Open Access Article
Open Access ArticleDyeLeS: a web platform for predicting and classifying fluorescence properties of bioactive molecules†
Jiangcheng Xu a,
Wenbo Yua,
Nan Zhoub,
Jiamin Zhongb,
Jintian Lyuc,
Zhihao Sud,
Yu Chen*a and
Kui Du
a,
Wenbo Yua,
Nan Zhoub,
Jiamin Zhongb,
Jintian Lyuc,
Zhihao Sud,
Yu Chen*a and
Kui Du *b
*b
aHangzhou Vocational & Technical College, Hangzhou 310014, P. R. China. E-mail: 2003010002@hzvtc.edu.cn
bChemistry and Chemical Engineering, Shaoxing University, Shaoxing 312000, P. R. China. E-mail: dkui@usx.edu.cn
cL.E.K. Consulting, 75 State Street 19th Floor Boston, MA 02109, USA
dCollege of Pharmaceutical Sciences, Zhejiang University of Technology, Hangzhou, Zhejiang 310014, P. R. China
First published on 27th June 2025
Abstract
Fluorescent drug molecules play a pivotal role in biomedical research and precision medicine. Their intrinsic fluorescence enables real-time tracking of drug distribution, target engagement, and metabolic pathways, while avoiding interference from external labeling. However, traditional fluorescent drug discovery relies heavily on trial-and-error approaches, which are inefficient and resource-intensive. To address this, we developed DyeLeS (Dye-Likeness Scoring), a web platform designed to rapidly evaluate molecular fluorescence potential and predict key photophysical properties such as Stokes shift and quantum yield. DyeLeS utilizes a curated dataset of fluorescent and non-fluorescent compounds, applies a Naive Bayes-inspired algorithm for fluorescence classification (AUC = 0.995), and employs a LightGBM model for quantitative prediction of fluorescence properties, achieving an R2 of 0.88 in absorption wavelength (λabs) prediction. Leveraging DyeLeS, this study constructed FluoBioDB, the first publicly available library of fluorescent bioactive compounds, encompassing 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 structurally diverse molecules, including kinase inhibitors and GPCR modulators. Case analyses indicate that FluoBioDB compounds typically possess polycyclic conjugated frameworks, donor–acceptor (D–A) structures, and rigid planar cores, endowing them with strong potential for applications in bioimaging, targeted therapy, and theranostics. This work presents a robust computational framework and a valuable molecular resource to facilitate the rapid discovery and optimization of fluorescent drug candidates. All source codes and datasets are available at https://github.com/MolAstra/DyeLeS, and the web server can be accessed at https://dyeles.molastra.com.
865 structurally diverse molecules, including kinase inhibitors and GPCR modulators. Case analyses indicate that FluoBioDB compounds typically possess polycyclic conjugated frameworks, donor–acceptor (D–A) structures, and rigid planar cores, endowing them with strong potential for applications in bioimaging, targeted therapy, and theranostics. This work presents a robust computational framework and a valuable molecular resource to facilitate the rapid discovery and optimization of fluorescent drug candidates. All source codes and datasets are available at https://github.com/MolAstra/DyeLeS, and the web server can be accessed at https://dyeles.molastra.com.
1 Introduction
Fluorescent drug molecules play a significant role in medical and biological research.1,2 Their inherent fluorescence enables real-time tracking and imaging in vivo, facilitating the observation of drug distribution,3 localization,4 and metabolism5 without the need for additional labeling, thereby simplifying experimental procedures. Moreover, fluorescent drugs allow for direct validation of targeting efficiency, confirming whether they have accurately reached specific biological targets such as tumor tissues or receptor sites.6,7 In drug discovery, fluorescent compounds can be directly applied in high-throughput screening (HTS), enhancing the efficiency of candidate selection.1 Furthermore, drugs with intrinsic fluorescence avoid the “labeling effect” often caused by external fluorescent probes, thus preserving their native bioactivity.7 Some fluorescent drugs also support theranostic applications, simultaneously enabling imaging and treatment, as exemplified by their use in photodynamic therapy (PDT).6 Therefore, the development of fluorescent drug molecules holds great promise for advancing pharmaceutical research and precision medicine.7The development of fluorescent drug molecules typically requires a complex, multi-step process, including lead identification, fluorescence and bioactivity characterization, structural optimization, mechanistic studies, pharmacokinetic evaluation, and safety assessment.8–10 Each stage demands extensive experimental effort and relies heavily on manual trial-and-error approaches, resulting in high resource consumption and limited efficiency. Streamlining this workflow is crucial for accelerating fluorescent drug discovery.11
With the advancement of artificial intelligence technologies and the expansion of pharmaceutical big data, the speed of drug development has significantly increased.12–14 For example, the development of machine learning (ML) models or algorithm for predicting drug-like properties has enabled the rapid construction of drug-like compound libraries,15–17 thereby streamlining the drug discovery process. Similarly, models for assessing natural product-likeness have facilitated the identification of compounds with natural origins and the creation of natural product libraries,18,19 improving development efficiency. Although databases such as ChEMBL,20 MedChemExpress,21 and ZINC22 offer extensive collections of bioactive molecules to support drug discovery, to the best of our knowledge, there have been no reports focusing on methods for the rapid assessment of molecular fluorescence properties or on the establishment of fluorescent bioactive molecule libraries.
To enable the rapid identification of fluorescent properties in drug molecules and to construct a dedicated library of fluorescent drug candidates, we developed DyeLeS, a web-based application for Dye-Likeness Scoring. DyeLeS allows for the efficient evaluation of molecular fluorescence potential and predicts key properties such as Stokes shift and fluorescence quantum yield. Furthermore, by applying DyeLeS to the virtual screening of known bioactive molecules, we constructed the first publicly available fluorescent drug molecule library, comprising 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 compounds annotated with detailed fluorescence characteristics, including maximum absorption wavelengths, quenching wavelengths, and fluorescence lifetimes. This study leverages ML to accelerate the development of fluorescent drugs, utilizing the inherent labeling effect of fluorescent molecules to advance drug pharmacokinetics research, target validation strategies, and the integration of therapeutic and diagnostic (theranostic) applications.
865 compounds annotated with detailed fluorescence characteristics, including maximum absorption wavelengths, quenching wavelengths, and fluorescence lifetimes. This study leverages ML to accelerate the development of fluorescent drugs, utilizing the inherent labeling effect of fluorescent molecules to advance drug pharmacokinetics research, target validation strategies, and the integration of therapeutic and diagnostic (theranostic) applications.
2 Result and discussion
2.1 Construction of training databases for fluorescence prediction
To enable the ML model to accurately recognize the characteristics of fluorescent drug molecules, we first constructed two major databases: a Fluorescent Molecule database (positive set) and a Non-Fluorescent Molecule database (negative set).The fluorescent molecule database was built from two primary sources: (a) approximately 3200 fluorescent compounds manually curated based on extensive experimental experience in the field of fluorescent molecules;23–25 (b) an additional ∼25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fluorescent compounds collected from the Dye database26 developed by the Song research group. After deduplication, RDKit standardization, and quality control, the final library contained 26
000 fluorescent compounds collected from the Dye database26 developed by the Song research group. After deduplication, RDKit standardization, and quality control, the final library contained 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255 unique fluorescent molecules represented by SMILES. Among them, 6703 were annotated with properties such as Stokes shift, quantum yield (Φfl), absorption (λabs), and emission wavelengths (λem).
255 unique fluorescent molecules represented by SMILES. Among them, 6703 were annotated with properties such as Stokes shift, quantum yield (Φfl), absorption (λabs), and emission wavelengths (λem).
The non-fluorescent molecule database was primarily sourced from the Collection of Open Natural Products27 (COCONUT) database. Based on our group's expertise in fluorescence research,23–25 we performed fluorescence screening and manually excluded molecules with potential fluorescent properties. This resulted in a non-fluorescent dataset containing 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991 compounds with no observable fluorescence activity.
991 compounds with no observable fluorescence activity.
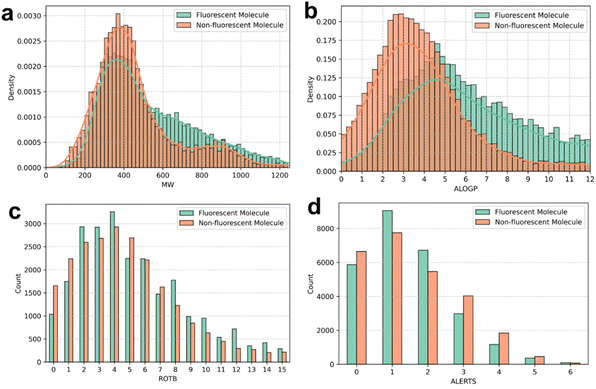
Comparative analyses of molecular weight (MW, Fig. 1a), atom-based octanol–water partition coefficient (ALOGP, Fig. 1b), number of rotatable bonds (ROTB, Fig. 1c), structural alerts (ALERTS, Fig. 1d), hydrogen bond acceptors (HBA), and hydrogen bond donors (HBD) (Fig. S1a and b, ESI†) between the fluorescent and non-fluorescent compound libraries revealed that the two datasets share comparable key physicochemical properties, such as molecular weight, thereby minimizing the risk of data-type bias. Moreover, both libraries exhibit approximately normal distributions with uniform data coverage, reducing the likelihood of bias arising from the overrepresentation of structurally similar fluorescent molecules.
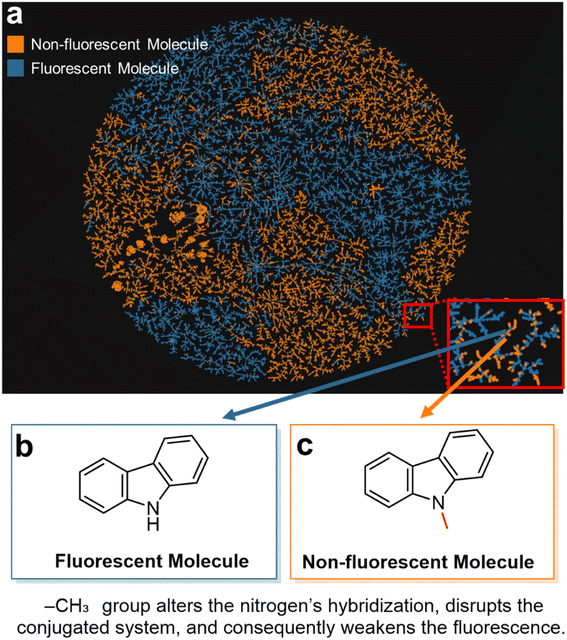
To further explore the data characteristics of the two databases, we employed the TMAP28 dimensionality reduction tool for analysis. Two key observations can be drawn from Fig. 2a: First, on a global scale, there is a clear distinction between fluorescent molecules (blue) and non-fluorescent molecules (orange), as evidenced by the separate clustering of the two colors. A possible explanation is that, while the molecules in both datasets share similar physical properties, the fluorescent molecule dataset contains a higher proportion of aromatic rings (Fig. S2, ESI†), which differentiates it from the non-fluorescent dataset. This distinct clustering in chemical space suggests that machine learning algorithms may be able to effectively distinguish between the two classes of molecules. Second, at a more detailed level, we observe several orange points (non-fluorescent molecules) scattered among the blue cluster (fluorescent molecules), particularly in the left-central region of the map, as well as in the upper right and left areas. This indicates that some fluorescent and non-fluorescent molecules are very close to each other in chemical space. A representative example is the pair of adjacent blue and orange dots located in the lower right of the plot. Upon examination, these correspond to carbazole, a fluorescent molecule (Fig. 2b), and N-methylcarbazole, a non-fluorescent molecule (Fig. 2c).
In carbazole, the nitrogen atom is sp2-hybridized, and its lone pair can participate in π-conjugation within the aromatic system, facilitating fluorescence. However, when the hydrogen on the nitrogen is replaced by a methyl group, the nitrogen's lone pair can no longer delocalize, thereby disrupting the electronic conjugation across the molecule. The only structural difference between the two molecules is the presence of a single methylene group, but this substitution increases the molecular flexibility and enhances non-radiative decay pathways in the excited state, leading to a significant reduction in fluorescence.29 In the TMAP dimensionality reduction plot, it is also easy to identify cases where molecules with highly similar structures exhibit markedly different fluorescence behaviors (Fig. S3, ESI†).
Based on structural analyses of molecules from the fluorescent and non-fluorescent molecule databases, we found that fluorescent compounds typically possess key structural features such as aromatic ring systems, donor–acceptor architectures, and specific functional group substitutions. Therefore, in designing DyeLeS for fluorescence property prediction, we first developed DyeLeS-DyeS to extract characteristic fluorescent substructures from the molecular datasets, enabling a coarse screening of fluorescence potential. Subsequently, we implemented DyeLeS-DyeP, a regression-based machine learning model, which considers additional structural factors—such as molecular rigidity and the extent of π-conjugation—to perform refined and quantitative predictions of fluorescence properties.
2.2 DyeLeS-DyeS: fluorescent dye-likeness score using Bayes-like algorithm
To efficiently identify characteristic molecular fragments of fluorescent compounds, First, we selected Morgan fingerprints30 to represent molecules in the positive (fluorescent molecule database) and negative (non-fluorescent molecule database) sample datasets. Given that fluorescence properties are often highly sensitive to minor structural changes31 (such as the presence or absence of small substituents), Morgan fingerprints—also known as Extended-Connectivity Fingerprints32 (ECFPs)—were chosen due to their ability to capture subtle structural differences that significantly impact molecular activity. This method enables the precise characterization of local chemical environments, including functional groups, branches, and ring systems. The Morgan fingerprints were generated using the RDKit toolkit.33Next, inspired by the Ertl18 and Sorokina19 research group, we adopted a log-likelihood ratio scoring algorithm (statistical fragments) combined with the Morgan Fingerprint approach, providing a highly efficient, interpretable, and scalable solution ideal for large-scale fluorescent molecule screening and early-stage library construction. We refer to this approach as DyeLeS-DyeS. This strategy has two key advantages: (1) it does not require modeling complex dependencies among features, instead focusing solely on the relationship between fragment presence and fluorescence properties; (2) it is resistant to overfitting in high-dimensional spaces because each molecular fragment is modeled independently.
After processing the positive and negative sample datasets, a log-likelihood ratio (F-score) is calculated by DyeLeS-DyeS for each fingerprint bit to identify features more common in dye molecules. This score is based on fragment frequencies in both datasets, with Laplace smoothing applied. A molecule's fluorescence score is then computed by summing the contributions of its fragments and normalizing by molecular size. DyeLeS-DyeS assigns a fluorescence-likeness score from −5 to +5, where higher scores indicate a greater likelihood of fluorescence.
To evaluate the performance of the DyeLeS-DyeS model, we conducted both classification and scoring tasks across multiple molecular datasets (Fig. 3). In the binary classification task distinguishing fluorescent molecules from non-fluorescent ones, the model achieved an area under the ROC curve (AUC) of 0.995 (Fig. 3a), indicating excellent predictive accuracy. To further assess the model's generalizability, we tested its classification ability on three additional datasets where fluorescence properties were uncertain: ZINC, NPAtlas, and ChEMBL. These were compared against a dye molecule database composed of known fluorescent compounds. The resulting AUC values were 0.911, 0.995, and 0.917, respectively (Fig. 3b), demonstrating the robustness and transferability of the model across chemically diverse datasets. Notably, SHapley Additive exPlanations (SHAP) analysis (Table S1, ESI;† additional cases available at https://github.com/MolAstra/DyeLeS) revealed that aromatic systems, donor–acceptor motifs, and functional group substituents (e.g., hydroxyl, amino, and carbonyl groups) consistently ranked as top-contributing features. This finding aligns with established fluorescence mechanisms, validating both the model's predictive accuracy and chemical interpretability.
In addition to classification, we also applied the DyeLeS-DyeS model to fluorescence scoring tasks across datasets of varying sizes (Fig. 3c). The scoring distributions reflected the expected fluorescence tendencies of each dataset. Specifically, the Dyes database, consisting of fluorescent molecules, showed score distributions primarily in the range of 0 to 5. The COCONUT database, generally regarded as containing non-fluorescent natural products, had scores concentrated between −5 and 0. Meanwhile, the ZINC database, which contains molecules with ambiguous or unknown fluorescence behavior, exhibited a broader score distribution from approximately −4 to 4 (Fig. 3d). These results confirm the effectiveness of the pseudo-Bayesian scoring strategy implemented in the model, which enables both quantitative ranking and qualitative screening of molecular fluorescence potential.
2.3 Prediction of photophysical properties using DyeLeS-DyeP
To enable quantitative prediction of fluorescence-related properties, we developed DyeLeS-DyeP, a regression model based on LightGBM.34,35 As shown in Fig. 4a, the model takes molecular structures as input and outputs four key photophysical parameters: absorption wavelength (λabs), Stokes shift, emission wavelength (λem), and fluorescence quantum yield (Φfl). The model demonstrated strong predictive performance on a curated dataset of fluorescent molecules. The R2 values reached 0.88 for λabs, 0.83 for λem, 0.66 for Stokes shift, and 0.48 for Φfl (Fig. 4b–e, The predicted mean squared error (MSE) and root mean squared error (RMSE) values are detailed in Table S2, ESI†). The high accuracy for λabs and λem suggests that the model can effectively learn structure–property relationships related to electronic transitions, which are strongly governed by conjugation length, aromatic systems, and electron-donating/withdrawing substituents. DyeLeS-DyeP accurately predicts optical wavelengths by evaluating key structural determinants of π–π* and charge transfer (CT) transitions. Our unified LightGBM framework effectively discriminates between these transition types by autonomously learning their distinct electronic characteristics, with particular sensitivity to Coulomb interactions in CT states. The current model demonstrates robust predictive accuracy, with future development pathways including transition-specific submodel architectures for further performance optimization. The moderate performance on Stokes shift prediction reflects its dependence not only on electronic structure but also on excited-state relaxation behavior, which is partially encoded in structural features.The relatively lower R2 value for Φfl indicates that fluorescence quantum yield remains a more difficult target to model. This is consistent with literature reports, as Φfl is affected by a variety of subtle and non-structural factors, including molecular rigidity, intramolecular motions, solvent polarity, and the presence of non-radiative decay pathways. Although the model's predictive accuracy for Φfl is limited, it still offers valuable first-pass screening capability, enabling prioritization of candidates for experimental validation.
Together, these results highlight the complementary strengths of DyeLeS-DyeP and DyeLeS-DyeS. While DyeLeS-DyeS enables rapid classification of fluorescent versus non-fluorescent candidates, DyeLeS-DyeP provides fine-grained and property-specific predictions, enhancing the overall utility of the DyeLeS platform for fluorescent molecule design and screening.
2.4 Construction of the fluorescent bioactive molecule database using DyeLeS
To develop a comprehensive library of bioactive compounds with potential fluorescence, we applied the DyeLeS platform to perform large-scale classification and property prediction based on data from ChEMBL. The resulting database, named FluBioDB (Fluorescent Bioactive Molecule Database), contains compounds predicted to possess both bioactivity and favorable fluorescence characteristics, providing a valuable resource for fluorescent probe development and imaging-related drug discovery.As illustrated in Fig. 5, the construction of FluBioDB involved three key steps: (1) fluorescence-Likeness Scoring: We first evaluated compounds from ChEMBL and NPAtlas using the DyeLeS-DyeS, which assigns a fluorescence-likeness score ranging from −5 to +5 based on fragment-level enrichment. As shown in Fig. 5a, only ChEMBL compounds with scores above 0.5 were selected. All NPAtlas compounds fell below this threshold and were excluded from the final dataset. (2) Fluorescence Property Annotation: The filtered ChEMBL compounds were then analyzed using DyeLeS-DyeP, which predicts four key photophysical parameters: absorption wavelength (λabs), emission wavelength (λem), Stokes shift, and fluorescence quantum yield (Φfl). Notably, DyeLeS-DyeP has been implemented as a web application, allowing users to conveniently obtain fluorescence-related predictions based on molecular structure (Fig. 5b). (3) Database compilation: all bioactive compounds in ChEMBL were first encoded as Morgan fingerprints (radius = 2, 2048 bits) using the RDKit cheminformatics toolkit. The selected subset of compounds—those scoring above 0.5 in DyeLeS-DyeS—were then assembled into the final dataset. This resulted in a total of 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 fluorescent bioactive molecules, each annotated with predicted fluorescence properties from DyeLeS-DyeP (Fig. 5c). The resulting FluBioDB serves as an open-access, structurally diverse resource for accelerating fluorescent drug discovery, enabling virtual screening, experimental validation, and downstream theranostic application development.
865 fluorescent bioactive molecules, each annotated with predicted fluorescence properties from DyeLeS-DyeP (Fig. 5c). The resulting FluBioDB serves as an open-access, structurally diverse resource for accelerating fluorescent drug discovery, enabling virtual screening, experimental validation, and downstream theranostic application development.
We selected nine representative compounds from the FluBioDB with high DyeLeS fluorescence-likeness scores for detailed analysis. These compounds exhibit promising potential for applications in bioimaging probes, drug delivery tracking, and related fields.
For instance, the compound shown in Fig. 6a is a potential kinase inhibitor or signaling pathway modulator. Its structure includes a pyrimidine ring, enone moiety, and urea group, forming an extended conjugated system. Molecules with similar scaffolds are often designed as fluorescent probes.36 The compound in Fig. 6b features a [1,2,4]triazolo[1,5-a]pyridine fluorophore—a conjugated core commonly found in fluorescent probes,37 structurally reminiscent of quinoline. It also includes a thiazole ring, chlorophenyl aromatic system, and a flexible linker combining a piperidine ring and fluorocyclopropylamine. The presence of a difluoromethoxy group, a strong electron-withdrawing unit, contributes to a donor–acceptor (D–A) configuration, promoting charge transfer (CT) fluorescence. The predicted fluorescence quantum yield is 0.37. The molecule shown in Fig. 6c features a structure commonly associated with kinase inhibitors or GPCR (G Protein-Coupled Receptor) modulators, characterized by a fused dibenzofuran, benzoyl, and methoxyphenyl moiety forming an extended π-conjugated system that facilitates efficient electronic transitions. Its rigid and planar architecture indicates strong potential for development as a fluorescent drug candidate.38,39 The compound in Fig. 6d is a multi-heterocyclic covalent-binding candidate with rich aromatic conjugation, significant intramolecular charge transfer (ICT) capability, and high structural rigidity. According to DyeLeS predictions, it exhibits a Stokes shift of 83.41 nm and a fluorescence quantum yield of 0.35.
 | ||
| Fig. 6 Case studies of fluorescent compounds from FluBioDB. Most FluBioDB compounds feature polycyclic aromatic or heterocyclic cores, extended π-conjugation, and donor–acceptor (D–A). | ||
Structural analysis of additional compounds in Fig. 6e–i further supports these findings. Most FMDB hits possess characteristic polycyclic aromatic or heterocyclic cores, extended π-conjugation, and donor–acceptor (D–A) architectures that facilitate intramolecular charge transfer (ICT) and enhance Stokes shift. These features validate the effectiveness of DyeLeS in enriching for fluorescence-active molecules and demonstrate that FMDB can significantly accelerate the discovery of fluorescent drug candidates and bioimaging probes.
3 Conclusion
The study developed DyeLeS, a web–based fluorescence scoring model, and constructed FluBioDB, the first publicly available database of fluorescent bioactive compounds. DyeLeS achieved high classification accuracy (AUC = 0.995) using a Naive Bayes-inspired algorithm and reliable regression performance (R2 = 0.88, 0.83) for predicting absorption wavelength (λabs) and emission wavelength (λem) via LightGBM. Applying DyeLeS to the ChEMBL database enabled the identification of 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 865 fluorescence-enriched molecules, including kinase inhibitors and GPCR modulators. Structural analysis revealed that FluBioDB compounds commonly feature polycyclic conjugated scaffolds and donor–acceptor architectures, supporting strong ICT fluorescence and biological relevance. Despite its promising performance, the current version of DyeLeS does not account for environmental conditions (e.g., pH or temperature), which may influence fluorescence properties in experimental settings. Future work will focus on integrating these factors to enhance the model's robustness and applicability.
865 fluorescence-enriched molecules, including kinase inhibitors and GPCR modulators. Structural analysis revealed that FluBioDB compounds commonly feature polycyclic conjugated scaffolds and donor–acceptor architectures, supporting strong ICT fluorescence and biological relevance. Despite its promising performance, the current version of DyeLeS does not account for environmental conditions (e.g., pH or temperature), which may influence fluorescence properties in experimental settings. Future work will focus on integrating these factors to enhance the model's robustness and applicability.
This work fills a critical gap in fluorescent drug discovery by providing a scalable computational framework and a curated molecular resource. DyeLeS and FluBioDB offer practical tools to accelerate the design of fluorescent probes and theranostic agents, facilitating the translation of fluorescent drugs toward real-world biomedical applications.
4 Methods
4.1 DyeLeS-DyeS
Naive Log-likelihood Ratio Scoring is a simplified form of Naive Bayes classification, where the log-likelihood ratio is used to distinguish between fluorescent and non-fluorescent molecules. Instead of directly calculating conditional probabilities as in traditional Naive Bayes, this approach focuses on the relative probabilities of molecular fragments (fingerprint bits) occurring in fluorescent versus non-fluorescent molecules. The log-likelihood ratio for each fragment is computed as:In this context, Fluoi denotes the number of occurrences of fragment i in fluorescent molecules, while Non-Fluoi refers to its occurrences in non-fluorescent molecules. Fluototal represents the total number of fragments across all fluorescent molecules, and Non-Fluototal corresponds to the total fragment count in the non-fluorescent molecule set.
4.2 DyeLeS-DyeP
To predict the photophysical properties of fluorescent molecules—including absorption and emission wavelengths, fluorescence quantum yield, and Stokes shift—we employed a multi-output regression model based on LightGBM. Molecular structures were encoded as 2048 bit Morgan fingerprints (radius = 2), capturing relevant substructural features. The model was implemented using a MultiOutputRegressor wrapper around LightGBM to enable simultaneous prediction of multiple continuous targets. Key hyperparameters were set as follows: 64 leaves, maximum depth of 10, learning rate of 0.05, and 2000 estimators with early stopping. Feature and data subsampling (0.8 each) and L1/L2 regularization (both 0.5) were used to enhance generalization. Performance was evaluated using MAE, RMSE, and R2 on a held-out validation set.4.3 Web-based implementation
To enhance accessibility and practical utility, we developed DyeLeS, a web-based application for fluorescent dye-likeness scoring and molecular property prediction. The platform accepts SMILES strings as input and provides predictions of key photophysical properties, along with molecular structure visualization and formula display. Users can interactively explore results and download outputs for further analysis. DyeLeS is designed to support a wide range of use cases, including computational chemistry research, high-throughput compound screening, and early-stage drug discovery. By integrating predictive modeling with a user-friendly interface, DyeLeS facilitates rapid evaluation of dye-like properties in both known and novel molecular structures.We have deployed a fully-featured standalone version of the tool on GitHub (https://github.com/MolAstra/DyeLeS), complete with comprehensive installation guidelines and usage documentation. The web interface offers a streamlined subset of functionalities designed for browser-based accessibility, while the GitHub repository provides the complete analytical toolkit, including extensively annotated Jupyter notebooks with step-by-step code explanations. Both platforms will receive continuous maintenance and updates.
Data availability
The datasets used in this study are available at https://github.com/MolAstra/DyeLeS. The web server can be accessed at https://dyeles.molastra.com.Code availability
The source code of DyeLeS and associated data preparation python scripts are available at https://github.com/MolAstra/DyeLeS.Author conributions
J. X. designed the research project; W. Y. and K. D. collected literature and established dataset; Z. N. and Z. J. designed and trained the models; J. X., Y. C. and K. D. analyzed data and wrote the manuscript. All authors discussed the results and approved the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This research was supported by the General Research Project of the Zhejiang Provincial Department of Education No. Y202456962 and Zhejiang Provincial Research Project on Chinese Vocational Education No. ZJCV2024B31.References
- T. Sun, H. Zhao, L. Hu, X. Shao, Z. Lu, Y. Wang, P. Ling, Y. Li, K. Zeng and Q. Chen, Acta Pharm. Sin. B, 2024, 14, 2428–2446 CrossRef CAS.
- G. Chen, J. Yu, L. Wu, X. Ji, J. Xu, C. Wang, S. Ma, Q. Miao, L. Wang, C. Wang, S. E. Lewis, Y. Yue, Z. Sun, Y. Liu, B. Tang and T. D. James, Chem. Soc. Rev., 2024, 53, 6345–6398 RSC.
- Y. Wei, L. Kong, H. Chen, Y. Liu, Y. Xu, H. Wang, G. Fang, X. Shao, F. Liu, Y. Wang and Q. Chen, Chem. Eng. J., 2022, 429, 132134 CrossRef CAS.
- Q.-J. Duan, Z.-Y. Zhao, Y.-J. Zhang, L. Fu, Y.-Y. Yuan, J.-Z. Du and J. Wang, Adv. Drug Delivery Rev., 2023, 196, 114793 CrossRef CAS.
- E. G. Shcherbakova, B. Zhang, S. Gozem, T. Minami, P. Y. Zavalij, M. Pushina, L. D. Isaacs and P. Jr. Anzenbacher, J. Am. Chem. Soc., 2017, 139, 14954–14960 CrossRef CAS PubMed.
- J. Ou, M. Fang, M. Chen, C. Wang, X. Xu, Q. Wang, Y. Feng and X. Meng, ACS Sens., 2025, 10, 3569–3578 CrossRef CAS PubMed.
- H. Li, Y. Kim, H. Jung, J. Y. Hyun and I. Shin, Chem. Soc. Rev., 2022, 51, 8957–9008 RSC.
- L. Zhou, M. Tian, B. Zhang, X. Cao, X. Huo, F. Yang, P. Cao, L. Feng, X. Ma and X. Tian, Int. J. Biol. Macromol., 2024, 263, 130307 CrossRef CAS.
- N. Banahene, D. M. Gepford, K. J. Biegas, D. H. Swanson, Y.-P. Hsu, B. A. Murphy, Z. E. Taylor, I. Lepori, M. S. Siegrist, A. Obregón-Henao, M. S. Van Nieuwenhze and B. M. Swarts, Angew. Chem., Int. Ed., 2023, 62, e202213563 CrossRef CAS.
- Z. Zhang and W. Tang, Acta Pharm. Sin. B, 2018, 8, 721–732 CrossRef.
- S. A. Dugger, A. Platt and D. B. Goldstein, Nat. Rev. Drug Discovery, 2018, 17, 183–196 CrossRef CAS.
- L. Qian, Z. Dong and T. Guo, Cell Research, 2025, 35, 319–321 CrossRef.
- D. A. Barnes, L. Ladeira and R. Masereeuw, Nat. Rev. Nephrol., 2025 DOI:10.1038/s41581-025-00962-1.
- C. Niu, Q. Lyu, C. D. Carothers, P. Kaviani, J. Tan, P. Yan, M. K. Kalra, C. T. Whitlow and G. Wang, Nat. Commun., 2025, 16, 1523 CrossRef CAS PubMed.
- H. Zhu, Annu. Rev. Pharmacol. Toxicol., 2020, 60, 573–589 CrossRef CAS PubMed.
- M. Duran-Frigola, A. Fernández-Torras, M. Bertoni and P. Aloy, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2019, 9, e1408 CAS.
- B. Li, Z. Wang, Z. Liu, Y. Tao, C. Sha, M. He and X. Li, Briefings Bioinf., 2024, 25, bbae321 CrossRef CAS PubMed.
- P. Ertl, S. Roggo and A. Schuffenhauer, J. Chem. Inf. Model., 2008, 48, 68–74 CrossRef CAS.
- M. Sorokina and C. Steinbeck, J. Cheminf., 2019, 11, 55 Search PubMed.
- D. Mendez, A. Gaulton, A. P. Bento, J. Chambers, M. De Veij, E. Félix, M. P. Magariños, J. F. Mosquera, P. Mutowo, M. Nowotka, M. Gordillo-Marañón, F. Hunter, L. Junco, G. Mugumbate, M. Rodriguez-Lopez, F. Atkinson, N. Bosc, C. J. Radoux, A. Segura-Cabrera, A. Hersey and A. R. Leach, Nucleic Acids Res., 2019, 47, D930–D940 CrossRef CAS.
- Q. M. Thai, T. H. Nguyen, H. T. T. Phung, M. Q. Pham, N. K. T. Pham, J.-T. Horng and S. T. Ngo, RSC Adv., 2024, 14, 18950–18956 RSC.
- J. J. Irwin, K. G. Tang, J. Young, C. Dandarchuluun, B. R. Wong, M. Khurelbaatar, Y. S. Moroz, J. Mayfield and R. A. Sayle, J. Chem. Inf. Model., 2020, 60, 6065 CrossRef CAS PubMed.
- D. Shen, J. Liu, L. Sheng, Y. Lv, G. Wu, P. Wang and K. Du, Spectrochim. Acta, Part A, 2020, 228, 117690 CrossRef CAS.
- K. Du, S. Niu, L. Qiao, Y. Dou, Q. Zhu, X. Chen and P. Zhang, RSC Adv., 2017, 7, 40615–40620 RSC.
- K. Du, J. Liu, R. Shen and P. Zhang, Luminescence, 2019, 34, 407–414 CrossRef CAS PubMed.
- W. Song, L. Xiong, X. Li, Y. Zhang, B. Wang, G. Liu, W. Li, Y. Yang and Y. Tang, J. Chem. Inf. Model., 2025, 65, 2854–2867 CrossRef CAS PubMed.
- V. Chandrasekhar, K. Rajan, S. R. S. Kanakam, N. Sharma, V. Weißenborn, J. Schaub and C. Steinbeck, Nucleic Acids Res., 2025, 53, D634–D643 CrossRef PubMed.
- TMAP, https://github.com/reymond-group/tmap, (accessed November 10, 2024).
- J. A. Ruiz-Santoyo, A. Y. Torres-Boy, J. A. Minguela-Gallardo, J. T. Yi, S. A. Romero-Servín, D. W. Pratt and L. Álvarez-Valtierra, J. Mol. Struct., 2020, 1217, 128282 CrossRef CAS.
- S. Zhong and X. Guan, Environ. Sci. Technol., 2023, 57, 18193–18202 CrossRef CAS PubMed.
- A. Liu, Q. Zhang, L. Pan, F. Yang, D. Lin and C. Jiang, Adv. Funct. Mater., 2025, 35, 2415250 CrossRef CAS.
- D. Rogers and M. Hahn, J. Chem. Inf. Model., 2010, 50, 742 CrossRef CAS PubMed.
- A. P. Bento, A. Hersey, E. Félix, G. Landrum, A. Gaulton, F. Atkinson, L. J. Bellis, M. De Veij and A. R. Leach, J. Cheminf., 2020, 12, 51 CAS.
- A. Shehadeh, O. Alshboul, R. E. Al Mamlook and O. Hamedat, Autom. Constr., 2021, 129, 103827 CrossRef.
- J. Zhong, X. Zhang, K. Gui, Y. Wang, H. Che, X. Shen, L. Zhang, Y. Zhang, J. Sun and W. Zhang, Natl. Sci. Rev., 2021, 8, nwaa307 CrossRef CAS PubMed.
- S. Songsri, A. H. Harkiss and A. Sutherland, J. Org. Chem., 2023, 88, 13214–13224 CrossRef CAS PubMed.
- C. Cao, W.-C. Chen, S. Tian, J.-X. Chen, Z.-Y. Wang, X.-H. Zheng, C.-W. Ding, J.-H. Li, J.-J. Zhu, Z.-L. Zhu, Q.-X. Tong and C.-S. Lee, Mater. Chem. Front., 2019, 3, 1071–1079 RSC.
- Y. S. Kim, J. Lim, J. Y. Lee, Y. Lee and C. Choo, Chem. Eng. J., 2022, 429, 132584 CrossRef CAS.
- M. Hu, Y. Liu, Y. Chen, W. Song, L. Gao, H. Mu, J. Huang and J. Su, RSC Adv., 2017, 7, 7287–7292 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra03164h |
| This journal is © The Royal Society of Chemistry 2025 |