 Open Access Article
Open Access ArticleHighly selective DNA aptamer sensor for intracellular detection of coenzyme A†
Yuan
Ma‡
abc,
Whitney
Lewis‡
 ab,
Peng
Yan
bd,
Xiangli
Shao
ab,
Quanbing
Mou
ab,
Peng
Yan
bd,
Xiangli
Shao
ab,
Quanbing
Mou
 abc,
Linggen
Kong
abc,
Linggen
Kong
 efg,
Weijie
Guo
efh and
Yi
Lu
efg,
Weijie
Guo
efh and
Yi
Lu
 *abefg
*abefg
aDepartment of Chemistry, The University of Texas at Austin, Austin, Texas 78712, USA
bDepartment of Chemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA
cDepartment of Chemistry, Rice University, Houston, TX 77005, USA
dSchool of Health and Life Sciences, University of Health and Rehabilitation Sciences, Qingdao, Shandong 266113, P. R. China
eDepartment of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, USA
fInterdisciplinary Life Sciences Graduate Program, The University of Texas at Austin, Austin, Texas 78712, USA
gCenter for Biophysics and Quantitative Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA
hDepartment of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA
First published on 28th March 2025
Abstract
Detecting Coenzyme A (CoA) in cells is vital for understanding its role in metabolism. DNA aptamers, though widely used for monitoring many other molecules, have not been effective for CoA detection, as previous attempts at obtaining DNA aptamers for CoA using SELEX resulted in aptamers that only recognize the adenine moiety of CoA. This “tyranny” of adenine dominating in SELEX has, therefore, hampered the SELEX of aptamers specific for CoA. To meet this challenge, we employed a capture SELEX method by incorporating rigorous counter selections against adenine, adenosine, ATP, pantetheine, and pantothenic acid, resulting in a highly specific DNA aptamer for CoA over adenosine, ATP and other related metabolites such as NADH, with a dissociation constant of 48.9 μM. This aptamer was then converted to a fluorescent sensor for CoA across pH 6.4–8.0. Confocal microscopy showed its ability to visualize CoA in living cells, with fluorescence changes observed upon manipulating CoA levels. This method broadens SELEX's application and presents a promising approach for studying and understanding CoA dynamics.
Introduction
Aptamers, single-stranded DNA (ssDNA) or RNA molecules can fold into specific three-dimensional structures to selectively bind their target molecules.1,2 Aptamers are smaller and more cost-effective than protein-based sensors, and their target binding can readily be translated into a DNA/RNA hybridization melting temperature change that is quite predictable and often results in a large signal change when they are labeled with a Förster resonance energy transfer (FRET) pair. Because of these advantages, aptamers have been increasingly applied in sensing and imaging in biological systems.3–22Another major advantage of the aptamers is that they can be selected to be highly specific to a wide range of targets using Systematic Evolution of Ligands by Exponential Enrichment (SELEX).23–32 In fact, the SELEX is so powerful, it is considered to be generally applicable to almost any target. However, despite the perception, applying SELEX to obtain aptamers for some targets remains a significant challenge. One example of challenging metabolites is coenzyme A (CoA), a molecule with a 3'-phosphoadenosine diphosphate structure that closely resembles adenine-containing metabolites like ATP (Fig. 1). This “tyranny” of adenine dominating in SELEX has, therefore, hampered the SELEX of aptamers specific for CoA, as previous attempts yielded only aptamers that bind the adenine moiety of CoA, resulting in poor selectivity over adenosine, ATP, and other adenosine-containing metabolites such as NADH.33–35 As a result, despite decades of research and advances in aptamer technology, there is no aptamer ideal for detecting CoA due to selectivity challenges.33–35
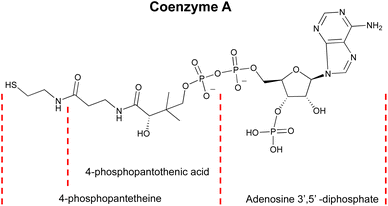 | ||
| Fig. 1 Chemical structure of Coenzyme A (CoA), showing key structural motifs including adenosine 3′,5′-diphosphate, 4-phosphopantetheine, and 4-phosphopantothenic acid. | ||
CoA is a vital metabolite central to acyl group transfer and enzymatic regulation.36,37 It functions both as a substrate and allosteric modulator in critical pathways, including post-translational modifications like acetylation of histones and other proteins, which influence gene expression and cellular function.38,39 Maintaining CoA homeostasis is essential for cellular health, as disruptions are linked to conditions including Pantothenate Kinase-Associated Neurodegeneration (PKAN),40 CoA Synthase Protein-Associated Neurodegeneration (CoPAN),41 cataracts, cardiomyopathy, and other CoA synthesis pathway diseases.37 Real-time monitoring of CoA in living cells is therefore essential to understanding its metabolic roles and disease implications.
Real-time detection of CoA in living cells remains challenging due to its structural complexity and similarity to other biologically relevant small molecules.37,42,43 Traditional methods, such as high-performance liquid chromatography (HPLC) and mass spectrometry, while sensitive, are unsuitable for real-time and spatially resolved measurements in living cells.44–46 Semisynthetic biosensors have been developed to couple a green fluorescent protein-HaloTag fusion protein with a CoA-dependent fluorescent ligand to produce Förster resonance energy transfer (FRET) signals.47 Despite progress, it is challenging to convert CoA binding into the FRET signals, as this conversion requires careful system design to translate the binding of the small CoA molecule into conformation changes in a relatively larger protein.48 As a result, the FRET signals are often weak, making CoA detection difficult in the complex cellular environment.49 Thus, a rapid, simple, sensitive, specific, and cost-effective method for monitoring CoA levels in cells is still needed.
In this study, we address the issue of selectivity in CoA detection via aptamers by utilizing a capture SELEX method specifically designed to isolate DNA aptamers that are highly selective for CoA. We incorporate rigorous counter-selections against a variety of adenine-containing molecules, allowing us to overcome previous limitations. The resulting aptamer was then converted into a fluorescent sensor for the real-time detection and visualization of intracellular CoA levels. This approach provides not only a sensitive, specific, and cost-effective tool for studying CoA metabolism, but also represents a significant advancement in the selection and development of aptamer-based detection technologies.
Results and discussion
SELEX of DNA aptamers for CoA against adenine or other molecules containing the adenine moiety
Previous SELEX efforts to isolate CoA-binding aptamers resulted in those that primarily bind the adenine portion of CoA. As a result, the isolated aptamers can also bind ATP.33,34 In these SELEX processes, CoA was attached to a solid support at a different attachment point, and a DNA library was then added to the solid support containing the CoA to isolate sequences that bind CoA. We hypothesize that the attachment of CoA to the solid support can significantly impact the selection outcome, often imposing steric constraints that eliminate desirable binding structures. To address this issue, we employed a capture SELEX method that leverages the aptamer's inherent ability to transition between two distinct conformations: a duplex formation with an antisense DNA strand and a complex structure binding its specific target, CoA.50–52 This SELEX method was achieved by immobilizing the structural-switching DNA library onto a solid support, followed by introducing CoA that remains free in its native state in solution. This library-immobilized SELEX strategy ensured that all the epitopes of CoA were exposed to the DNA library and could participate in aptamer binding. The DNA molecules that bound to CoA were subsequently released from the support. This approach aimed to remove steric constraints and facilitate the isolation of a more diverse and selective population of CoA-binding aptamers. More importantly, we enriched the DNA pools with an affinity for the (R)-pantetheine, a subunit of CoA, and then employed in SELEX for CoA. Moreover, starting at the 6th round of selection, we incorporated rigorous counter-selection steps against other molecules, including adenine and molecules with adenine as a component of their structures, such as ATP. Specifically, the CoA aptamer was selected using a DNA library containing 82 nucleotides (Fig. 2a). This library includes 40-nucleotide randomized sequences (red) flanked by two constant sequences (blue) on each end that include and
and  , which serve as primers for PCR amplification. To ensure successful selection without altering the chemical properties of CoA, we chose not to covalently link the CoA to a solid support as in previous CoA SELEX studies.33–35 Instead, we immobilized the DNA library on microbeads through a capture strand DNA that can hybridize to the underlined sequences in one of the two flanking regions,
, which serve as primers for PCR amplification. To ensure successful selection without altering the chemical properties of CoA, we chose not to covalently link the CoA to a solid support as in previous CoA SELEX studies.33–35 Instead, we immobilized the DNA library on microbeads through a capture strand DNA that can hybridize to the underlined sequences in one of the two flanking regions,  . Only the DNA molecules with sequences that bound to CoA and subsequently altered their secondary structure were released from the microbeads into the solution (Fig. 2a).
. Only the DNA molecules with sequences that bound to CoA and subsequently altered their secondary structure were released from the microbeads into the solution (Fig. 2a).
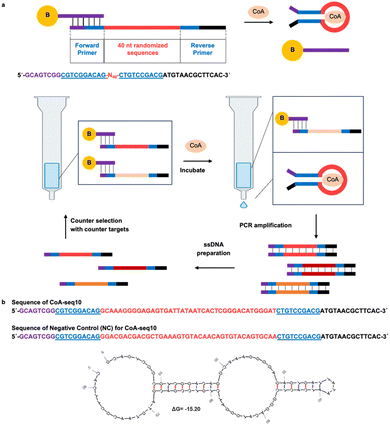 | ||
| Fig. 2 Selection of CoA aptamer. (a) In vitro selection process for obtaining the CoA aptamer. Each DNA library strand contains a 40-nucleotide randomized sequence (red) flanked by two constant sequences (purple, blue, and black) at both ends, serving as PCR primers. The binding region between the capture strand and the DNA library is highlighted in purple. The forward primer consists of purple and blue regions, and the reverse primer is composed of blue and black regions. After immobilizing the DNA library onto beads (yellow), CoA is introduced into the solution. DNA strands that bind to CoA undergo a structural switch, detaching from the capture strand and entering the solution, forming a stem-loop structure with sequences represented by blue color. These eluted strands are then PCR amplified, and ssDNA libraries are prepared. After re-immobilizing the DNA library onto the beads, counter-selection molecules are added to the solution, and the non-specific DNA strands are removed. The DNA library is then incubated with CoA, and the eluted CoA-aptamer DNA is collected. This selection process is repeated multiple times. (b) CoA-seq10 aptamer sequence and its predicted secondary structure, generated using UNAfold software.56 The sequence is illustrated with different colors, whose function is explained in legends of (a). The UNAfold prediction illustrates the secondary structure of the CoA aptamer upon binding to CoA.56 | ||
To ensure high selectivity for CoA against adenine and other molecules containing the adenine moiety, the first four SELEX rounds were initially carried out using (R)-pantetheine, a subunit of CoA, to enrich the DNA pools with an affinity for the (R)-pantetheine portion of CoA (Table S1†). At round 5, the selection target was switched to CoA to isolate DNA aptamers that bind CoA. The concentrations of CoA used in the selection were gradually decreased from 5 mM to 0.1 mM to enrich aptamers with a strong binding affinity for CoA. To achieve high selectivity, counter-selections against adenine, adenosine triphosphate (ATP), pantetheine, and pantothenic acid were introduced from the 6th round of selection, with gradually increasing concentrations. Specifically, from the 6th round of selection, 1 mM adenosine and 9 mM ATP were introduced as counter-selection targets and any DNA molecules eluted with adenosine or ATP were discarded from the selection. Additionally, 150 μM pantetheine was introduced as a counter-selection target starting from round 8 through round 13 and 150 μM of pantothenic acid was introduced as a counter-selection target starting from round 12. These counter-selections were introduced to remove any aptamers that interacted with molecules that bear similarities to only a component of the CoA, rather than CoA itself. To monitor the selection progress, we employed qPCR to determine the elution yield from each selection round.53 The elution yield is defined as the amount of ssDNA bound to CoA and eluted from the microbeads into solution, divided by the total amount of ssDNA added to the microbeads (Fig. S1†). When the elution yield stopped increasing, high-throughput sequencing (HTS) was utilized to analyze the sequences from round 9 to round 13, enabling us to track the enrichment of individual sequences. After sorting the sequences by sequencing quality and identifying those with intact forward and reverse primer sequences, we used the FASTAptamer analysis toolkit to find sequences or clusters that were conserved across all rounds.54,55 Specifically, we used the FASTAptamer-count and FASTAptamer-cluster functions to analyze the abundance and similarity of individual sequences, tracking changes across different SELEX rounds.54 We then applied the FASTAptamer-enrich function to monitor changes in sequence distribution across selection rounds and identified representative enriched aptamer sequences (Table S2†).54 We then tested their binding affinity to CoA by labeling the aptamer candidates with a 5′-FAM fluorophore and the capture strand that attached to the microbeads, as shown in Fig. 2a, with a 3′-Black Hole Quencher®-1 (BHQ1), measuring the increase in fluorescence after the candidates bind CoA, which causes its strand displacement from the capture strand (Fig. 3d). CoA-seq10, a most active sequence and continuously enriched during SELEX, was chosen for sensor development (Fig. 2b and S2†). As illustrated in Fig. 2b, CoA-seq10 is predicted to form a stem-loop secondary structure, as determined using UNAfold software.56
Characterization of the CoA-seq10 aptamer and its conversion into a fluorescent sensor for CoA
To characterize the binding of CoA-seq10 to CoA and its affinity, isothermal titration calorimetry (ITC) was employed, revealing a dissociation constant (Kd) of 39.8 μM (Fig. 3a–c), which is comparable to other small molecule aptamers, like the ATP aptamer with a Kd of 6 μM.3 To convert the CoA aptamer into a fluorescent sensor, we utilized an intramolecular strand displacement fluorescence sensor design (Fig. 3d). Specifically, a FAM fluorophore was conjugated to the 5′ end of the aptamer CoA-seq10, and a Black Hole Quencher®-1 (BHQ1) was attached to the 3′ end of the quencher strand. When the aptamer strand hybridizes with the quencher strand (Tm = 62.3 °C), it brings the quencher next to the fluorophore, resulting in the quenching of the FAM fluorophore by BHQ1. The binding to CoA by the CoA-seq10 aptamer weakens this hybridization (Tm decreases to 0 °C), allowing the quencher strand to dehybridize from the aptamer strand, causing a significant increase in fluorescence.To minimize background fluorescence, we optimized the ratio between aptamer and quencher and found an aptamer:quencher ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to be optimal (Fig. 3e). With this aptamer:quencher ratio, the fluorescence of the aptamer sensor increased with increasing concentrations of CoA, with no significant further increase observed at 10 mM. The limit of detection (LOD) was determined to be 0.126 mM (Fig. 3f), based on 3σb/slope, where σb is the standard deviation of three blank samples. Additionally, replacing the CoA aptamer with a DNA of the same length but with a scrambled sequence did not increase fluorescence signals in the presence of CoA. This result underscores the specificity of the aptamer in sensor development. The fluorescent sensor can also serve as an alternative method to determine Kd values for the aptamer-CoA interaction. In this method, a Kd for the quencher with the aptamer in the absence of CoA was determined to be 55.38 nM (Fig. 3e), while a Kd for the CoA-induced aptamer–quencher dissociation was found to be 1.133 mM (Fig. 3f). Therefore, using a formula reported previously,57 the Kd for the aptamer-CoA interaction was calculated from these two Kd's to be 48.9 μM. This fluorescent method provides an independent method to evaluate the Kd, which is similar to the Kd (39.8 μM) obtained by ITC. Additionally, additional mutations were introduced into the CoA-seq10 aptamer to identify the critical regions involved in its binding and fluorescence response to CoA. The data in Fig. S3† show that these mutations in the aptamer sequence resulted in a decrease in the fluorescence response to CoA, indicating that these specific regions of the aptamer are crucial for both CoA recognition and the conformational change needed to trigger the fluorescence response.
5 to be optimal (Fig. 3e). With this aptamer:quencher ratio, the fluorescence of the aptamer sensor increased with increasing concentrations of CoA, with no significant further increase observed at 10 mM. The limit of detection (LOD) was determined to be 0.126 mM (Fig. 3f), based on 3σb/slope, where σb is the standard deviation of three blank samples. Additionally, replacing the CoA aptamer with a DNA of the same length but with a scrambled sequence did not increase fluorescence signals in the presence of CoA. This result underscores the specificity of the aptamer in sensor development. The fluorescent sensor can also serve as an alternative method to determine Kd values for the aptamer-CoA interaction. In this method, a Kd for the quencher with the aptamer in the absence of CoA was determined to be 55.38 nM (Fig. 3e), while a Kd for the CoA-induced aptamer–quencher dissociation was found to be 1.133 mM (Fig. 3f). Therefore, using a formula reported previously,57 the Kd for the aptamer-CoA interaction was calculated from these two Kd's to be 48.9 μM. This fluorescent method provides an independent method to evaluate the Kd, which is similar to the Kd (39.8 μM) obtained by ITC. Additionally, additional mutations were introduced into the CoA-seq10 aptamer to identify the critical regions involved in its binding and fluorescence response to CoA. The data in Fig. S3† show that these mutations in the aptamer sequence resulted in a decrease in the fluorescence response to CoA, indicating that these specific regions of the aptamer are crucial for both CoA recognition and the conformational change needed to trigger the fluorescence response.
To further validate the specificity of the CoA-aptamer against other intracellular metabolites, we tested its fluorescence response to various concentrations of CoA and compared it to the responses elicited by other structurally similar molecules, such as acetyl coenzyme A (acetyl-CoA), ATP, and D-pantothenic acid. As shown in Fig. 4a, other metabolites, including acetyl-CoA, ATP, and D-pantothenic acid, did not cause any significant increase in fluorescence intensity. In contrast, the sensor exhibited a marked increase in fluorescence intensity as the concentration of CoA increased. Further testing involved incubating the CoA aptamer sensor to a variety of intracellular metabolites at physiologically relevant concentrations (Fig. 4b). The metabolites include: (1) acetyl-CoA, (2) D-pantothenic acid, (3) nucleotides (ATP, CTP, GTP, UTP), (4) other adenine-based metabolites ADP, nicotinamide adenine dinucleotide (NAD), NADH, NADP, NADPH, (5) CoA derivates, such as propionyl-CoA, butyryl-CoA, hexanoyl-CoA, malonyl-CoA, succinyl-CoA, 3-hydroxy-3-methylglutaryl (HMG-CoA), lauroyl-CoA, myristoyl-CoA, and oleoyl-CoA, and (6) divalent cations like Ca2+ and Mg2+. Additionally, to rule out the possibility that the CoA aptamer can still bind the competing targets of CoA without structural changes and thus no detachment of the quencher-labeled oligo, we used ITC to measure the Can10 aptamer's binding with ATP and found no evidence of binding (Fig. S4†). At physiologically relevant concentrations, none of these metabolites induced an increase in the fluorescence signal, confirming the high specificity for CoA aptamer sensor against all other metabolites, including those that share a similar component (e.g., adenine) as CoA.
Cellular environments vary significantly in pH, ranging from acidic compartments such as lysosomes to more alkaline areas like the mitochondrial matrix. To demonstrate that the CoA aptamer sensor can work across different pH levels in diverse physiological conditions, we evaluated its pH sensitivity across a range of physiologically relevant pH values. As shown in Fig. S5,† the CoA aptamer sensor maintained a consistent fluorescence response to CoA, with fluorescence intensity increasing when CoA concentration increases, across the tested pH range (pH 6.4–8.0). These results indicate that the aptamer can function and generate fluorescence signals across these physiological pH conditions.
Application of the aptamer sensor to monitor CoA in living cells
To demonstrate the applicability of the CoA aptamer sensor in visualizing CoA distribution and abundance in living cells, we delivered it into HeLa cells as a representative cell line and visualized it using confocal laser-scanning microscopy (CLSM). Although FAM is a commonly used fluorophore for studies in test tubes, we did not use it to label the CoA aptamer sensor for cellular studies since FAM is sensitive to different pH environments inside the cells. Instead, we used Cy5.5 to label the CoA aptamer strand and Iowa Black® RQ to label the quencher strand and then delivered them into HeLa cells using lipofectamine 3000. As shown in Fig. 5, S6 and S7,† bright signals from the CoA aptamer sensor were observed in all HeLa cells. In contrast, using a negative control (NC) of a DNA oligo with the same length as the CoA aptamer but the aptamer sequence replaced with a scrambled sequence (see Fig. 2b) resulted in a reduced fluorescence signal. This negative control (/5Cy5/ATGCAGTCGGCGTCGGACAGGGACGACGACGCTGAAAGTGTACAACAGTGTACAGTGCAACTGTCCGACGATGTAACGCTTCAC) highlights the essential role of the aptamer in recognizing CoA using a DNA sequence of the same length as the CoA aptamer but with a scrambled sequence—where the region that binds to the quencher strand is preserved while the CoA-binding portion is disrupted—resulted in a reduced fluorescence signal. This negative control highlights the essential role of the aptamer in recognizing CoA. To assess the duration over which the fluorescence signal from the CoA-seq10 aptamer sensor can be distinguished from the negative control (NC) sensor after transfection, a time-course study was performed. As shown in Fig. S6 and S7,† 3.5 hours after the transfection, the fluorescence signal from the CoA aptamer is significantly higher than that of the NC sensor for up to 8 hours. Although the fluorescence signal from the CoA-seq10 aptamer decreases over time, it remains distinguishable from the NC sensor. To ensure our sensor can monitor CoA selectively, we added either Pantazine 2891 (PZ-2891),58 which is an allosteric PANK activator that increases free CoA concentration inside cells, presumably by disrupting pantothenate kinase (PanK) feedback regulation59,60 or hopantenate (HoPan), an analog of pantothenate, which is used to chemically inhibit CoA biosynthesis.61,62 After treating the HeLa cells with either 3 μM PZ-2891 or 1 mM HoPan to regulate the cellular CoA levels, we added Cy5.5-labeled aptamer sensors and compared them with untreated cells. As shown in Fig. 5, compared with untreated cells, PZ-2891 treated cells exhibited a significant fluorescence increase in the red channel, while HoPan-treated samples showed a lower red fluorescence signal. Furthermore, the fluorescence intensity of the negative control with scrambled sequence produced relatively weak fluorescence signals compared to untreated cells (Fig. 5). In addition, we conducted a time-course drug treatment with 3 μM PZ-2891 in HeLa cells to assess the dynamic response of the CoA-seq10 aptamer sensor under drug exposure, measuring the fluorescence signal at multiple time points post-treatment. As shown in Fig. S7 and S8,† the fluorescence intensity of the CoA-seq10 aptamer sensor is significantly higher than the no-treatment control after 6 hours of incubation with 3 μM PZ-2891. A statistically significant difference is observed between the CoA-seq10 aptamer sensor and the NC sensor at each drug treatment time point. These results collectively demonstrate that the CoA aptamer sensor can be applied to monitor intracellular CoA.Conclusions
CoA is an essential metabolite involved in key biological processes, including the synthesis and oxidation of fatty acids and the citric acid cycle for energy production. Despite its critical role, sensors for CoA are limited. While SELEX has been a widely used method for generating DNA and RNA aptamers for many targets, efforts to isolate aptamers for CoA have largely failed, with most aptamers binding only the adenine moiety of CoA, highlighting a major limitation of the SELEX methods to obtain aptamers with high selectivity. To claim that SELEX can be generally applied to almost any target, we need to develop methods to overcome this major limitation.To meet this challenge, we have improved the SELEX process by incorporating rigorous counter-selections against adenine, ATP, pantetheine, and pantothenic acid. This approach led to the successful identification of a highly selective DNA aptamer, CoA-seq10, making this aptamer capable of monitoring CoA at the physiological concentration of CoA in the cytoplasm (0.02 to 0.14 mM) and mitochondria (2.2 to over 5 mM).36 These results were confirmed using ITC and fluorescence assays, with a limit of detection (126 μM). The aptamer's specificity allowed it to distinguish CoA from other intracellular metabolites and function effectively across different physiological pH levels. Confocal microscopy further demonstrated the aptamer sensor's ability to visualize CoA in living cells, with significant fluorescence changes upon regulating cellular CoA levels. In summary, this work contributes to SELEX methodology, overcoming some of its limitations and enabling the development of a CoA-selective aptamer that opens new opportunities for studying CoA metabolism in living cells.
Data availability
All data are available in the main text or the ESI†.Author contributions
Conceptualization: Yuan Ma, Whitney Lewis, Yi Lu. Methodology: Yuan Ma, Whitney Lewis, Peng Yan, Linggen Kong, Quanbing Mou, Xiangli Shao, Weijie Guo. Investigation: Yuan Ma, Whitney Lewis, Peng Yan, Linggen Kong (in vitro selection of CoA aptamers); Yuan Ma, Whitney Lewis, Quanbing Mou (aptamer characterization experiments and data analysis); Yuan Ma, Whitney Lewis, Xiangli Shao, Weijie Guo (cell experiments and data analysis). Writing – original Draft: Yuan Ma, Whitney Lewis, Yi Lu. Writing – review & editing: all authors. Supervision: Yi Lu. Equal Contribution: Yuan Ma and Whitney Lewis contributed equally to this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based on work supported by the U.S. National Institutes of Health (GM141931) and the Allen Distinguished Investigator Award (to Y. L.), a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Family Foundation. We also acknowledge the Robert A. Welch Foundation (grant F-0020) for supporting the Lu group research program at the University of Texas at Austin. We thank Dr Sandy McMasters at the Department of Chemistry at the University of Illinois Urbana-Champaign for providing custom-made DMEM without vitamin B5. Confocal imaging was performed at the Center for Biomedical Research Support Microscopy and Imaging Facility at the University of Texas at Austin (RRID: SCR_021756). We thank Anna Webb and Paul Oliphint at UT Austin for their valuable advice on confocal imaging.References
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 Search PubMed.
- W. Zhou, P.-J. J. Huang, J. Ding and J. Liu, Analyst, 2014, 139, 2627–2640 RSC.
- C. A. Dougherty, W. Cai and H. Hong, Curr. Top. Med. Chem., 2015, 15, 1138–1152 CrossRef CAS PubMed.
- M. Dunn, R. Jimenez and J. Chaput, Nat. Rev. Chem, 2017, 1, 415700 Search PubMed.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- J. Zhang, L. P. Smaga, N. S. R. Satyavolu, J. Chan and Y. Lu, J. Am. Chem. Soc., 2017, 139, 17225–17228 CrossRef CAS PubMed.
- A. Moutsiopoulou, D. Broyles, E. Dikici, S. Daunert and S. K. Deo, Small, 2019, 15, 1902248 CrossRef PubMed.
- Y. Song, J. Song, X. Wei, M. Huang, M. Sun, L. Zhu, B. Lin, H. Shen, Z. Zhu and C. Yang, Anal. Chem., 2020, 92, 9895–9900 CrossRef CAS.
- S. Hong, X. Zhang, R. J. Lake, G. T. Pawel, Z. Guo, R. Pei and Y. Lu, Chem. Sci., 2020, 11, 713–720 RSC.
- H. Yu, O. Alkhamis, J. Canoura, Y. Liu and Y. Xiao, Angew. Chem., Int. Ed., 2021, 60, 16800–16823 CrossRef CAS PubMed.
- Q. Mou, X. Xue, Y. Ma, M. Banik, V. Garcia, W. Guo, J. Wang, T. Song, L.-Q. Chen and Y. Lu, Sci. Adv., 2022, 8, eabo0902 CrossRef CAS PubMed.
- A. M. Downs and K. W. Plaxco, ACS Sens., 2022, 7(10), 2823–2832 CrossRef CAS PubMed.
- Y. Ding and J. Liu, J. Am. Chem. Soc., 2023, 145, 7540–7547 CrossRef CAS PubMed.
- B. Lin, F. Xiao, J. Jiang, Z. Zhao and X. Zhou, Chem. Sci., 2023, 14, 14039–14061 Search PubMed.
- O. Alkhamis, J. Canoura, Y. Wu, N. A. Emmons, Y. Wang, K. M. Honeywell, K. W. Plaxco, T. E. Kippin and Y. Xiao, J. Am. Chem. Soc., 2024, 146, 3230–3240 Search PubMed.
- M. Banik, A. P. Ledray, Y. Wu and Y. Lu, ACS Cent. Sci., 2024, 10(20), 1585–1593 CrossRef CAS PubMed.
- H. Wang, H. Cheng, J. Wang, L. Xu, H. Chen and R. Pei, Talanta, 2016, 154, 498–503 CrossRef CAS PubMed.
- Y. Chen, H. Li, T. Gao, T. Zhang, L. Xu, B. Wang, J. Wang and R. Pei, Sens. Actuators, B, 2018, 254, 214–221 CrossRef CAS.
- J. Wang, Y. Liu, X. Li, H. Lei and J. Liu, Chem. Commun., 2024, 60, 14272–14275 Search PubMed.
- Y. Liu, X. Wang and J. Liu, Chem. Commun., 2024, 60, 6280–6283 RSC.
- K. Sefah, D. Shangguan, X. Xiong, M. B. O'Donoghue and W. Tan, Nat. Protoc., 2010, 5, 1169–1185 Search PubMed.
- R. Stoltenburg, N. Nikolaus and B. Strehlitz, J. Anal. Methods Chem., 2012, 2012, 415697 Search PubMed.
- K.-A. Yang, M. Barbu, M. Halim, P. Pallavi, B. Kim, D. M. Kolpashchikov, S. Pecic, S. Taylor, T. S. Worgall and M. N. Stojanovic, Nat. Chem., 2014, 6, 1003–1008 CrossRef CAS PubMed.
- K.-A. Yang, H. Chun, Y. Zhang, S. Pecic, N. Nakatsuka, A. M. Andrews, T. S. Worgall and M. N. Stojanovic, ACS Chem. Biol., 2017, 12, 3103–3112 CrossRef CAS PubMed.
- H. Yu, Y. Luo, O. Alkhamis, J. Canoura, B. Yu and Y. Xiao, Anal. Chem., 2021, 93, 3172–3180 CrossRef CAS PubMed.
- D. Wu, C. K. L. Gordon, J. H. Shin, M. Eisenstein and H. T. Soh, Acc. Chem. Res., 2022, 55, 685–695 CrossRef CAS PubMed.
- Y. Zhao, S. Ong, Y. Chen, P.-J. Jimmy Huang and J. Liu, Anal. Chem., 2022, 94, 10175–10182 Search PubMed.
- Y. Zhao, A. Z. Li and J. Liu, Environ. Health, 2023, 1, 102–109 CAS.
- A. Brown, J. Brill, R. Amini, C. Nurmi and Y. Li, Angew. Chem., Int. Ed., 2024, 63, e202318665 CAS.
- L. Wang, J. Canoura, C. Byrd, T. Nguyen, O. Alkhamis, P. Ly and Y. Xiao, ACS Cent. Sci., 2024, 10(12), 2213–2228 CrossRef CAS PubMed.
- D. H. Burke and D. C. Hoffman, Biochemistry, 1998, 37, 4653–4663 CAS.
- D. Saran, J. Frank and D. H. Burke, BMC Evol. Biol., 2003, 3, 26 Search PubMed.
- D. Hu, Y. Hu, T. Zhan, Y. Zheng, P. Ran, X. Liu, Z. Guo, W. Wei and S. Wang, Biosens. Bioelectron., 2020, 150, 111934 Search PubMed.
- A. Czumaj, S. Szrok-Jurga, A. Hebanowska, J. Turyn, J. Swierczynski, T. Sledzinski and E. Stelmanska, Int. J. Mol. Sci., 2020, 21, 9057 CAS.
- S. A. Barritt, S. E. DuBois-Coyne and C. C. Dibble, Nat. Metab., 2024, 6, 1008–1023 Search PubMed.
- R. Leonardi, Y.-M. Zhang, C. O. Rock and S. Jackowski, Prog. Lipid Res., 2005, 44, 125–153 Search PubMed.
- D. L. Martinez, Y. Tsuchiya and I. Gout, Biochem. Soc. Trans., 2014, 42, 1112–1117 Search PubMed.
- B. Zhou, S. K. Westaway, B. Levinson, M. A. Johnson, J. Gitschier and S. J. Hayflick, Nat. Genet., 2001, 28, 345–349 CAS.
- S. Dusi, L. Valletta, T. B. Haack, Y. Tsuchiya, P. Venco, S. Pasqualato, P. Goffrini, M. Tigano, N. Demchenko, T. Wieland, T. Schwarzmayr, T. M. Strom, F. Invernizzi, B. Garavaglia, A. Gregory, L. Sanford, J. Hamada, C. Bettencourt, H. Houlden, L. Chiapparini, G. Zorzi, M. A. Kurian, N. Nardocci, H. Prokisch, S. Hayflick, I. Gout and V. Tiranti, Am. J. Hum. Genet., 2014, 94, 11–22 Search PubMed.
- B. Srinivasan, M. Baratashvili, M. van der Zwaag, B. Kanon, C. Colombelli, R. A. Lambrechts, O. Schaap, E. A. Nollen, A. Podgoršek, G. Kosec, H. Petković, S. Hayflick, V. Tiranti, D.-J. Reijngoud, N. A. Grzeschik and O. C. M. Sibon, Nat. Chem. Biol., 2015, 11, 784–792 Search PubMed.
- C. C. Dibble, S. A. Barritt, G. E. Perry, E. C. Lien, R. C. Geck, S. E. DuBois-Coyne, D. Bartee, T. T. Zengeya, E. B. Cohen, M. Yuan, B. D. Hopkins, J. L. Meier, J. G. Clohessy, J. M. Asara, L. C. Cantley and A. Toker, Nature, 2022, 608, 192–198 CrossRef CAS PubMed.
- L. L. Bieber, Anal. Biochem., 1992, 204, 228–230 CAS.
- Y. Tsuchiya, U. Pham and I. Gout, Biochem. Soc. Trans., 2014, 42, 1107–1111 CAS.
- Y. I. Shurubor, M. D'Aurelio, J. Clark-Matott, E. P. Isakova, Y. I. Deryabina, M. F. Beal, A. J. L. Cooper and B. F. Krasnikov, Molecules, 2017, 22, 1388 Search PubMed.
- L. Xue, P. Schnacke, M. S. Frei, B. Koch, J. Hiblot, R. Wombacher, S. Fabritz and K. Johnsson, Nat. Chem. Biol., 2023, 19, 346–355 CAS.
- D. M. Chudakov, S. Lukyanov and K. A. Lukyanov, Trends Biotechnol., 2005, 23, 605–613 CAS.
- S. J. Leavesley, N. Annamdevula, S. Johnson, D. J. Pleshinger and T. C. Rich, in cAMP Signaling: Methods and Protocols, ed. M. Zaccolo, Springer US, New York, NY, 2022, pp. 167–180 Search PubMed.
- R. Nutiu and Y. Li, Angew. Chem., Int. Ed., 2005, 44, 1061–1065 Search PubMed.
- M. Rajendran and A. D. Ellington, Nucleic Acids Res., 2003, 31, 5700–5713 CrossRef CAS PubMed.
- K.-A. Yang, R. Pei and M. N. Stojanovic, Methods, 2016, 106, 58–65 CrossRef CAS PubMed.
- Z. Luo, L. He, J. Wang, X. Fang and L. Zhang, Analyst, 2017, 142, 3136–3139 Search PubMed.
- K. K. Alam, J. L. Chang and D. H. Burke, Mol. Ther.--Nucleic Acids, 2015, 4, e230 CrossRef CAS PubMed.
- M. R. Gotrik, T. A. Feagin, A. T. Csordas, M. A. Nakamoto and H. T. Soh, Acc. Chem. Res., 2016, 49, 1903–1910 CrossRef CAS PubMed.
- N. R. Markham and M. Zuker, Methods Mol. Biol., 2008, 453, 3–31 CrossRef CAS PubMed.
- N. Nakatsuka, K.-A. Yang, J. M. Abendroth, K. M. Cheung, X. Xu, H. Yang, C. Zhao, B. Zhu, Y. S. Rim, Y. Yang, P. S. Weiss, M. N. Stojanović and A. M. Andrews, Science, 2018, 362, 319–324 CrossRef CAS PubMed.
- L. K. Sharma, C. Subramanian, M.-K. Yun, M. W. Frank, S. W. White, C. O. Rock, R. E. Lee and S. Jackowski, Nat. Commun., 2018, 9, 4399 CrossRef PubMed.
- R. Leonardi, Y.-M. Zhang, M.-K. Yun, R. Zhou, F.-Y. Zeng, W. Lin, J. Cui, T. Chen, C. O. Rock, S. W. White and S. Jackowski, Chem. Biol., 2010, 17, 892–902 Search PubMed.
- L. K. Sharma, R. Leonardi, W. Lin, V. A. Boyd, A. Goktug, A. A. Shelat, T. Chen, S. Jackowski and C. O. Rock, J. Med. Chem., 2015, 58, 1563–1568 CrossRef CAS PubMed.
- Y.-M. Zhang, S. Chohnan, K. G. Virga, R. D. Stevens, O. R. Ilkayeva, B. R. Wenner, J. R. Bain, C. B. Newgard, R. E. Lee, C. O. Rock and S. Jackowski, Chem. Biol., 2007, 14, 291–302 CrossRef CAS PubMed.
- K. J. Mostert, N. Sharma, M. van der Zwaag, R. Staats, L. Koekemoer, R. Anand, O. C. M. Sibon and E. Strauss, ACS Chem. Biol., 2021, 16, 2401–2414 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc00332f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |



