX) smFRET (Fig. 1b and S1†).33–35 This method allows measuring the FRET efficiency of each freely diffusing double stranded DNA (dsDNA) molecule at the single-molecule level. Moreover, ALEX allows the sorting of FRET pairs from heterogeneous subpopulations of molecules within a buffer solution (Fig. S1†). The FRET efficiency of the selected FRET pairs was calculated by obtaining the FRET histogram (Fig. 1c). The low-FRET peak corresponds to Z-DNA and the high-FRET peak corresponds to B-DNA (Fig. 1c).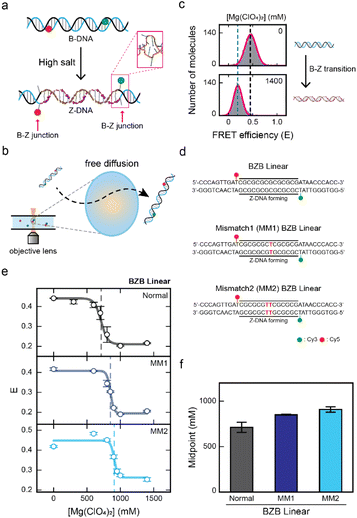 | ||
| Fig. 1 Observing the effect of a mismatch in the CG repeat region for Z-DNA formation by single-molecule FRET measurement. (a) Illustration of the B–Z transition in the linear dsDNA that consists of a Z-DNA forming region that is flanked by two random sequences at both ends. Z-DNA formation is accompanied by two B–Z junctions with an extruded base pair (pink rectangle, PDB code 2ACJ). (b) Brief schematic illustration of the ALEX measurement, which detects the FRET efficiency of freely diffusing single molecules that pass through the confocal excitation volume. (c) The FRET histograms of the linear dsDNA undergoing the B–Z transition. The low-FRET peak represents Z-DNA and the high-FRET peak represents B-DNA of the BZB linear sample. (d) DNA sequences used for the experiments. The BZB linear sample consists of the CG repeat, a Z-DNA forming region, with two random sequences at both ends. Mismatched base pairs are substituted within the CG repeat region and indicated in red color. Cy3 and Cy5 were used as a FRET pair. (e) The average FRET efficiency (E) of each linear dsDNA sample (BZB linear, MM1 BZB linear, and MM2 BZB linear) plotted against the concentration of Mg(ClO4)2. The dotted lines denote the midpoint of each sample. Error bars were obtained from three independent measurements. (f) B–Z transition midpoints for each sample were obtained from (e). As the number of the mismatches in the CG repeat increases, the B–Z transition midpoint increases. Error bars were obtained from three independent measurements. | ||
The effect of mismatched base pairs on Z-DNA formation
First, we investigated the effect of a mismatch in the CG repeat region on Z-DNA formation. As a control, we prepared a linear dsDNA consisting of seven CG repeats, capable of forming Z-DNA flanked by random sequences on both ends that remain in B-DNA conformation (Fig. 1d, top panel). We denote this dsDNA as BZB linear, which consists of 34 base pairs (bp). Next, we substituted a T–T mismatch at the 7th base pair within the CG repeat region, which we denote as MM1 BZB linear (Fig. 1d, middle panel). We also substituted two T–T mismatches in the 7th and 8th base pairs within the CG repeat region, which we denote as MM2 BZB linear (Fig. 1d, bottom panel). For a single-molecule FRET experiment, the donor (Cy3) and acceptor (Cy5) dyes were labeled at both ends of the Z-DNA forming region (10th and 25th base pairs). Upon Z-DNA formation, all three dsDNA samples were accompanied by two B–Z junctions (Fig. 1a). Magnesium perchlorate (Mg(ClO4)2) is known to induce the formation of Z-DNA.32 We gradually increased the concentration of Mg(ClO4)2 to observe the conformational changes of DNA upon the B–Z transition for all three dsDNA samples (Fig. 1e and see S2† for the FRET efficiency histograms). Without Mg(ClO4)2, all three dsDNA samples had a similar average FRET efficiency (E) of approximately 0.4, indicating B-DNA conformation. However, at high concentration of Mg(ClO4)2, the E for all three dsDNA samples decreased to approximately 0.2, indicating Z-DNA conformation (Fig. 1e). The E of MM2 BZB linear increased slightly at lower salt concentrations, possibly due to enhanced DNA flexibility resulting from the increased salt concentrations.36–38 To quantitatively evaluate the effect of the mismatch on Z-DNA formation upon the salt gradient, the B–Z transition midpoints, where B- and Z-DNA were equally probable, were obtained for each sample. The midpoint for BZB linear was approximately 710 mM Mg(ClO4)2 and increased to approximately 850 mM and 910 mM Mg(ClO4)2 as the number of mismatches in the CG repeat region increased from one to two, suggesting that mismatches in the CG repeat region inhibit Z-DNA formation (Fig. 1f).Previous studies have provided insights into the energetic costs22,26,27 associated with mismatches in Z-DNA regions. Despite these findings, the impact of such energetic costs caused by mismatches on the B–Z transition has not been previously demonstrated. Our results address this gap by clearly showing that mismatches hinder Z-DNA formation, consistent with earlier prediction. The insertion of a mismatch in the alternating purine and pyrimidine sequences disrupts the base-stacking interactions, which may hinder the formation of Z-DNA.
Next, we investigated the effect of a mismatch in the B–Z junction on Z-DNA formation (Fig. 2). We have used a linear dsDNA that forms one B–Z junction upon formation of Z-DNA, which consists of seven CG repeats, capable of forming Z-DNA, with one random sequence that remains in the B-DNA conformation (Fig. 2a, left panel).21 The total length of the dsDNA was 29 bp, with an equal number of CG repeats as in the BZB linear sample shown in Fig. 1d. We denote this dsDNA as a BZ linear. We prepared a linear dsDNA denoted as MM1 BZ linear containing a mismatch in the B–Z junction, where thymine is substituted in place of adenine to form a T–T mismatch (Fig. 2a, right panel). For a single-molecule FRET experiment, an acceptor (Cy5) was end-labeled at the 1st base pair and a donor (Cy3) was labeled at the B–Z junction in the 15th base pair. The B–Z transition midpoint for BZ linear was observed at approximately 450 mM Mg(ClO4)2 in previous work.21 The linear dsDNA that forms two B–Z junctions (Fig. 1, top panel) requires more salt concentrations (710 mM Mg(ClO4)2) to form Z-DNA, which is in agreement with results from the previous work.39
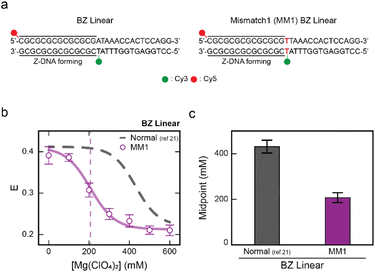 | ||
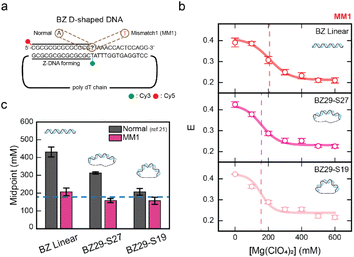
| Fig. 2 Mismatch at the B–Z junction facilitates Z-DNA formation. (a) DNA sequences used for the experiments. The BZ linear sample consists of the CG repeat, a Z-DNA forming region, and one random sequence. Z-DNA formation is accompanied by one B–Z junction. A mismatched base pair is substituted in the B–Z junction and indicated in red color (right panel). Cy3 and Cy5 were used as a FRET pair. (b) The E of each linear dsDNA sample (BZ linear and MM1 BZ linear) plotted against the concentration of Mg(ClO4)2. Data for E of the BZ linear sample was obtained from the previous work for comparison.21 The dotted line denotes the midpoint of the MM1 BZ linear sample. Error bars were obtained from three independent measurements. (c) B–Z transition midpoints of the BZ linear sample and MM1 BZ linear sample. The midpoints for each sample were obtained from (b). Mismatch at the B–Z junction significantly decreases the B–Z transition midpoint. Error bars were obtained from three independent measurements. | ||
Fig. 2b presents the E values for MM1 BZ linear at various Mg(ClO4)2 concentrations (see Fig. S2† for the FRET efficiency histograms). The B–Z transition midpoint for MM1 BZ linear was observed at approximately 200 mM Mg(ClO4)2 (Fig. 2c). This result indicates that the introduction of mismatch in the B–Z junction reduces the transition midpoint by 2.3-fold, facilitating Z-DNA formation under lower salt conditions. This finding markedly differs from cases involving a mismatch within the CG repeat region (Fig. 1f). Peck et al. reported that the formation of a B–Z junction requires a free-energy of +5 kcal mol−1.40 Indeed, Ha et al. revealed that a B–Z junction has the structure of the extrusion of a single base pair (Fig. 1a).41 The free-energy difference between A–T and T–T (mismatch) pairs including the nearest-neighbor effect is approximately 3 kcal mol−1.42 Hence, the mismatch in the B–Z junction may reduce the energy required for the base-pair extrusion, thereby facilitating Z-DNA formation at lower salt concentrations. Our results provide clear evidence that the local distortion induced by the mismatch in the B–Z junction helps to overcome the energy barrier of forming the B–Z junction by facilitating base-pair extrusion, as supported by the 2.3-fold decrease in the transition midpoint in the presence of the mismatch.
The combined effect of mismatch and bending force on Z-DNA formation
Then, how does the mismatch affect Z-DNA formation under DNA bending force? To explore this, we used D-shaped DNA nanostructures (Fig. 3a).43,44 No bending force is exerted on the linear dsDNA (Fig. 3a, left panel). D-shaped DNA consists of a dsDNA portion and a ssDNA string (Fig. 3a, middle panel). It is formed by annealing the ring-ssDNA with a partially complementary linear ssDNA (Fig. S3†).21,44–46 The dsDNA portion is bent due to the entropic stretching force exerted by the ssDNA string,45 which consists entirely of thymine. A longer ssDNA string length results in a weaker applied bending force on the dsDNA portion (Fig. 3a, middle panel), and the shorter ssDNA string length results in a stronger applied bending force on the dsDNA portion (Fig. 3a, right panel). The length of the dsDNA of the D-shaped DNA was fixed to 34 bp, using the same sequence as the BZB linear sample and MM1 BZB linear sample, while the ssDNA string lengths were varied at 30 nucleotides (nt) and 22 nt (Fig. 3b). We denote a D-shaped DNA as BZB34-S30, where 34 represents the dsDNA length and S30 indicates the 30 nt ssDNA string. As S22 is shorter than S30, a greater bending force is applied in BZB34-S22 than BZB34-S30. The bending force applied to the dsDNA portion increased in the following order: BZB linear, BZB34-S30, and BZB34-S22. Fig. 3c presents the decrease in E values for the normal samples of BZB linear, BZB34-S30, and BZB34-S22 (without a mismatch), due to the formation of Z-DNA as increased Mg(ClO4)2 concentrations (see Fig. S4† for the FRET efficiency histograms). These data clearly show that the B–Z transition midpoint decreases, i.e., facilitating Z-DNA formation, as the bending force increases. Fig. 3d presents the decrease in E values for the MM1 samples of BZB linear, BZB34-S30, and BZB34-S22. Notably, unlike the normal samples, we observed a smaller decrease in the B–Z transition midpoint for the MM1 samples despite equivalent bending force (Fig. 3e). As a result, the difference in B–Z transition midpoint values of the two samples becomes larger as the bending force increases. This implies that the effect of bending in inducing Z-DNA formation becomes less significant in the presence of mismatch in the middle of CG repeats.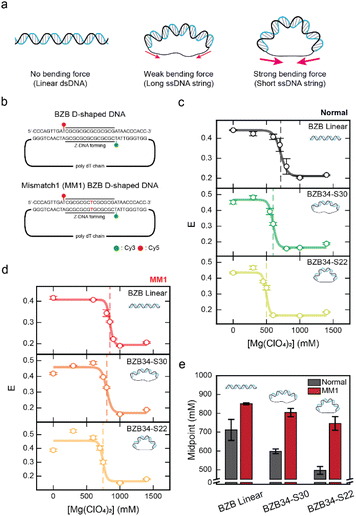 | ||
| Fig. 3 The effect of DNA bending on the B–Z transition with a mismatch in the Z-DNA forming region. (a) Illustration of linear dsDNA and D-shaped DNA nanostructures. The linear dsDNA has no bending force. In the D-shaped DNA nanostructures, the long ssDNA string length exerts a weaker bending force on the dsDNA portion, whereas the short ssDNA string length exerts a stronger bending force. (b) Schematic of the D-shaped DNA sequences used for the experiments. The dsDNA portion has the same sequence as the linear dsDNA samples shown in Fig. 1d. The bending force on the dsDNA portion was controlled by using different lengths of the ssDNA string that consists only of thymine. (c) The E of the normal linear dsDNA sample (BZB linear) and each normal D-shaped DNA sample (BZB34-S30 and BZB34-S22) plotted against the concentration of Mg(ClO4)2. The dotted lines denote the midpoint of each sample. Error bars were obtained from three independent measurements. (d) The E of the MM1 linear dsDNA sample (MM1 BZB linear) and MM1 D-shaped DNA samples (MM1 BZB34-S30 and MM1 BZB34-S22) plotted against the concentration of Mg(ClO4)2. The dotted lines denote the midpoint of each sample. Error bars were obtained from three independent measurements. (e) Comparison of the B–Z transition midpoint of the BZB linear and BZB D-shaped DNAs of normal and MM1 samples. B–Z transition midpoints for each sample were obtained from (c) and (d). As the bending force increases, the difference between the B–Z transition midpoint of the normal and MM1 samples becomes larger. Error bars were obtained from three independent measurements. | ||
Next, we investigated the effect of Z-DNA formation when a mismatch is in the B–Z junction under the bending force. The length of the dsDNA of the D-shaped DNA was fixed to 29 bp, using the same sequence as the BZ linear and MM1 BZ linear samples in Fig. 2a, while the ssDNA string lengths were varied at 27 nt and 19 nt (Fig. 4a). Again, the bending force applied to the dsDNA portion increased in the following order: BZ linear, BZ29-S27, and BZ29-S19. Fig. 4b presents the decrease in E values of the MM1 samples (BZ linear, BZ29-S27, BZ29-S19), as the concentration of Mg(ClO4)2 increased (MM1 BZ linear in Fig. 2b is also presented here for comparison; see Fig. S5† for the FRET efficiency histograms). The B–Z transition midpoints are shown in Fig. 4c (pink bars). For the purpose of comparison, the B–Z transition midpoint values of the normal dsDNA samples, reported in our previous work,21 were presented (Fig. 4c, gray bars). For the normal dsDNA samples, a substantial decrease in the B–Z transition midpoint values was observed, as the bending force increases.21 Interestingly, as for the MM1 dsDNA samples, there was a slight decrease in the B–Z transition midpoint value upon weak bending when the ssDNA string length was 27 nt (Fig. 4c). Even under stronger bending force by the 19 nt ssDNA string length, the B–Z transition midpoint value remained unchanged. These results suggest that the facile formation of a B–Z junction with an extruded base pair due to the mismatch induces the overall dsDNA structure to remain in the lower energy state under bending force. As a result, the bending force does not facilitate Z-DNA formation in the presence of a mismatch at the B–Z junction. This observation further supports our proposition that the formation of a B–Z junction with an extruded base pair effectively releases the bending stress, making Z-DNA with a B–Z junction thermodynamically more stable than B-DNA under high bending stress.21 Interestingly, the B–Z transition midpoints of MM1 BZ linear and MM1 BZ29-S19 are comparable. This suggests that under strong bending force, the extrusion of a base pair fully releases bending stress in BZ29-S19, similar to the effect of a mismatch at the B–Z junction. As a result, the bending force no longer facilitates the B–Z transition when a mismatch is present at the B–Z junction.
 | ||
| Fig. 4 The effect of DNA bending on the B–Z transition with a mismatch at the B–Z junction. (a) Schematic of the D-shaped DNA sequences used for the experiments. The dsDNA portion has the same sequence as the linear dsDNA samples shown in Fig. 2a. The bending force on the dsDNA portion was controlled by using different lengths of the ssDNA string that consists only of thymine. (b) The E of the MM1 linear dsDNA sample (MM1 BZ linear) and MM1 D-shaped DNA samples (MM1 BZ29-S27 and MM1 BZB29-S19) plotted against the concentration of Mg(ClO4)2. The dotted lines denote the midpoint of each sample. Error bars were obtained from three independent measurements. (c) Comparison of the B–Z transition midpoint of the BZ linear and BZ D-shaped DNAs of normal and MM1 samples. The B–Z transition midpoints for the normal samples were obtained from the previous work for comparison.21 For MM1 samples, a weak bending force slightly decreases the B–Z transition midpoint, but increasing the bending force has a negligible effect on the B–Z transition. Error bars were obtained from three independent measurements. | ||
Proposed model of Z-DNA formation
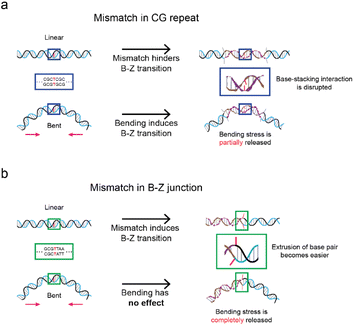
Fig. 5 summarizes the proposed mechanisms demonstrating how the mismatch position affects the B–Z transition both with and without bending force. When the mismatch is within the Z-DNA forming region, Z-DNA formation is hindered due to the disruptions in base-stacking energy (Fig. 5a). Under the bending force, Z-DNA formation is still induced, but to a lesser extent, as the local distortion by the mismatch in the CG repeat region partially releases the bending stress. Hence, we propose that the mismatch in the CG repeat region lacks the base-stacking interactions which mitigates the role of bending stress in inducing the formation of Z-DNA. Conversely, when the mismatch is in the B–Z junction (Fig. 5b), the local melting by the mismatch at the B–Z junction lowers the energy barrier of the base-pair extrusion, thereby assisting the B–Z transition. Under the bending force, the mismatch at the B–Z junction has nearly no effect in inducing Z-DNA formation, as the extrusion of the mismatched base pair fully releases the bending stress.Conclusions
In this work, we report a novel insight into the role of DNA mismatches and bending force in regulating the formation of Z-DNA. The mismatch in the CG repeat region hinders Z-DNA formation, while a mismatch in the B–Z junction facilitates the B–Z transition. Sequences capable of adopting Z-DNA conformations are abundant in the human genome, occurring approximately once every 3000 base pairs.47 Beyond the perfectly alternating purine–pyrimidine sequences, natural DNA also contains abundant cases of potential Z-DNA forming sequences that contain defects, such as mismatches, within the alternating sequence.22 Our work also revealed the combined effect of mismatch location and DNA bending on the B–Z transition. DNA bending is one of the most common types of mechanical deformations induced by DNA–protein interactions. Proteins that bind to Z-DNA, such as Z-DNA binding protein 1 (ZBP1) and adenosine deaminase acting on RNA (ADAR), stabilize the left-handed helical structure and may also contribute to DNA bending.48 Z-DNA-forming sequences are known to act as mutagenic hotspots in both yeast and human cells, leading to genetic instability through activation of the mismatch repair pathway. In this pathway, the MSH2–MSH3 complex binds to Z-DNA regions that form loop structures, which are often associated with DNA bending.13,49 Under bending force, a mismatch in the CG repeat partially releases the bending stress exerted on the dsDNA portion, which reduces the role of bending stress in promoting the formation of Z-DNA. The extrusion of the mismatched base pair at the B–Z junction effectively releases the strong bending force on the dsDNA, causing the bending to no longer facilitate Z-DNA formation. Our results provide a deeper understanding of the interplay between DNA mismatches and bending force in Z-DNA formation, shedding light on the conditions that induce the B–Z transition.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
SJ. P., J. Y., and N. K. L. designed the experiments. SJ. P. and J. Y. prepared the DNA samples, performed the experiments and analysed the results. SJ. P., J. Y., and N. K. L. wrote and revised the manuscript. N. K. L. supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Creative-Pioneering Researchers Program of Seoul National University and RS-2023-NR077154 and RS-2020-NR049542 of the National Research Foundation of Korea grant funded by the Korea government (MSIT).Notes and references
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893–5909 RSC.
- F. M. Pohl and T. M. Jovin, J. Mol. Biol., 1972, 67, 375–396 CrossRef CAS PubMed.
- A. H. J. Wang, G. J. Quigley, F. J. Kolpak, J. L. Crawford, J. H. van Boom, G. van der Marel and A. Rich, Nature, 1979, 282, 680–686 CrossRef CAS PubMed.
- A. Rich, Gene, 1993, 135, 99–109 CrossRef CAS PubMed.
- A. Rich and S. Zhang, Nat. Rev. Genet., 2003, 4, 566–572 CrossRef CAS PubMed.
- B. Wittig, S. Wölfl, T. Dorbic, W. Vahrson and A. Rich, EMBO J., 1992, 11, 4653–4663 CrossRef CAS PubMed.
- I. M. A. del Mundo, M. Zewail-Foote, S. M. Kerwin and K. M. Vasquez, Nucleic Acids Res., 2017, 45, 4929–4943 CrossRef CAS PubMed.
- M. M. Garner and G. Felsenfeld, J. Mol. Biol., 1987, 196, 581–590 CrossRef CAS PubMed.
- B. Wong, S. Chen, J.-A. Kwon and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2229 CrossRef CAS PubMed.
- N. Mulholland, Y. Xu, H. Sugiyama and K. Zhao, Cell Biosci., 2012, 2, 3 CrossRef CAS PubMed.
- G. Wang, L. A. Christensen and K. M. Vasquez, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 2677 CrossRef CAS PubMed.
- G. Wang and K. M. Vasquez, DNA Repair, 2014, 19, 143–151 CrossRef CAS PubMed.
- J. A. McKinney, G. Wang, A. Mukherjee, L. Christensen, S. H. S. Subramanian, J. Zhao and K. M. Vasquez, Nat. Commun., 2020, 11, 236 CrossRef CAS PubMed.
- T. J. Thamann, R. C. Lord, A. H. J. Wang and A. Rich, Nucleic Acids Res., 1981, 9, 5443–5458 CrossRef CAS PubMed.
- A. Herbert, J. Alfken, Y.-G. Kim, I. S. Mian, K. Nishikura and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 8421 CrossRef CAS PubMed.
- S. Bae, D. Kim, K. K. Kim, Y.-G. Kim and S. Hohng, J. Am. Chem. Soc., 2011, 133, 668–671 CrossRef CAS PubMed.
- A.-R. Lee, N.-H. Kim, Y.-J. Seo, S.-R. Choi and J.-H. Lee, Molecules, 2018, 23 Search PubMed.
- C. K. Singleton, J. Klysik, S. M. Stirdivant and R. D. Wells, Nature, 1982, 299, 312–316 CrossRef CAS PubMed.
- A. R. Rahmouni and D. Wells Robert, Science, 1989, 246, 358–363 CrossRef CAS PubMed.
- B. Wittig, T. Dorbic and A. Rich, J. Cell Biol., 1989, 108, 755–764 CrossRef CAS PubMed.
- J. Yi, S. Yeou and N. K. Lee, J. Am. Chem. Soc., 2022, 144, 13137–13145 CrossRef CAS PubMed.
- B. H. Johnston, G. J. Quigley, A. Rich and M. J. Ellison, Biochemistry, 1991, 30, 5257–5263 CrossRef CAS PubMed.
- M. de Rosa, D. de Sanctis, A. L. Rosario, M. Archer, A. Rich, A. Athanasiadis and M. A. Carrondo, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9088 CrossRef CAS PubMed.
- G. Rossetti, P. D. Dans, I. Gomez-Pinto, I. Ivani, C. Gonzalez and M. Orozco, Nucleic Acids Res., 2015, 43, 4309–4321 CrossRef CAS PubMed.
- X.-L. Yang and A. H. J. Wang, Biochemistry, 1997, 36, 4258–4267 CrossRef CAS PubMed.
- M. J. Ellison, R. J. Kelleher, A. H. Wang, J. F. Habener and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1985, 82, 8320–8324 CrossRef CAS PubMed.
- S. M. Mirkin, V. I. Lyamichev, V. P. Kumarev, V. F. Kobzev, V. V. Nosikov and A. V. Vologodskii, J. Biomol. Struct. Dyn., 1987, 5, 79–88 CrossRef CAS PubMed.
- P. J. Nichols, S. Bevers, M. Henen, J. S. Kieft, Q. Vicens and B. Vögeli, Nat. Commun., 2021, 12, 793 CrossRef CAS PubMed.
- T. Forster, Naturwissenschaften, 1946, 33, 166–175 CrossRef CAS.
- L. Stryer and R. P. Haugland, Proc. Natl. Acad. Sci. U. S. A., 1967, 58, 719–726 CrossRef CAS PubMed.
- M. Lee, S. H. Kim and S.-C. Hong, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4985 CrossRef CAS PubMed.
- S. Bae, H. Son, Y.-G. Kim and S. Hohng, Phys. Chem. Chem. Phys., 2013, 15, 15829–15832 RSC.
- A. N. Kapanidis, N. K. Lee, T. A. Laurence, S. Doose, E. Margeat and S. Weiss, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8936 Search PubMed.
- N. K. Lee, A. N. Kapanidis, Y. Wang, X. Michalet, J. Mukhopadhyay, R. H. Ebright and S. Weiss, Biophys. J., 2005, 88, 2939–2953 CrossRef CAS PubMed.
- B. Hellenkamp, S. Schmid, O. Doroshenko, O. Opanasyuk, R. Kühnemuth, S. Rezaei Adariani, B. Ambrose, M. Aznauryan, A. Barth, V. Birkedal, M. E. Bowen, H. Chen, T. Cordes, T. Eilert, C. Fijen, C. Gebhardt, M. Götz, G. Gouridis, E. Gratton, T. Ha, P. Hao, C. A. Hanke, A. Hartmann, J. Hendrix, L. L. Hildebrandt, V. Hirschfeld, J. Hohlbein, B. Hua, C. G. Hübner, E. Kallis, A. N. Kapanidis, J.-Y. Kim, G. Krainer, D. C. Lamb, N. K. Lee, E. A. Lemke, B. Levesque, M. Levitus, J. J. McCann, N. Naredi-Rainer, D. Nettels, T. Ngo, R. Qiu, N. C. Robb, C. Röcker, H. Sanabria, M. Schlierf, T. Schröder, B. Schuler, H. Seidel, L. Streit, J. Thurn, P. Tinnefeld, S. Tyagi, N. Vandenberk, A. M. Vera, K. R. Weninger, B. Wünsch, I. S. Yanez-Orozco, J. Michaelis, C. A. M. Seidel, T. D. Craggs and T. Hugel, Nat. Methods, 2018, 15, 669–676 CrossRef CAS PubMed.
- A. Brunet, C. Tardin, L. Salomé, P. Rousseau, N. Destainville and M. Manghi, Macromolecules, 2015, 48, 3641–3652 Search PubMed.
- S. Guilbaud, L. Salomé, N. Destainville, M. Manghi and C. Tardin, Phys. Rev. Lett., 2019, 122, 028102 Search PubMed.
- S. Cruz-León, W. Vanderlinden, P. Müller, T. Forster, G. Staudt, Y.-Y. Lin, J. Lipfert and N. Schwierz, Nucleic Acids Res., 2022, 50, 5726–5738 CrossRef.
- H. Son, S. Bae and S. Lee, Biochem. Biophys. Res. Commun., 2021, 583, 142–145 CrossRef CAS PubMed.
- L. J. Peck and J. C. Wang, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 6206 CrossRef CAS.
- S. C. Ha, K. Lowenhaupt, A. Rich, Y.-G. Kim and K. K. Kim, Nature, 2005, 437, 1183–1186 CrossRef CAS PubMed.
- N. Peyret, P. A. Seneviratne, H. T. Allawi and J. SantaLucia, Biochemistry, 1999, 38, 3468–3477 CrossRef CAS PubMed.
- H. Shroff, B. M. Reinhard, M. Siu, H. Agarwal, A. Spakowitz and J. Liphardt, Nano Lett., 2005, 5, 1509–1514 CrossRef CAS PubMed.
- C. Kim, O. C. Lee, J. Y. Kim, W. Sung and N. K. Lee, Angew Chem. Int. Ed. Engl., 2015, 54, 8943–8947 CrossRef CAS PubMed.
- O. c. Lee, C. Kim, J.-Y. Kim, N. K. Lee and W. Sung, Sci. Rep., 2016, 6, 28239 CrossRef CAS PubMed.
- S. Yeou and N. K. Lee, Mol. Cells, 2022, 45, 33–40 CrossRef CAS PubMed.
- G. P. Schroth, P. J. Chou and P. S. Ho, J. Biol. Chem., 1992, 267, 11846–11855 CrossRef CAS.
- R. Karki and T.-D. Kanneganti, Trends Immunol., 2023, 44, 201–216 CrossRef CAS PubMed.
- J.-M. Oh, Y. Kang, J. Park, Y. Sung, D. Kim, Y. Seo, E. A. Lee, J. S. Ra, E. Amarsanaa, Y.-U. Park, S. Y. Lee, J. M. Hwang, H. Kim, O. Schärer, S. W. Cho, C. Lee, K.-I. Takata, J. Y. Lee and K. Myung, Nucleic Acids Res., 2023, 51, 5584–5602 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc00749f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |