Open Access Article
Open Access ArticleInterrogation of mirror-image L-RNA–protein interactions reveals key mechanisms of single-stranded G-rich L-RNA cytotoxicity and a potential mitigation strategy†
Chen-Hsu
Yu
a,
Xiaomei
He
 bd,
Rosemarie Elloisa P.
Acero‡
a,
Xuan
Han‡
a,
Yinsheng
Wang
b and
Jonathan T.
Sczepanski
bd,
Rosemarie Elloisa P.
Acero‡
a,
Xuan
Han‡
a,
Yinsheng
Wang
b and
Jonathan T.
Sczepanski
 *ac
*ac
aDepartment of Chemistry, Texas A&M University, College Station, Texas 77843, USA. E-mail: jon.sczepanski@chem.tamu.edu
bDepartment of Chemistry, University of California Riverside, Riverside, California 92521-0403, USA
cDepartment of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, USA
dDepartment of Biology, East Carolina University, Greenville, North Carolina 27858, USA
First published on 20th March 2025
Abstract
L-Oligonucleotides (ONs), the synthetic enantiomers of native D-nucleic acids, are being increasingly utilized in the development of diverse biomedical technologies, including molecular imaging tools, diagnostic biosensors, and aptamer-based therapeutics. Nevertheless, our understanding of how L-ONs behave in living systems falls far short of native D-ONs. In particular, despite the potential for an abundant L-ON-protein interactome, the extent to which L-ONs bind to endogenous proteins and the consequences of these interactions are unknown, posing a major hurdle towards engineering functional L-ONs with predictable intracellular behaviours. Towards closing this knowledge gap, we now report the first L-ON–protein interactome, revealing that a wide-range of nuclear proteins have the potential to bind L-RNA. Importantly, by focusing our study on cytotoxic single-stranded G-rich L-RNA sequences, our data reveal key protein interactions that contribute to the cytotoxicity of these sequences. Furthermore, we show that introducing 2′-O-methyl modifications into single-stranded G-rich L-RNA can decrease its cytotoxicity through reducing L-RNA–protein interactions, thereby demonstrating that a well-established strategy for mitigating the cytotoxic effects of antisense ONs may translate across the chiral mirror. Overall, these findings greatly deepen our understanding of the intracellular behavior of L-ONs and provide valuable guidance for the future development of safe and effective L-ON-based biomedical technologies.
Introduction
Nucleic acids, as chiral molecules, have enantiomers known as L-nucleic acids (Fig. 1). L-Oligonucleotides (ONs) offer unique advantages compared to their native counterparts. One of the most notable features is their resistance to nucleases, providing remarkable biostability even under harsh biological conditions, including within living organisms.1–3 Additionally, L-ONs exhibit identical hybridization thermodynamics and kinetics to their native counterparts, allowing for easy adaptation without further optimization.4,5 Furthermore, L-ONs cannot form Watson–Crick (WC) base pairs with native nucleic acids, significantly reducing the potential for in vivo off-target effects.2,6,7 These properties have led to the increasing use of L-ONs in various biomedical technologies, including diagnostic biosensors,8,9 aptamers,10–14 live cell imaging probes,15–18 and drug delivery agents.19,20 Notably, L-aptamers (also referred to as Spiegelmers) have shown promise as drugs, with several examples currently undergoing clinical trials.21Despite the enormous promise of L-ONs in research and medicine, we are still just beginning to understand how L-ONs behave in living systems. In particular, while many biological interactions are stereospecific, a large number of proteins are capable of binding nucleic acids in a “promiscuous” manner that may be independent of chirality.22 Indeed, several nucleic acid-binding proteins have been shown to bind L-ONs, and in some cases, they bind equally well to both enantiomers of their targets.23–25 For example, we showed that polycomb repressive complex 2 (PRC2), a promiscuous RNA-binding protein (RBP) with important gene regulatory functions, can bind G-rich RNA sequences irrespective of chirality.23 It is therefore reasonable to predict that L-ON–protein interactions could be widespread in vivo, potentially leading to undesired effects.
Consistent with this notion, we recently investigated the impact of chirality on the behavior of ONs in living cells, revealing that L-ONs have the potential to be highly cytotoxic.26 Notably, we found that L-RNA sequences rich in guanines (e.g., L-r(GA)20) were substantially more cytotoxic than other sequence contexts tested. Furthermore, D-RNA, L-DNA, and D-DNA versions of the same guanine (G)-rich sequences showed mild or no cytotoxicity, suggesting that G-rich L-RNA sequences have unique cytotoxic potential. Importantly, the cytotoxicity of G-rich L-RNA sequences was dependent on their structure. Potent cytotoxicity was observed for single-stranded G-rich sequences regardless of their potential to form G-quadruplexes (G4s), indicating that the cytotoxic effects were mostly independent of G4s. However, a G-rich duplex structure wherein the majority of G residues were base paired was nontoxic. Thus, we concluded that the deleterious interactions require unpaired Gs. Although the majority of cytotoxic sequences tested were 30–40 nt in length and contained 40–50% G, single-stranded L-RNA sequences with as little as 25% G-content and as short as 10 nt (e.g., L-r(GA)5) exhibit some adverse cellular effects. We showed that the cytotoxic effects of single-stranded G-rich L-RNAs were accompanied by dramatic perturbations in gene expression levels and stimulation of an innate immune response. Because L-ONs are believed to be incapable of hybridizing with endogenous nucleic acids, these effects are most likely the results of yet to be revealed L-RNA–protein interactions.
If L-ONs are to be routinely employed in living systems, especially as drugs, then it is imperative that we characterize their protein interactomes. Indeed, efforts to define the protein interactome of D-ON-based drugs, such as antisense oligonucleotides (ASOs),27–31 have greatly benefited the clinical translation of these reagents. However, the extent to which L-ONs bind to endogenous proteins and the consequences of these interactions have not been carefully studied. This knowledge gap represents a major roadblock towards engineering functional L-oligonucleotides with predictable intracellular behaviours and the future development of safe and efficacious L-ON-based therapeutics, such as Spiegelmers. Towards closing this knowledge gap, we now report the first L-ON–protein interactome, revealing that a wide range of nuclear proteins have the potential to bind L-RNA, including Spiegelmers. Importantly, by focusing our study on cytotoxic single-stranded G-rich L-RNA, our data reveal key mechanisms of cytotoxicity for these sequences and insights into mitigating their deleterious effects. Overall, these findings provide valuable guidance for the future development of L-ON-based therapeutics and other biomedical technologies.
Results and discussion
A nuclear protein interactome for G-rich L-RNA
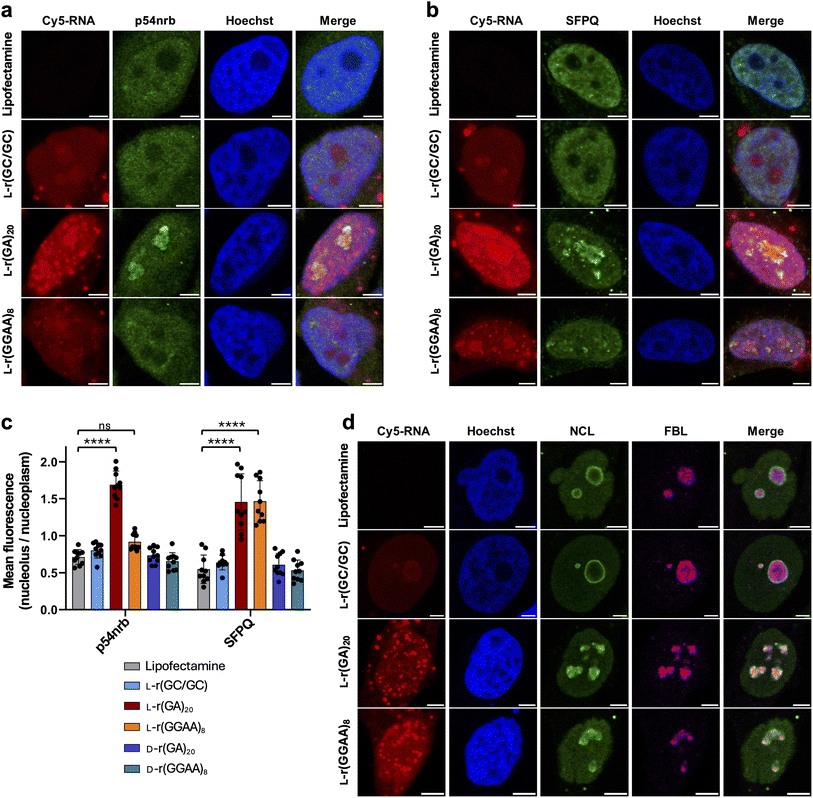
We previously reported that single-stranded G-rich L-RNAs are cytotoxic and immunogenic when transfected into human cells, effects that were attributed to protein binding.26 Thus, we chose to focus our initial L-ON–protein interaction studies on single-stranded G-rich L-RNA. Not only did we expect these sequences to have an abundant protein interactome, but we also envisioned that the identities of bound proteins would shed light on potential mechanisms of cytotoxicity, as well as strategies for mitigating these effects. To this end, we employed stable isotope labeling by amino acids in cell culture (SILAC)-based32 quantitative proteomics to identify the proteins capable of interacting selectively with cytotoxic single-stranded G-rich L-RNAs (Fig. 2a). We prepared 3′-biotin labelled L-r(GA)20 (EC50 = 28 nM (ref. 26)) as a representative cytotoxic G-rich sequence (Table S1†). We and others have previously shown that this sequence, regardless of its chirality, is essentially unstructured in solution.23,26,33 We also prepared L-r(GC/GC) as the nontoxic control bait (Fig. S1a†). L-r(GC/GC) contains the same fraction of G residues as L-r(GA)20, but it forms a tight double-stranded hairpin structure and shows no adverse cellular effects, which we attribute to fewer protein interactions.26 By comparing protein interactomes between cytotoxic L-r(GA)20 and the non-toxic control, we expected to identify proteins that bound selectively to the cytotoxic, single-stranded, G-rich sequence, which could point to potential mechanisms of cytotoxicity and provide further insights into the role of L-RNA structure. We obtained nuclear lysates from HeLa cells that had been cultured separately in a heavy or light SILAC medium. Nuclear lysates were used because cytotoxic L-RNAs were previously shown to localize strongly to the nucleus upon transfection.26 Equal amounts of heavy and light extracts were incubated with L-r(GA)20 or the L-r(GC/GC) control, respectively, followed by pull-down with streptavidin beads. Three biological replicates were carried out, as well as a fourth replicate done in the reverse orientation to minimize experimental bias associated with SILAC labeling. Proteins that remained bound to the beads following extensive washing were eluted, digested with trypsin, and subjected to LC-MS/MS analysis. The proteomics results showed a strong correlation in SILAC protein ratios between forward and reverse experiments, with a Pearson correlation coefficient of 0.8601 (Fig. S1b†). In addition, we showed previously that similar experimental conditions yielded a labeling efficiency of at least 99%.34 These results suggest a nearly complete heavy isotope labeling in our SILAC experiments and the reliability of our affinity pull-down procedures. We identified 86 nuclear proteins that were significantly enriched for cytotoxic L-r(GA)20 over the hairpin control, indicating that a wide range of nuclear proteins interact selectively with the cytotoxic, single-stranded sequence (Fig. 2b, S2–S8, and ESI† File 1). Gene ontology analysis for molecular function revealed nucleic acid binding and, specifically, RNA binding, among the top five enriched terms for proteins bound selectively to cytotoxic L-r(GA)20 (Fig. 2c). Moreover, biological process analysis revealed that proteins enriched by L-r(GA)20 were primarily involved in mRNA processing and splicing (Fig. 2d). Interestingly, a large portion of the proteins enriched by L-r(GA)20 (25 out of 86) contained one or more RNA recognition motifs (RRMs), suggesting that proteins containing this common RNA-binding domain may be prone to deleterious interactions with single-stranded G-rich L-RNA (Fig. 2e).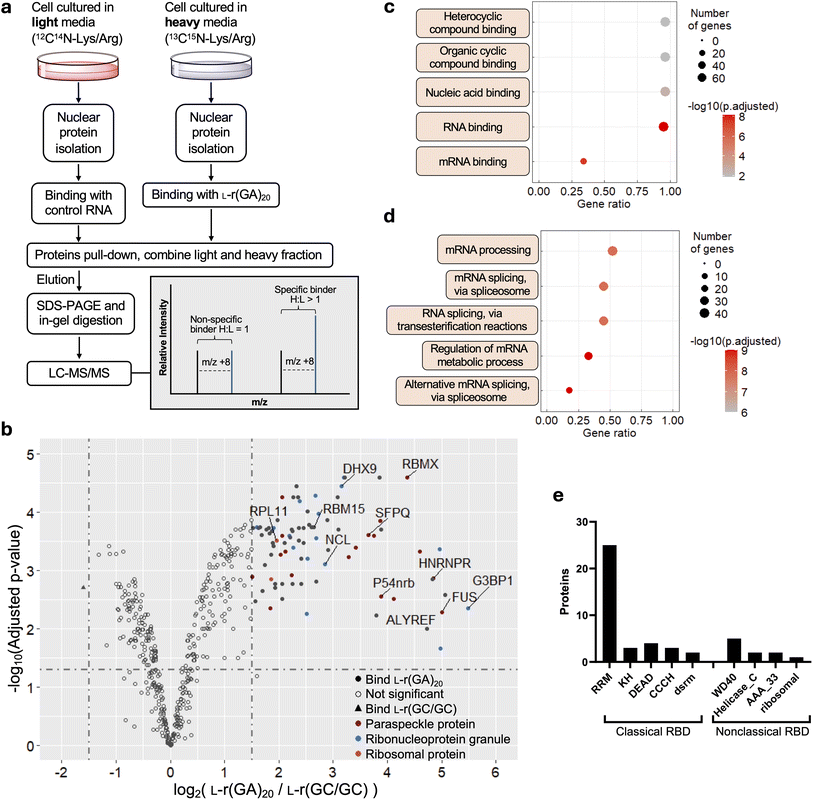 | ||
| Fig. 2 An L-RNA–protein interactome. (a) Schematic of the SILAC-based proteomics experiment employed in this study. (b) A volcano plot showing proteins enriched by L-r(GA)20 over the L-r(GC/GC) hairpin control (n = 4). Proteins with log2 fold change >1.5 are shown as solid circles. (c) and (d) Gene ontology (GO) analysis of 86 proteins enriched for L-r(GA)20 over L-r(GC/GC). The top five GO terms in molecular function (c) and biological process (d) are listed. (e) RNA-binding domain (RBD) analysis of proteins enriched for L-r(GA)20 over L-r(GC/GC). RBD analysis was conducted using NCBI batch-CD search and referenced to the Pfam database.38 RRM: RNA recognition motif; KH: K-homology; DEAD: DEAD/DEAH box helicase domain; CCCH: zinc finger, CCCH-type; dsrm: double-stranded RNA-binding motif; WD-40: WD40 domain; Helicase_C: helicase, C-terminal domain-like; AAA_33: AAA domain. | ||
A closer look at the proteins enriched by L-r(GA)20 revealed that 19 of the 86 (22%) are associated with nuclear paraspeckles (Fig. 2b), including the essential paraspeckle proteins p54nrb/NONO, splicing factor proline and glutamine rich (SFPQ/PSF), and fused in sarcoma (FUS).35,36 Enrichment of L-r(GA)20 by paraspeckle-associated proteins is consistent with the prior observation that this sequence localizes into discrete nuclear foci upon transfection into HeLa cells, whereas hairpin L-r(GC/GC) does not.26 Moreover, paraspeckle-associated RNAs often contain single-stranded GA-rich domains arranged in tandem,37 indicating that such sequences, regardless of chirality, may have an intrinsic affinity for paraspeckle proteins. Importantly, several studies have shown that off-target interactions of phosphorothioate antisense oligonucleotides (PS-ASOs) involve paraspeckle proteins, resulting in their localization to nuclear paraspeckles.29,31 In particular, the interaction of PS-ASOs with p54nrb, SFPQ and FUS has been associated with the cytotoxic effects of these reagents. Binding of L-r(GA)20 to these same proteins suggests a common mechanism of cytotoxicity, which we explore in detail below.
Overall, these results demonstrate that a wide range of nuclear proteins have the potential to interact with L-RNA despite their inverted sugar backbones and, for L-r(GA)20, reveal an interactome reminiscent of cytotoxic ASOs. To the best of our knowledge, this is the first reported protein interactome of an L-ON.
Validation of L-RNA–protein interactions in vitro
To directly characterize these newly identified L-RNA–protein interactions, we performed electrophoretic mobility shift assays (EMSAs) for several readily available proteins that were enriched by L-r(GA)20 in the SILAC experiment, including FUS, nucleolin (NCL) and heterogeneous nuclear ribonucleoprotein R (HNRNPR) (Fig. S9a†). Consistent with our proteomics data, all three proteins bound much more tightly to L-r(GA)20 (Kd < 10 nM) compared to the control L-r(GC/GC) hairpin (Fig. 3a, b and S9b†). In addition to L-r(GA)20, we also examined binding to another cytotoxic sequence L-r(GGAA)8.26 All three proteins bound tightly to L-r(GGAA)8 as well (Fig. 3a and b), suggesting that the protein interactome for L-r(GA)20 extends to other G(A)-rich sequences. Interestingly, the affinity of these proteins towards L-r(GA)20 and L-r(GGAA)8 was found to be similar to that of their native enantiomers, D-r(GA)20 and D-r(GGAA)8 (Fig. 3a and b), indicating that they have a promiscuous mode of binding with respect to chirality.39,40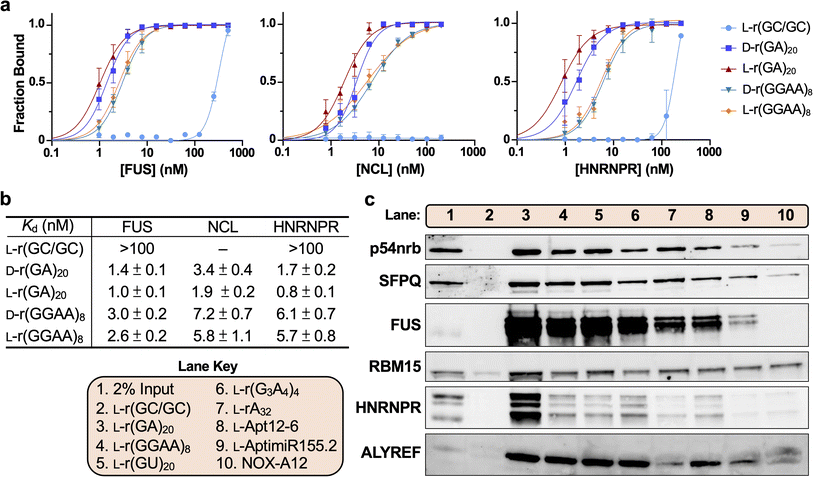 | ||
| Fig. 3 The diversity of L-RNA–protein interactions. (a) Saturation plot for FUS, NCL, and HNRNPR binding to different D- and L-RNAs. Data are mean ± S.D. (n = 3). Representative EMSA data are shown in Fig. S9b.† (b) Equilibrium dissociation constants (Kd) determined for FUS, NCL and HNRNPR binding to different RNAs. Data are mean ± S.D. (n = 3). (c) Representative Western blot analysis of the indicated protein following pull-down by different L-RNAs. The lane key is offset to the left. 2% of the input lysate was loaded as the control (lane 1). | ||
To further probe the generality of the interactions observed in the proteomics experiment, we carried out a more extensive survey of L-RNA-protein interactions via pull-down assays using biotinylated RNAs in combination with Western blotting. Nine L-RNA sequences were tested, including several additional G-rich L-RNAs, poly[A], and three published Spiegelmers, L-Apt12-6, AptamiR-155.2, and NOX-A12 which contain 59%, 41% and 33% G, respectively (Fig. 3c and Table S1†).10,13L-r(GGAA)8 and L-r(G3A4)4 were previously shown to form G-quadruplex (G4) structures in solution,26,33 whereas the Spiegelmers are predicated to adopt diverse secondary structures (Fig. S10†). Each L-RNA was separately incubated with HeLa cell nuclear lysates and bound proteins were captured by a biotin pull-down using streptavidin beads, followed by elution and immunoblotting for six proteins enriched by L-r(GA)20 in the proteomics experiment (Fig. 3c). As expected, all proteins tested were strongly precipitated by L-r(GA)20 and exhibited little to no interactions with the L-r(GC/GC) control, consistent with the results of our proteomics study. FUS bound strongly to all G-rich sequences tested (3–6), but less so to L-rA32 and the more strongly structured Spiegelmers (8–10). While transcriptome-wide binding maps for FUS indicate a preference for binding short G-rich motifs, in vitro binding assays show that FUS RNA-binding is dependent on the structure, including among different G4 RNAs.41–43 Thus, our results likely reflect this nuanced mode of RNA recognition. RBM15 and ALYREF bound all sequences tested to some extent (except the hairpin control (L-r(GC/GC))), consistent with their broad RNA-binding potential previously observed for D-RNA.43–45 HNRNPR also bound all sequences tested to some extent, but had a clear preference for L-r(GA)20. SFPQ and p54nrb are considered homologues, and thus, it is not surprising that they had similar interaction profiles with the L-RNA sequences tested herein (Fig. 3c). Both proteins bound all G-rich sequences (3–6), as well as L-rA32. However, weaker binding was observed for the L-RNA Spiegelmers (8–10), which was correlated with G content (i.e., greater G content led to stronger binding). Prior work has shown that SFPQ and p54nrb have a general G-rich RNA binding capacity, including G-quadruplexes, but bind weakly to double-stranded RNA.46–49 Given the inability of these proteins to bind the hairpin control and their comparably weaker interactions with the Spiegelmers, our results with L-RNA are mostly consistent with these prior observations.
Taken together, these results demonstrate that interactions between L-RNA and nuclear proteins are not limited to the sequences employed in our proteomics study, but extend to other G-rich sequences, as well as Spiegelmers having mixed sequences and diverse secondary structures. While G-rich L-RNAs appear capable of binding a wide range of nuclear proteins, these data also show that their precise protein interactomes will depend on their sequence and structure. Moreover, L-rA32 and L-Apt12-6 were reported to have mild or no cytotoxicity despite our finding that they bind tightly to several of the proteins identified by cytotoxic L-r(GA)20 (Fig. 3c).10,26 Thus, the cytotoxicity of single-stranded G-rich L-RNA relative to nontoxic sequences is likely dependent on their specific interaction profiles, including with proteins that were not examined discretely here. Interestingly, for most of the proteins tested, their preferences for binding L-RNA align with previous studies involving D-RNA. Indeed, we found that FUS, NCL, and HNRNPR bound the D-RNA and L-RNA versions of r(GA)20 and r(GGAA)8 with similar affinity when measured by EMSA (Fig. 3b). Considering that similar observations have been made for other proteins not identified herein (e.g., PRC2),23–25 the stereochemical promiscuity among RNA-binding proteins may be wide-spread, especially for those with RRMs. Knowledge of these “ambidextrous” proteins and their substrate preferences will undoubtably aid the design of future L-ON-based biomedical technologies.
Cytotoxic G-rich L-RNAs delocalize paraspeckle proteins to nucleoli
The studies above show that G-rich L-RNAs bind to essential paraspeckle proteins p54nrb and SFPQ. PS-ASOs have been shown to bind these same proteins, and in the case of cytotoxic sequences, these interactions cause them to delocalize into nucleoli, resulting in nucleolar stress and eventual apoptosis.29,31 Therefore, we asked whether cytotoxic G-rich L-RNAs cause a similar delocalization of p54nrb and SFPQ. HeLa cells were transfected with Cy5-labled L-r(GGAA)8 or L-r(GA)20 (200 nM) and protein localization was visualized by immunofluorescence two hours later. As observed previously, both L-r(GGAA)8 and L-r(GA)20 formed discrete nuclear foci and accumulated in the nucleolus (Fig. 4a and b). Importantly, compared to the lipofectamine only controls, cells treated with L-r(GA)20 showed strong nucleolar staining for both p54nrb and SFPQ, indicating that treatment with cytotoxic L-RNA resulted in delocalization of these proteins to the nucleolus (Fig. 4c and S11†). For SFPQ, this effect was concentration dependent, with increasing concentrations of L-r(GA)20 leading to greater accumulation of SFPQ in the nucleolus (Fig. S12†). Treatment of cells with L-r(GGAA)8 also resulted in significant nucleolar accumulation of SFPQ, but not p54nrb. This is inconsistent with the pull-down assay above (Fig. 3c), which shows that both proteins bind L-r(GGAA)8, suggesting that other factors (e.g., protein–protein interactions, cellular localization, etc.) contribute to the L-RNA interactome within the complex environment of the cell. Notably, p54nrb and SFPQ localization was unaffected by D-r(GA)20 and D-r(GGAA)8 (Fig. 4c and S13†), both of which are far less toxic to cells than their L-RNA versions.26 Thus, the cytotoxicity of L-r(GA)20 and L-r(GGAA)8 may be associated with their unique ability to delocalize these (and possibly other) paraspeckle proteins to the nucleolus. The nontoxic hairpin L-r(GC/GC), which localizes to the nucleolus but does not bind tightly to p54nrb and SFPQ in vitro, also had no effect on p54nrb and SFPQ localization (Fig. 4c), indicating that a direct interaction between these proteins and the L-RNA is necessary for nucleolar accumulation. Delocalization of p54nrb and SFPQ to nucleoli by L-r(GGAA)8 and/or L-r(GA)20 was not the result of apoptosis, as pre-treating cells with the apoptosis inhibitor Z-VAD-FMK29 did not prevent their nucleolar accumulation in the presence of the cytotoxic L-RNA (Fig. S14†). Nucleolar disruption by cytotoxic L-RNA was also observed. The subnucleolar localization of key nucleolar proteins fibrillarin (FBL) and NCL was highly altered in cells treated with L-r(GGAA)8 and L-r(GA)20 compared to the controls (Fig. 4d), indicating a loss of nucleolar structural integrity upon treatment with cytotoxic L-RNAs.50 Nucleolar reorganization and the redistribution of FBL are hallmarks of nucleolar stress.51 Taken together, these data show that cytotoxic G-rich L-RNA sequences, but not their D-RNA equivalents, can recruit paraspeckle proteins to the nucleolus and cause the loss of nucleolar integrity.Cytotoxic G-rich L-RNAs cause splicing defects
The subcellular mislocalization of proteins can result in a loss of function.52 Our results above show that p54nrb and SFPQ are delocalized to the nucleolus following treatment with cytotoxic L-RNAs. These proteins play an essential role in pre-mRNA processing,53,54 suggesting that splicing could be disrupted upon delocalization. Additionally, RNA splicing terms were highly enriched in the gene ontology analyses of our proteomics data (Fig. 2d), further indicating that splicing patterns may be affected by cytotoxic L-RNAs. We previously performed RNA-seq analysis of the total RNA isolated from HeLa cells 12 hours after treatment with either L-r(GA)20, L-r(GGAA)8, or L-r(GC/GC).26 Therefore, we reanalyzed these data by rMATs55 for altered splicing. Indeed, compared to the nontoxic sequence L-r(GC/GC), cells treated with the cytotoxic sequences L-r(GA)20 and L-r(GGAA)8 had a much higher number of altered splicing events, with skipped exon (SE) events occurring most frequently (Fig. 5a). The percent spliced-in (PSI) score was decreased for most SE events, indicating that L-r(GA)20 and L-r(GGAA)8 promote exon skipping. Sashimi plots generated for several SE events confirmed this observation (Fig. S15†). Among the 411 and 381 SE events induced by L-r(GA)20 and L-r(GGAA)8, respectively, approximately 40% (154 events) were shared between the two (Fig. 5b). While some differences between L-r(GA)20 and L-r(GGAA)8 were expected given their unique sequences and structures—L-r(GGAA)8 forms G4s, whereas L-r(GA)20 does not26—the high degree of common SE events suggests that these sequences share a large number of overlapping interactions, which may extend to other G(A)-rich L-RNA sequences. Gene ontology analysis of transcripts with common SE events revealed that the corresponding proteins were primarily involved in cell cycle regulation and checkpoints, the disruption of which could lead to apoptosis (Fig. S16†). For example, MDM2 exon 4, which shows a high level of SE events for both L-RNAs, codes for part of the p53 binding domain.56 The loss of the p53 binding to MDM2 activates p53, leading to cell cycle arrest and apoptosis.57 Nevertheless, whether splicing defects at individual genes contributes to the observed cytotoxicity of these L-RNAs requires further investigation.Exon skipping at three highly altered transcripts (MDM2 exon 4, EIF4A2 exon 4, and UTP15 exon 2) was further validated by RT-PCR using primers that span the up- and downstream exons. Consistent with the rMATs analysis, treatment of HeLa cells with L-r(GA)20 and L-r(GGAA)8 led to increased exon skipping at MDM2 exon 4 and UTP15 exon 2 (Fig. 5c and S17†). Increased exon skipping was not observed at these transcripts following treatment with arsenic or camptothecin, which induce cell stress and apoptosis, respectively. This suggests that exon skipping at MDM2 exon 4 and UTP15 exon 2 is the result of direct interference of the L-RNA with the spliceosome at these sites and is not due to a general stress response. In contrast, increased exon skipping at EIF4A2 exon 4 was observed under all conditions tested, including the lipofectamine only control, indicating that the observed alteration is due to a general response to stress.
In addition to direct interactions of G-rich L-RNAs with spliceosomal proteins, another possible explanation for exon skipping is binding of the L-RNA to the pre-mRNA at or near the splice site. While it is generally accepted that L-ONs of mixed sequences do not form stable duplexes with complementary D-ONs, earlier studies into heterochiral interaction between ONs of opposite stereochemistry revealed the formation of stable complexes between homopolymers of L-DNA/RNA (e.g., L-poly[A]) and their complementary natural nucleic acids.58–60 Given that our L-RNAs contained only G and A, we wondered whether the observed splicing defects could arise from their interaction with U-rich sequences within the pre-mRNA. To explore this possibility, we carried out a k-mer analysis (k = 5) of intron sequences 150 bps upstream and downstream of exons showing increased skipping in the presence of L-r(GA)20 and L-r(GGAA)8 (Fig. 5d). As a control, the same analysis was carried out on all known exon skipping events from the exon skipping database ExonSkipDB.61 While introns are typically AU-rich,62–64 we found that intron sequences adjacent to skipped exons affected by L-RNA treatment were significantly enriched in U compared to the control, with 5′-r(U)5 being the most enriched k-mer for both L-RNAs. Consistently, the sequence distribution among enriched k-mers for L-RNA-affected exons was less than that of the control (Fig. S18†). Together, this analysis supports the notion that direct interactions between the G/A-rich L-RNAs tested herein and U-rich sequences near the splice site contribute to the observed splicing defects.
2′-OMe modifications reduce G-rich L-RNA cytotoxicity
Previous work has shown that incorporation of 2′-modifications into cytotoxic PS-ASOs can reduce protein interactions, attenuate nucleolar delocalization of paraspeckle proteins (e.g., p54nrb), and mitigate cytotoxicity in vitro and in vivo.29 Given the similarities between cytotoxic L-RNAs and PS-ASOs, such as their propensity for binding nuclear paraspeckle proteins, we hypothesized that incorporation of 2′-OMe modifications into the L-ribose backbone of cytotoxic L-RNAs may similarly reduce protein interactions and mitigate cytotoxicity. To test this, we synthesized modified versions of the cytotoxic sequence L-r(GA)10 to contain either 50% (L-(mGrA)10) or 100% (L-m(GA)10) L-2′-OMe ribonucleotides (Fig. 6a). We first confirmed by CD spectroscopy that, like L-r(GA)20, L-r(GA)10 is unstructured in solution and that this is unaltered by methylation (Fig. S19†). We also showed that the 2′-OMe modification does not negatively impact cellular uptake (Fig. S20a†). We then determined the EC50 values for the modified L-RNA via the CCK-8 assay. Compared to the unmodified L-r(GA)10 control, EC50 values for L-(mGrA)10 and L-m(GA)10 increased 5-fold and 7.5-fold, respectively (Fig. 6a and S20b†), indicating that the 2′-OMe modifications reduced the cytotoxicity of the L-RNA. Moreover, EMSA of the modified L-RNAs with HeLa cell nuclear lysates showed that the 2′-OMe modifications reduced overall protein interactions (Fig. 6b, c and S21†), further supporting a correlation between protein-binding and toxicity. 2′-OMe modifications also reduced the nucleolar localization of L-r(GA)10 (Fig. 6d and e) and led to noticeably less nuclear foci formation (Fig. S20c†), consistent with the decreased interactions of the 2′-OMe modified L-RNA with paraspeckle proteins. Taken together, these results demonstrated that incorporation of 2′-OMe modifications into a cytotoxic L-RNA can decrease their cytotoxicity, possibly through reducing L-RNA–protein interactions, providing a promising solution for overcoming the off-target effects of these reagents.Conclusions
The routine use of L-ONs in basic research and in clinical settings will require an improved understanding of how they interact with biology and the potential consequences. Indeed, investigations into the mechanisms of action and off-target interactions of D-ONs, such as ASOs27–31 and aptamers,65,66 have greatly benefited the development and broader adoption of these reagents. Although Spiegelmer therapeutics (i.e., L-aptamers) have been the focus of several in vivo pharmacological studies,21 the majority of these reagents have been developed to target extracellular proteins. Consequently, little attention has been paid to the behavior of L-ONs inside cells. As the field of L-ONs continues to mature and therapeutic applications shift towards intracellular targets, it will become increasingly important to understand how L-ONs behave in this complex environment and, importantly, the extent to which they interact with proteins.Towards this goal, we have established the first L-ON–protein interactome, revealing that a broad range of nuclear proteins have the capacity to interact tightly with L-RNA. Our study focused on single-stranded G-rich L-RNAs due to their cytotoxic effects and prior evidence of protein interactions.23–25 In particular, by comparing protein enrichment by the cytotoxic, single-stranded sequence L-r(GA)20 to that of a non-toxic hairpin control, we identified 86 nuclear proteins that were significantly enriched for the cytotoxic sequence. These proteins are involved in a variety of important RNA-related processes, including ribosomal biogenesis, mRNA processing, and nuclear architecture. Interestingly, we found that a large portion of proteins selectively enriched by cytotoxic L-r(GA)20 contained RRMs, suggesting that proteins containing this common RNA-binding domain are prone to deleterious interactions with G-rich, single-stranded L-RNA. RRMs commonly interact with short stretches (2–8 nt) of single-stranded RNA through base-stacking and other hydrophobic interactions that have been shown to exhibit versatile molecular recognition activities.67,68 However, future experiments are required to determine whether RRMs are the structural domain responsible for binding L-RNA. It is important to note that both the toxic and nontoxic sequences used in this study are G-rich, with the key difference being their structures. While the cytotoxic sequence L-r(GA)20 is mostly unstructured, the nontoxic sequence L-r(GC/GC) forms a double-stranded hairpin structure such that the majority of G residues are base paired (Fig. S1a†).26 In general, hairpin L-r(GC/GC) was found to bind more weakly to the nuclear proteome, and especially to key paraspeckle proteins, compared to L-r(GA)20 and several other single-stranded G-rich L-RNAs tested (Fig. 3). These observations suggest that double-stranded structures can hinder deleterious protein interactions with G-rich L-RNA sequences, leading to reduced cytotoxicity, and further highlight the importance of structure in the behavior of G-rich L-RNAs in cells. While we acknowledge that the results of our proteomics experiment should not be generalized across all L-RNA sequences, many of the proteins enriched by L-r(GA)20 bound tightly to other single-stranded L-RNAs, including those with the potential to form G4 structures (e.g., L-r(GGAA)8), as well as Spiegelmers having mixed sequences and diverse secondary structures. It is therefore reasonable to predict that proteins enriched by L-r(GA)20 have the capacity to bind a large variety of L-RNA sequences, and potentially L-DNA. In the future, it will be important to expand our proteomics study to include a more diverse set of L-RNA sequences and structures, as well as L-DNA, in order to determine the generality of the G-rich L-RNA–protein interactions observed herein. Furthermore, acquiring the structure of an L-RNA-protein complex, especially with an RRM, could provide crucial information about chirality-independent binding modes, while also providing additional mitigation strategies.
A key goal of the study was to identify mechanisms underlying the cytotoxicity of single-stranded G-rich L-RNAs. Our results now point to nucleolar stress induced by L-RNA–protein interactions as a key source of cytotoxicity for these sequences. Nucleolar stress typically involves dysregulation of ribosome biogenesis, which involves the disruption of ribosomal RNA (rRNA) synthesis and processing. Indeed, a significant portion of the proteins identified in our proteomics study (10 out of 86) is involved in ribosomal biogenesis. One such protein is RPL11, which together with RPL5 and the 5S rRNA, forms the 5S ribonucleoprotein particle (RNP), an essential component of the large ribosomal subunit.69 Impairment of RPL11 abrogates ribosome biogenesis and protein synthesis, leading to nucleolar stress signaling.70,71 RPL11 is also a key contributor to nucleolar integrity, and knockdown of RPL11 leads to nucleolar morphological defects,72 a hallmark of nucleolar stress.51 Consistently, HeLa cells treated with cytotoxic L-RNA sequences showed extensive nucleolar reorganization, which was accompanied by the L-RNA-mediated delocalization of key paraspeckle proteins SFPQ and p54nrb to the nucleolus (Fig. 4). Very similar observations have been made for cytotoxic PS-ASOs, for which nucleolar stress is a major pathway of cytotoxicity.29,31 Given that cytotoxic PS-ASOs tend to be short (<20 nt), are diversely modified, and contain relatively few Gs (<25%),29 it is currently unclear what features (e.g., topology, electronic structure, and hydrophobicity) might be shared between cytotoxic G-rich L-RNA and PS-ASOs that give rise to their binding to similar proteins. Again, future structural studies aimed at characterizing L-RNA-protein interfaces will be critical for answering this question. Importantly, D-RNA versions of r(GGAA)8 and r(GA)20, which are far less toxic to cells compared to their L-RNA counterparts, did not affect p54nrb and SFPQ localization. This observation further points to the unique ability of G-rich L-RNAs to drive the delocalization of paraspeckle proteins to the nucleolus as a key mechanism of cytotoxicity. Furthermore, while several different L-RNAs were localized to the nucleolus upon transfection, only those that bound SFPQ and/or p54nrb in vitro resulted in their nucleolar accumulation and altered nucleolar morphology in cells, suggesting that these L-RNA–protein interactions contribute to nucleolar disruption. We acknowledge that nucleolar stress may not be the only cause of toxicity, and it is likely that no single mechanism of action can completely account for all pertinent observations. For example, we observed that cytotoxic G-rich L-RNAs caused significant splicing defects, including exon skipping within the key p53 regulator MDM2.57 Interestingly, our results suggest that the altered splicing could be due, in part, to direct L-RNA–pre-mRNA interactions near the splice site (Fig. 5d). Future pull-down experiments aimed at profiling the interactions of L-RNA with endogenous nucleic acids are required to confirm this mechanism. Other mechanisms for toxicity may involve protein dysfunction due to unfavorable binding. For example, p54nrb is a multi-functional RBP that is involved in transcription, splicing, RNA stability, and DNA repair, and loss of p54nrb has been shown to promote apoptosis.54,73
An important outcome of this work was demonstrating that modification of the 2′-position can decrease single-stranded G-rich L-RNA cytotoxicity through reduced protein interactions. To the best of our knowledge, this is the first time the behavior of 2′-OMe modified L-ONs has been explored inside live cells. This approach was inspired by the observation that 2′-modifications can reduce protein binding and attenuate nucleolar delocalization of p54nrb when incorporated into cytotoxic PS-ASOs, which behave similarly to cytotoxic G-rich L-RNAs in terms of protein interactions and nucleolar disruption.29,31 This highlights how a better understanding of the behavior of L-ONs in cells and the identification of underlying protein interactions can inform the development of safer L-ON reagents. Moreover, this work suggests that a well-established strategy for mitigating the cytotoxic effects of therapeutic D-ONs (i.e., 2′-modifications) can be translated across the chiral mirror.74,75 The ability to borrow proven design strategies from the D-ON therapeutics field is encouraging for the future development of therapeutic L-ONs. While we acknowledge that the reduction in toxicity for G-rich L-RNA was modest in response to 20 methyl groups, this is the first demonstrated approach for mitigating the cytotoxicity of L-ONs and represents an important starting point for the development of more effective strategies based on chemical modifications. Future studies using a more diverse set of 2′-modifications (e.g., 2′-O-methoxyethyl) and a broader range of cytotoxic L-RNA sequences are likely to reveal more effective chemistries and may guide the rational design of functional L-ONs with minimal cytotoxic effects. Indeed, it may not always be possible to avoid single-stranded G-rich sequences. Notably, the majority of published L-aptamers and Spiegelmer therapeutics contain high G content, several of which were shown herein to bind proteins.13,14,21,76–79 While selecting L-RNA aptamers using reduced G-content libraries is a potential solution, it may not be suitable for all applications. For example, nearly all L-RNA aptamers raised against D-nucleic acid targets contain G-rich motifs, including those that form G4 structures, suggesting that guanines play an important role in these “cross-chiral” interactions.77,78,80 Therefore, incorporating 2′-modifications, either during the in vitro selection process or in a site-specific manner post-selection, provides a potential strategy for mitigating the cytotoxic effects of G-rich motifs, which may prove valuable as the field of Spiegelmer therapeutics advances towards intracellular applications.
Overall, by revealing key L-RNA–protein interactions that contribute to L-RNA cytotoxicity, this work represents a major advance in our understanding of the intracellular behavior of L-ONs. These insights also provide a mechanism-based approach towards reducing L-RNA–protein interactions and the accompanying deleterious effects. These findings provide valuable guidance for the future development of L-ON-based therapeutics and other biomedical technologies.
Data availability
Raw proteomics data were submitted to ProteomeXchange under the accession number PXD054780. The complete list of enriched proteins can be accessed in ESI† File 1.Author contributions
Chen-Hsu Yu: methodology, investigation, formal analysis, visualization, writing – review & editing. Xiaomei He: methodology, investigation, formal analysis. Rosemarie Elloisa P. Acero: investigation, formal analysis, visualization. Xuan Han: investigation, formal analysis, visualization. Yinsheng Wang: formal analysis, visualization, writing – review & editing. Jonathan Sczepanski: conceptualization, writing – original draft, supervision, project administration, funding acquisition.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the National Institute of General Medical Sciences (R35GM124974 to J. T. S.) and the National Institute of Environmental Health Sciences (R35ES031707 to Y. W.) of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the Texas A&M University Microscopy and Imaging Center (RRID: SCR_022128) for usage of instrumentation. Portions of this research were conducted using the advanced computing resources provided by the Texas A&M University High Performance Research Computing group. We thank Dr Wen Zhang for the thoughtful discussions regarding the synthesis of 2′-OMe modified L-RNA.References
- H. Urata, E. Ogura, K. Shinohara, Y. Ueda and M. Akagi, Synthesis and properties of mirror-image DNA, Nucleic Acids Res., 1992, 20(13), 3325–3332 Search PubMed.
- N. C. Hauser, R. Martinez, A. Jacob, S. Rupp, J. D. Hoheisel and S. Matysiak, Utilising the left-helical conformation of L-DNA for analysing different marker types on a single universal microarray platform, Nucleic Acids Res., 2006, 34(18), 5101–5111 CrossRef CAS PubMed.
- W. Zhong and J. T. Sczepanski, Direct Comparison of D-DNA and L-DNA Strand-Displacement Reactions in Living Mammalian Cells, ACS Synth. Biol., 2021, 10(1), 209–212 Search PubMed.
- M. Szabat, D. Gudanis, W. Kotkowiak, Z. Gdaniec, R. Kierzek and A. Pasternak, Thermodynamic Features of Structural Motifs Formed by beta-L-RNA, PLoS One, 2016, 11(2), e0149478 CrossRef PubMed.
- M. J. Damha, P. A. Giannaris and P. Marfey, Antisense L/D-oligodeoxynucleotide chimeras: nuclease stability, base-pairing properties, and activity at directing ribonuclease H, Biochemistry, 1994, 33(25), 7877–7885 Search PubMed.
- K. Hoehlig, L. Bethge and S. Klussmann, Stereospecificity of oligonucleotide interactions revisited: no evidence for heterochiral hybridization and ribozyme/DNAzyme activity, PLoS One, 2015, 10(2), e0115328 CrossRef PubMed.
- A. Garbesi, M. L. Capobianco, F. P. Colonna, L. Tondelli, F. Arcamone, G. Manzini, C. W. Hilbers, J. M. Aelen and M. J. Blommers, L-DNAs as potential antimessenger oligonucleotides: a reassessment, Nucleic Acids Res., 1993, 21(18), 4159–4165 Search PubMed.
- N. Spurlock, W. E. Gabella, D. J. Nelson, D. T. Evans, M. E. Pask, J. E. Schmitz and F. R. Haselton, Implementing L-DNA analogs as mirrors of PCR reactant hybridization state: theoretical and practical guidelines for PCR cycle control, Anal. Methods, 2024, 16(18), 2840–2849 RSC.
- N. M. Adams, W. E. Gabella, A. N. Hardcastle and F. R. Haselton, Adaptive PCR Based on Hybridization Sensing of Mirror-Image L-DNA, Anal. Chem., 2017, 89(1), 728–735 CrossRef CAS PubMed.
- D. Ji, J. H. Yuan, S. B. Chen, J. H. Tan and C. K. Kwok, Selective targeting of parallel G-quadruplex structure using L-RNA aptamer, Nucleic Acids Res., 2023, 51(21), 11439–11452 CrossRef PubMed.
- A. M. Kabza and J. T. Sczepanski, An L-RNA Aptamer with Expanded Chemical Functionality that Inhibits MicroRNA Biogenesis, ChemBioChem, 2017, 18(18), 1824–1827 CrossRef CAS PubMed.
- S. Klussmann, A. Nolte, R. Bald, V. A. Erdmann and J. P. Fürste, Mirror-image RNA that binds D-adenosine, Nat. Biotechnol., 1996, 14, 1112–1115 Search PubMed.
- J. T. Sczepanski and G. F. Joyce, Specific Inhibition of MicroRNA Processing Using L-RNA Aptamers, J. Am. Chem. Soc., 2015, 137(51), 16032–16037 Search PubMed.
- J. T. Sczepanski and G. F. Joyce, Binding of a Structured D-RNA by an L-RNA Aptamer, J. Am. Chem. Soc., 2013, 135(36), 13290–13293 Search PubMed.
- W. Zhong and J. T. Sczepanski, Chimeric D/L-DNA Probes of Base Excision Repair Enable Real-Time Monitoring of Thymine DNA Glycosylase Activity in Live Cells, J. Am. Chem. Soc., 2023, 145(31), 17066–17074 CrossRef CAS PubMed.
- W. Zhong and J. T. Sczepanski, A Mirror Image Fluorogenic Aptamer Sensor for Live-Cell Imaging of MicroRNAs, ACS Sens., 2019, 4(3), 566–570 CrossRef CAS PubMed.
- L. Cui, R. Peng, T. Fu, X. Zhang, C. Wu, H. Chen, H. Liang, C. J. Yang and W. Tan, Biostable L-DNAzyme for sensing of metal ions in biological systems, Anal. Chem., 2016, 88(3), 1850–1855 CrossRef CAS PubMed.
- G. Ke, C. Wang, Y. Ge, N. Zheng, Z. Zhu and C. J. Yang, L-DNA Molecular Beacon: A Safe, Stable, and Accurate Intracellular Nano-thermometer for Temperature Sensing in Living Cells, J. Am. Chem. Soc., 2012, 134(46), 18908–18911 CrossRef CAS PubMed.
- K. R. Kim, H. Y. Kim, Y. D. Lee, J. S. Ha, J. H. Kang, H. Jeong, D. Bang, Y. T. Ko, S. Kim, H. Lee and D. R. Ahn, Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs, J. Controlled Release, 2016, 243, 121–131 CrossRef CAS PubMed.
- K. R. Kim, D. Hwang, J. Kim, C. Y. Lee, W. Lee, D. S. Yoon, D. Shin, S. J. Min, I. C. Kwon, H. S. Chung and D. R. Ahn, Streptavidin-mirror DNA tetrahedron hybrid as a platform for intracellular and tumor delivery of enzymes, J. Controlled Release, 2018, 280, 1–10 Search PubMed.
- A. Vater and S. Klussmann, Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics, Drug Discovery Today, 2015, 20(1), 147–155 Search PubMed.
- M. Corley, M. C. Burns and G. W. Yeo, How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms, Mol. Cell, 2020, 78(1), 9–29 Search PubMed.
- C. E. Deckard and J. T. Sczepanski, Polycomb repressive complex 2 binds RNA irrespective of stereochemistry, Chem. Commun., 2018, 54(85), 12061–12064 Search PubMed.
- L. A. McGregor, B. Zhu, A. M. Goetz and J. T. Sczepanski, Thymine DNA glycosylase is an RNA-binding protein with high selectivity for G-rich sequences, J. Biol. Chem., 2023, 299(4), 104590 CAS.
- J. C. Lai, B. D. Brown, A. M. Voskresenskiy, S. Vonhoff, S. Klussman, W. Tan, M. Colombini, R. Weeratna, P. Miller, L. Benimetskaya and C. A. Stein, Comparison of D-G3139 and Its Enantiomer L-G3139 in Melanoma Cells Demonstrates Minimal In Vitro but Dramatic In Vivo Chiral Dependency, Mol. Ther., 2007, 15(2), 270–278 CAS.
- C.-H. Yu and J. T. Sczepanski, The influence of chirality on the behavior of oligonucleotides inside cells: revealing the potent cytotoxicity of G-rich L-RNA, Chem. Sci., 2023, 14(5), 1145–1154 Search PubMed.
- F. Wang, E. Calvo-Roitberg, J. M. Rembetsy-Brown, M. Fang, J. Sousa, Z. J. Kartje, P. M. Krishnamurthy, J. Lee, M. R. Green, A. A. Pai and J. K. Watts, G-rich motifs within phosphorothioate-based antisense oligonucleotides (ASOs) drive activation of FXN expression through indirect effects, Nucleic Acids Res., 2022, 50(22), 12657–12673 Search PubMed.
- A. J. Pollak, L. Zhao and S. T. Crooke, Systematic Analysis of Chemical Modifications of Phosphorothioate Antisense Oligonucleotides that Modulate Their Innate Immune Response, Nucleic Acid Ther., 2023, 33(2), 95–107 CAS.
- W. Shen, C. L. De Hoyos, M. T. Migawa, T. A. Vickers, H. Sun, A. Low, T. A. Bell, M. Rahdar, S. Mukhopadhyay, C. E. Hart, M. Bell, S. Riney, S. F. Murray, S. Greenlee, R. M. Crooke, X.-h. Liang, P. P. Seth and S. T. Crooke, Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index, Nat. Biotechnol., 2019, 37(6), 640–650 Search PubMed.
- S. T. Crooke, S. Wang, T. A. Vickers, W. Shen and X.-H. Liang, Cellular uptake and trafficking of antisense oligonucleotides, Nat. Biotechnol., 2017, 35(3), 230–237 CAS.
- S. T. Crooke, T. A. Vickers and X.-H. Liang, Phosphorothioate modified oligonucleotide–protein interactions, Nucleic Acids Res., 2020, 48(10), 5235–5253 Search PubMed.
- X. Chen, S. Wei, Y. Ji, X. Guo and F. Yang, Quantitative proteomics using SILAC: principles, applications, and developments, Proteomics, 2015, 15(18), 3175–3192 Search PubMed.
- X. Wang, K. J. Goodrich, A. R. Gooding, H. Naeem, S. Archer, R. D. Paucek, D. T. Youmans, T. R. Cech and C. Davidovich, Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines, Mol. Cell, 2017, 65(6), 1056–1067 CAS.
- W. Miao, L. Li, Y. Zhao, X. Dai, X. Chen and Y. Wang, HSP90 inhibitors stimulate DNAJB4 protein expression through a mechanism involving N6-methyladenosine, Nat. Commun., 2019, 10(1), 3613 Search PubMed.
- F. McCluggage and A. H. Fox, Paraspeckle nuclear condensates: global sensors of cell stress?, BioEssays, 2021, 43(5), 2000245 Search PubMed.
- A. H. Fox, S. Nakagawa, T. Hirose and C. S. Bond, Paraspeckles: Where Long Noncoding RNA Meets Phase Separation, Trends Biochem. Sci., 2018, 43(2), 124–135 CAS.
- J. A. West, M. Mito, S. Kurosaka, T. Takumi, C. Tanegashima, T. Chujo, K. Yanaka, R. E. Kingston, T. Hirose, C. Bond, A. Fox and S. Nakagawa, Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization, J. Cell Biol., 2016, 214(7), 817–830 CAS.
- J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G. A. Salazar, E. L. L. Sonnhammer, S. C. E. Tosatto, L. Paladin, S. Raj, L. J. Richardson, R. D. Finn and A. Bateman, Pfam: the protein families database in 2021, Nucleic Acids Res., 2021, 49(D1), D412–D419 CAS.
- L. Ghisolfi-Nieto, G. Joseph, F. Puvion-Dutilleul, F. Amalric and P. Bouvet, Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA, J. Mol. Biol., 1996, 260(1), 34–53 CAS.
- X. Wang, J. C. Schwartz and T. R. Cech, Nucleic acid-binding specificity of human FUS protein, Nucleic Acids Res., 2015, 43(15), 7535–7543 Search PubMed.
- D. Dominguez, P. Freese, M. S. Alexis, A. Su, M. Hochman, T. Palden, C. Bazile, N. J. Lambert, E. L. Van Nostrand, G. A. Pratt, G. W. Yeo, B. R. Graveley and C. B. Burge, Sequence, Structure, and Context Preferences of Human RNA Binding Proteins, Mol. Cell, 2018, 70(5), 854–867 Search PubMed.
- A. Ishiguro, A. Katayama and A. Ishihama, Different recognition modes of G-quadruplex RNA between two ALS/FTLD-linked proteins TDP-43 and FUS, FEBS Lett., 2021, 595(3), 310–323 Search PubMed.
- E. L. Van Nostrand, P. Freese, G. A. Pratt, X. Wang, X. Wei, R. Xiao, S. M. Blue, J.-Y. Chen, N. A. L. Cody, D. Dominguez, S. Olson, B. Sundararaman, L. Zhan, C. Bazile, L. P. B. Bouvrette, J. Bergalet, M. O. Duff, K. E. Garcia, C. Gelboin-Burkhart, M. Hochman, N. J. Lambert, H. Li, M. P. McGurk, T. B. Nguyen, T. Palden, I. Rabano, S. Sathe, R. Stanton, A. Su, R. Wang, B. A. Yee, B. Zhou, A. L. Louie, S. Aigner, X.-D. Fu, E. Lécuyer, C. B. Burge, B. R. Graveley and G. W. Yeo, A large-scale binding and functional map of human RNA-binding proteins, Nature, 2020, 583(7818), 711–719 CrossRef CAS PubMed.
- N. Viphakone, I. Sudbery, L. Griffith, C. G. Heath, D. Sims and S. A. Wilson, Co-transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome, Mol. Cell, 2019, 75(2), 310–323 Search PubMed.
- M. Shi, H. Zhang, X. Wu, Z. He, L. Wang, S. Yin, B. Tian, G. Li and H. Cheng, ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo, Nucleic Acids Res., 2017, 45(16), 9640–9653 CrossRef CAS PubMed.
- C. Bruelle, M. Bédard, S. Blier, M. Gauthier, A. M. Traish and M. Vincent, The mitotic phosphorylation of p54nrb modulates its RNA binding activity, Biochem. Cell Biol., 2011, 89(4), 423–433 CAS.
- M. Hallier, A. Tavitian and F. Moreau-Gachelin, The Transcription Factor Spi-1/PU.1 Binds RNA and Interferes with the RNA-binding Protein p54nrb, J. Biol. Chem., 1996, 271(19), 11177–11181 CAS.
- A. Basu, B. Dong, A. R. Krainer and C. C. Howe, The Intracisternal A-Particle Proximal Enhancer-Binding Protein Activates Transcription and Is Identical to the RNA- and DNA-Binding Protein p54nrb/NONO, Mol. Cell Biol., 1997, 17(2), 677–686 CAS.
- E. A. J. Simko, H. Liu, T. Zhang, A. Velasquez, S. Teli, A. R. Haeusler and J. Wang, G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA, Nucleic Acids Res., 2020, 48(13), 7421–7438 CAS.
- D. L. J. Lafontaine, J. A. Riback, R. Bascetin and C. P. Brangwynne, The nucleolus as a multiphase liquid condensate, Nat. Rev. Mol. Cell Biol., 2021, 22(3), 165–182 CAS.
- K. Yang, J. Yang and J. Yi, Nucleolar Stress: hallmarks, sensing mechanism and diseases, Cell Stress, 2018, 2(6), 125–140 Search PubMed.
- X. Wang and S. Li, Protein mislocalization: mechanisms, functions and clinical applications in cancer, Biochim. Biophys. Acta., 2014, 1846(1), 13–25 Search PubMed.
- D. Yu, C. J. Huang and H. O. Tucker, Established and Evolving Roles of the Multifunctional Non-POU Domain-Containing Octamer-Binding Protein (NonO) and Splicing Factor Proline- and Glutamine-Rich (SFPQ), J. Dev. Biol., 2024, 12(1), 3 CAS.
- P. Feng, L. Li, T. Deng, Y. Liu, N. Ling, S. Qiu, L. Zhang, B. Peng, W. Xiong, L. Cao, L. Zhang and M. Ye, NONO and tumorigenesis: more than splicing, J. Cell Mol. Med., 2020, 24(8), 4368–4376 Search PubMed.
- S. Shen, J. W. Park, Z.-x. Lu, L. Lin, M. D. Henry, Y. N. Wu, Q. Zhou and Y. Xing, rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(51), E5593–E5601 Search PubMed.
- T. Iwakuma and G. Lozano, MDM2, an introduction, Mol. Cancer Res., 2003, 1(14), 993–1000 CAS.
- S. de Rozieres, R. Maya, M. Oren and G. Lozano, The loss of mdm2 induces p53 mediated apoptosis, Oncogene, 2000, 19(13), 1691–1697 CAS.
- G. W. Ashley, Modeling, synthesis, and hybridization properties of (L)-ribonucleic acid, J. Am. Chem. Soc., 1992, 114(25), 9732–9736 Search PubMed.
- Y. Hashimoto, N. Iwanami, S. Fujimori and K. Shudo, Enantio- and meso-DNAs: preparation, characterization, and interaction with complementary nucleic acids, J. Am. Chem. Soc., 1993, 115(22), 9883–9887 CAS.
- S. Fujimori, K. Shudo and Y. Hashimoto, Enantio-DNA recognizes complementary RNA but not complementary DNA, J. Am. Chem. Soc., 1990, 112(20), 7436–7438 Search PubMed.
- P. Kim, M. Yang, K. Yiya, W. Zhao and X. Zhou, ExonSkipDB: functional annotation of exon skipping event in human, Nucleic Acids Res., 2020, 48(D1), D896–D907 CAS.
- M. Amit, M. Donyo, D. Hollander, A. Goren, E. Kim, S. Gelfman, G. Lev-Maor, D. Burstein, S. Schwartz, B. Postolsky, T. Pupko and G. Ast, Differential GC content between exons and introns establishes distinct strategies of splice-site recognition, Cell Rep., 2012, 1(5), 543–556 CrossRef CAS PubMed.
- T. Bakheet, E. Hitti, M. Al-Saif, W. N. Moghrabi and K. S. A. Khabar, The AU-rich element landscape across human transcriptome reveals a large proportion in introns and regulation by ELAVL1/HuR, Biochim. Biophys. Acta, Gene Regul. Mech., 2018, 1861(2), 167–177 CrossRef CAS PubMed.
- E. Miriami, H. Margalit and R. Sperling, Conserved sequence elements associated with exon skipping, Nucleic Acids Res., 2003, 31(7), 1974–1983 Search PubMed.
- L. Kelly, K. E. Maier, A. Yan and M. Levy, A comparative analysis of cell surface targeting aptamers, Nat. Commun., 2021, 12(1), 6275 CrossRef CAS PubMed.
- S. M. Nimjee, R. R. White, R. C. Becker and B. A. Sullenger, Aptamers as Therapeutics, Annu. Rev. Pharmacol. Toxicol., 2017, 57, 61–79 CrossRef CAS PubMed.
- C. Maris, C. Dominguez and F. H. T. Allain, The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression, FEBS J., 2005, 272(9), 2118–2131 CrossRef CAS PubMed.
- Y. Muto and S. Yokoyama, Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems, Wiley Interdiscip. Rev.:RNA, 2012, 3(2), 229–246 CrossRef CAS PubMed.
- A. Chakraborty, T. Uechi and N. Kenmochi, Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway, Wiley Interdiscip. Rev.:RNA, 2011, 2(4), 507–522 Search PubMed.
- J. Kang, N. Brajanovski, K. T. Chan, J. Xuan, R. B. Pearson and E. Sanij, Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy, Signal Transduction Targeted Ther., 2021, 6(1), 323 CrossRef CAS PubMed.
- L. Golomb, S. Volarevic and M. Oren, p53 and ribosome biogenesis stress: the essentials, FEBS Lett., 2014, 588(16), 2571–2579 Search PubMed.
- E. Nicolas, P. Parisot, C. Pinto-Monteiro, R. de Walque, C. De Vleeschouwer and D. L. J. Lafontaine, Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress, Nat. Commun., 2016, 7(1), 11390 Search PubMed.
- Y. Shav-Tal and D. Zipori, PSF and p54nrb/NonO – multi-functional nuclear proteins, FEBS Lett., 2002, 531(2), 109–114 Search PubMed.
- T. C. Roberts, R. Langer and M. J. A. Wood, Advances in oligonucleotide drug delivery, Nat. Rev. Drug Discovery, 2020, 19(10), 673–694 Search PubMed.
- M. Egli and M. Manoharan, Chemistry, structure and function of approved oligonucleotide therapeutics, Nucleic Acids Res., 2023, 51(6), 2529–2573 CAS.
- J. Li and J. T. Sczepanski, Targeting a Conserved Structural Element from the SARS-CoV-2 Genome using L-DNA Aptamers, RSC Chem. Biol., 2021, 3(1), 79–84 Search PubMed.
- S. Dey and J. T. Sczepanski, In vitro selection of L-DNA aptamers that bind a structured D-RNA molecule, Nucleic Acids Res., 2020, 48(4), 1669–1680 CAS.
- M. I. Umar and C. K. Kwok, Specific suppression of D-RNA G-quadruplex–protein interaction with an L-RNA aptamer, Nucleic Acids Res., 2020, 48(18), 10125–10141 CAS.
- C.-Y. Chan and C. K. Kwok, Specific Binding of a D-RNA G-Quadruplex Structure with an L-RNA Aptamer, Angew. Chem., Int. Ed., 2020, 59(13), 5293–5297 CAS.
- H. L. Lau, H. Zhao, H. Feng and C. K. Kwok, Specific Targeting and Imaging of RNA G-Quadruplex (rG4) Structure Using Non-G4-Containing L-RNA Aptamer and Fluorogenic L-Aptamer, Small Methods, 2024, 2401097, DOI:10.1002/smtd.202401097.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and methods; experimental details; supplementary figures; oligonucleotide mass spectrometry data; DNA sequences; protein interactome data. See DOI: https://doi.org/10.1039/d5sc00596e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |